Abstract
Affective disorders are frequently associated with disrupted circadian rhythms. The existence of rhythmic secretion of central serotonin (5-hydroxytryptamine, 5-HT) pattern has been reported; however, the functional mechanism underlying the circadian control of 5-HTergic mood regulation remains largely unknown. Here, we investigate the role of the circadian nuclear receptor REV-ERBα in regulating tryptophan hydroxylase 2 (Tph2), the rate-limiting enzyme of 5-HT synthesis. We demonstrate that the REV-ERBα expressed in dorsal raphe (DR) 5-HTergic neurons functionally competes with PET-1—a nuclear activator crucial for 5-HTergic neuron development. In mice, genetic ablation of DR 5-HTergic REV-ERBα increases Tph2 expression, leading to elevated DR 5-HT levels and reduced depression-like behaviors at dusk. Further, pharmacological manipulation of the mice DR REV-ERBα activity increases DR 5-HT levels and affects despair-related behaviors. Our findings provide valuable insights into the molecular and cellular link between the circadian rhythm and the mood-controlling DR 5-HTergic systems.
Similar content being viewed by others
Introduction
The mammalian self-sustainable circadian clock is entrained for an approximate 24-h period and comprises positive and negative transcriptional–translational feedback loops that are tightly regulated and maintain the internal timekeeping system1. The master clock is located in the suprachiasmatic nucleus (SCN) of the anterior hypothalamus, which is considered the central pacemaker for subsidiary clocks in the extra-SCN brain regions of the central nervous system (CNS) and the peripheral clocks2,3. Recently, peripheral (local) clocks have gained attention because of their distinctive networks and regulatory functions, highlighting the importance of understanding their time-keeping mechanisms in various biological systems4,5,6.
Animal models with disrupted circadian clock genes display affective disorder-like phenotypes7,8,9. For instance, the circadian locomotor output cycle kaput (CLOCK) mutant, ClockΔ1910, and REV-ERBα knockout (KO)6 showed hyperdopaminergic state and mania-like phenotypes, basic helix-loop-helix ARNT like 1 (Bmal1) KO mice exhibits depression-like behaviors7, while those with period circadian clock 2 (Per2) KO showed alterations in depression-like phenotypes9. These studies have suggested that circadian clock genes play a role in both circadian rhythm and mood regulation, but differences in mood phenotypes in each model indicate the existence of different regulatory mechanisms. Circadian rhythms and the monoamine systems, such as dopamine (DA) and serotonin (5-hydroxytryptamine, 5-HT), have been extensively discussed in mood regulation in human and animal models6,11. A previous study showed that increased tyrosine hydroxylase (TH) and DA in REV-ERBα KO mice resulted in dysregulation of circadian DA expression6. However, there are limited studies on circadian regulations in the 5-HT system.
Central 5-HTergic neurotransmission involves several steps, including 5-HT biosynthesis, 5-HT transporter, SERT (encoded by Slc6a4), various serotonin receptors, and turnover. Central serotonin is synthesized from tryptophan by the following two enzymes: tryptophan hydroxylase 2 (TPH2) and aromatic amino acid decarboxylase (AADC). TPH2 is the rate-limiting enzyme in 5-HT biosynthesis and is regulated by the PET-1 nuclear activator12. Central 5-HT is mainly synthesized in the dorsal and the median raphe nuclei (DR and MR, respectively). Among these, DR 5-HTergic neurons play crucial roles in regulating mental states such as depression, anxiety, sociability, aggression, reward, and decision-making13,14,15,16. In our recent work, we demonstrated that the ablation of REV-ERBα in DR 5-HTergic neurons led to disrupted Tph2 expression and altered 5-HT levels. This, in turn, resulted in the loss of social preference, but not social recognition, in animal models at dawn and dusk16. This suggests that fine-tuning of 5-HT signaling by REV-ERBα in the DR 5-HTergic neuronal population is an important mechanism for modulating social behaviors16,17. Clinical evidence also supports the hypothesis that dysregulation of central 5-HTergic neurotransmission causes neuropsychiatric disorders, including depression, bipolar disorder, schizophrenia, and autism14,17,18. Genetic variations in human Tph2 loci are associated with the onset of major depressive disorder (MDD)19.
Despite growing recognition of the anatomical diversity and specific functions of the DR 5-HTergic system, the diurnal regulation of the 5-HT synthesis and transmission remains unresolved. Understanding the temporal dynamics and regulation of 5-HT signaling is crucial for understanding its complex role in various physiological processes and its implications in neuropsychiatric disorders. Limited evidence suggests that the rhythmic production of 5-HT results in 5-HT-derived circadian variations in physiological and behavioral phenotypes20,21,22. We hypothesized that the DR circadian nuclear receptor, REV-ERBα, a transcriptional repressor, may antagonistically crosstalk with PET-1, a nuclear activator of Tph2 transcription11, thereby driving the circadian expression of Tph2. In the present study, we aimed to clarify the roles and modes of the local circadian clockworks expressed in DR neurons in the rhythmic controls of 5-HT biosynthesis and its subsequent effects on mood-related phenotypes in mice.
Results
Nuclear receptor REV-ERBα binds to Tph2 promoter regions and represses Tph2 expression in functional competition with PET-1
We screened three in silico databases using Profiler of Multi-Omic data (PROMO), Genomatix, and TFsitescan which identified two putative REV-ERBα-binding sites in the upstream of the mouse Tph2 coding region. In the proximal region, two putative REV-ERBα-binding sites were positioned alternately with two PET-1-binding sites within 200-bp of the Tph2 promoter region12. This suggests that the rhythmic expression of Tph2 may be regulated by the circadian nuclear receptor, REV-ERBα, at its transcriptional level. Putative REV-ERBα and PET-1 binding sites were also identified in rat and human Tph2 promoter regions (Fig. 1a), suggesting the conservation of these transcription binding sites for circadian oscillation of the 5-HT system. To validate whether REV-ERBα regulates mouse Tph2 (mTph2) or human Tph2 (hTph2)19 expression, we performed a 2.7-kb mTph2 promoter- or a 1.9-kb hTph2 promoter-driven luciferase reporter assay to demonstrate the transcriptional regulation of Tph2 expression by REV-ERBα and PET-1, using a differentiated PC12 cell line, derived from rat pheochromocytoma. When REV-ERBα and PET-1 were overexpressed at the indicated molar ratios, REV-ERBα repressed, whereas PET-1-induced mTph2 promoter expression in a dose-dependent manner (Fig. 1b; F(5,18) = 130.1, P < 0.0001) and hTph2 promoter expression (Fig. 1b; F(5,18) = 22.49, P < 0.0001). Moreover, we performed a quantitative chromatin immunoprecipitation (ChIP) assay, which showed that REV-ERBα competed with PET-1 for the binding sites on the mTph2- or hTph2-promoter, and that their relative binding depended on the molar ratio of REV-ERVα and PET-1 cDNA. When the same molar ratio of REV-ERBα and PET-1 were overexpressed, the binding affinities of REV-ERBα to mTph2 promoter regions were stronger than those of PET-1 in vitro (Fig. 1c). For hTph2p ChIP assay, we quantified proximal REV-ERBα and distal REV-ERBα/PET-1 binding sites to demonstrate the plausible REV-ERBα repression on hTph2 expression, amplicon 1 and amplicon 2, respectively. Indeed, REV-ERBα binds to both putative REV-ERBα binding sites on the hTph2 promoter. To determine which cis-element serves as a recognition site for each transcription factor, we designed site-directed deletion mutants using 330-bp-long mTph2 promoter-driven luciferase reporter vectors (Fig. 1d). REV-ERBα was successfully repressed, whereas PET-1 activated wildtype (WT) mTph2 promoter reporter with 330-bp in length (Fig. 1e; F(3, 8) = 42.80, P < 0.0001) similar to the 2.7-kb full-length mTph2 promoter reporter (Fig. 1b). We then performed the mTph2 promoter assay with REV-ERBα responsive element (RRE) mut 1 or RRE mut 2 reporters, each lacking a REV-ERBα-binding site at the distal or proximal motif, respectively. The overexpressed REV-ERBα could not repress the RRE mut 1 or RRE mut 2 reporters, suggesting that both distal and proximal Rev-erbα cis-elements are necessary to repress Tph2 expression by the REV-ERBα transcription factor (Fig. 1f; F(3, 8) = 12.04, P = 0.0025 for RRE mut 1; F(3, 8) = 24.53, P = 0.0002 for RRE mut 2). However, PET-1 was able to induce the luciferase activity regardless of the mutants of REV-ERBα binding sites. As previously reported12, we confirmed that both the distal (PET-1 responsive element; PRE mut 3) and proximal (PRE mut 4) PET-1-binding sites are required for Tph2 activation (Fig. 1g; F(3, 8) = 6.214, P = 0.0174 for PRE mut 3; F(3, 8) = 4.228, P = 0.0457 for PRE mut 4). REV-ERBα overexpression in either PRE mut 3 or PRE mut 4 effectively repressed the mTph2 promoter-driven luciferase activity, suggesting that point mutations on PRE did not alter the effect of REV-ERBα on Tph2 expression. Based on these in vitro results, we speculated that REV-ERBα repression of Tph2 expression is greater than that of PET-1 activation at a similar molar ratio. These findings suggest that REV-ERBα binds to two Rev-erbα recognition sites on the Tph2 promoter and represses Tph2 expression in functional competition with the PET-1 nuclear activator.
a Schematic representation of the mouse Tph2 (mTph2), rat Tph2, and human Tph2 (hTph2) promoter regions with PET-1 binding sites (blue bars)12 and putative REV-ERBα binding sites (red bars), which are predicted using the PROMO, Genomatix, and TFsitescan databases. b Effects of REV-ERBα and/or PET-1 on the 2.7-kb mTph2 promoter or 1.9-kb hTph2 promoter activity. Dose-dependent repression of the each Tph2 promoter by REV-ERBα in functional competition with a PET-1 nuclear activator at the indicated ratios was assessed. c Assessment of target amplicon(s) on the Tph2 promoter for relative binding of REV-ERBα or PET-1 on the mTph2p and hTph2p via a ChIP-qPCR analysis. Differentiated PC12 cells were transfected with EGFP-REV-ERBα- and/or HA-PET-1-expressing constructs at the indicated ratios (n = 3). d Schematic representation of WT and designed site-directed deletions of REV-ERBα or PET-1 binding sites on mTph2 promoter regions (target deletion sites are indicated with underlines). Transcriptional activities of WT (e), RRE mut 1, RRE mut 2 (f), PRE mut 3, and PRE mut 4 (g) of mTph2 promoters upon REV-ERBα and/or PET-1 expression were measured (n = 3). Functional studies of RORα transcription factors on mTph2- (h) or mBmal1- (i) promoter-driven luciferase reporter in competition with REV-ERBα. For the promoter assays, the Tph2 promoter-driven luciferase reporter indicated that the transcription factor(s) and pRL-TK were co-transfected into differentiated PC12 cell lines (REV: REV-ERBα, PET: PET-1). Luciferase activity (A.U.) was normalized using the renilla activity (pRL-TK), which was used as the internal control. For promoter assays, data are presented as mean ± SEM, and the significance was assessed by one-way ANOVA followed by Dunnett’s post-hoc test; *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, and ns = not significant.
In the canonical circadian clock machinery, REV-ERBα is known to compete with retinoic acid receptor-related orphan receptor alpha (RORα) for REV-ERBα/RORα responsive element and regulates the transcription of circadian clock genes, such as Bmal1. This transcriptional regulation, in addition to BMAL1:CLOCK transcriptional activation, generates the circadian oscillations of the molecular clockwork23. We examined whether the REV-ERBα binding sites on the Tph2 promoter region are also affected by the competitive binding of RORα for Tph2 expression. According to the results of the Tph2 promoter assay, in differentiated PC12, RORα overexpression did not elicit the expression of Tph2 promoter-driven luciferase activities (Fig. 1h; F(3, 12) = 48.29, P < 0.0001), whereas the Bmal1 promoter-driven luciferase activity levels were increased by the RORα overexpression in competition with REV-ERBα in differentiated PC12 (Fig. 1i; F(3, 12) = 100.3, P < 0.0001).
REV-ERBα binds to the Tph2 promoter region in the DR 5-HT system
Considering that REV-ERBα represses Tph2 expression in vitro, we further investigated whether endogenous DR REV-ERBα and PET-1 are the key transcription regulators for rhythmic TPH2 expression in DR 5-HTergic neurons. To examine the binding affinities of REV-ERBα and PET-1 to the Tph2 promoter region over the circadian time (CT) points, we performed a DR ChIP assay at 6-h intervals. Interestingly, the binding affinities of REV-ERBα to the Tph2 promoter region changed over the CT points, whereas those of PET-1 remained constant (F(3, 8) = 49.68, P < 0.0001 for REV-ERBα; F(3, 8) = 1.473, P = 0.2934 for PET-1). Notably, the highest binding affinity of REV-ERBα on the Tph2 promoter was observed at CT12 (Fig. 2a). This finding is consistent with the mRNA profiles of DR Rev-erbα, which showed the elevated mRNA levels at CT08 (Supplementary Fig. 1).
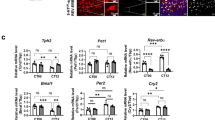
a Schematic representation of target amplicon of the Tph2 promoter region with the PET-1 and REV-ERBα binding sites (left). DR ChIP-qPCR analyses with REV-ERBα and PET-1 antibodies in WT at the indicated circadian time (CT) points. b mRNA profiles of 5-HT-related genes at CT points in the DR regions (Tph2, Pet-1, Rev-erbα, and Ddc). Each mRNA was normalized using the level of a housekeeping gene, TATA-box-binding protein (Tbp; n = 3 for each measurement, two mice pooled per sample). c 5-HT levels in the raphe region exhibit circadian oscillation at the CT points. Data are presented as mean ± SEM (n = 3–4), and significance was assessed and compared to that of CT00 using unpaired Student t test; **P < 0.01, **** P < 0.0001, ns = not significant. Ddc; aromatic L-amino acid decarboxylase.
Canonical clock genes, such as Bmal1, Rev-erbα, Rev-erbβ, RORα, RORβ, Cry 1 and Cry2 (Cryptochromes 1 and 2), and Per1 and Per2 (Periods 1 and 2) in DR, showed circadian rhythmic patterns (Supplementary Fig. 1). Compared to the molecular clockwork in the SCN, the DR core clock machinery, particularly Rev-erbα, and Bmal1, showed different peak times in their circadian rhythmicity, indicating that the DR local clock maintains its distinctive rhythmic patterns. In addition to analyzing the REV-ERBα and PET-1 binding affinities with the Tph2 promoter, we investigated the chromatin landscapes of the Tph2 promoter region at different zeitgeber time (ZT) points in the DR using an assay for transposase-accessible chromatin with high-throughput sequencing (ATAC-seq) followed by a motif-based analysis for REV-ERBα- and PET-1-binding motifs. The chromatin landscape of the Tph2 promoter region was more accessible at ZT00 than at other time points (Supplementary Fig. 2a), indicating that the transcription factors bind to the Tph2 promoter region in a circadian manner, implying the circadian regulation of Tph2 in the DR 5-HT system (Fig. 2b). Motif-based analysis revealed that there were no significant differences in the motif binding for DR REV-ERBα or PET-1 throughout the day (Supplementary Fig. 2b). We next compared the mRNA expression patterns of core clock machinery in DR and SCN at various CT points to demonstrate the presence of a 5-HTergic local clock in the DR. In addition to the circadian pattern of Tph2 mRNA, the mRNA expression profile of Pet-1, a nuclear activator of Tph2, did not show any circadian pattern. Aromatic L-amino acid decarboxylase (Ddc) showed no circadian patterns (Fig. 2b). Together, the regulation of rhythmic DR Tph2 expression is controlled by REV-ERBα at the transcriptional level, which subsequently leads to the circadian oscillation of 5-HT (Fig. 2c; F(5, 15) = 45.11, P < 0.0001).
Collectively, our results suggest that the binding affinities of DR REV-ERBα to the Tph2 promoter region are more favorable than those of PET-1; thus, the rhythmic Tph2 expression is due to REV-ERBα rather than the transcription factor PET-1 in DR, resulting in the diurnal variation in the 5-HT levels. However, whether REV-ERBα and PET-1 bind to the Tph2 promoter region in a competitive manner in DR remains unclear because the ChIP assay cannot discriminate between competitive binding and competitive action of the two transcription factors, REV-ERBα and PET-1 on their cis-elements. We speculate that the competitive action is more likely because RREs and PREs do not physically overlap in the Tph2 promoter region.
DR 5-HTergic REV-ERBα plays a role in generating circadian mood regulation
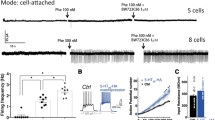
We further investigated the specific function of REV-ERBα in the DR 5-HT neurons and mood regulation, and we generated Rev-erbα conditional KO (cKO) mice using the Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)/Cas9 gene-editing system. To eliminate only DR 5-HTergic REV-ERBα, we injected an adeno-associated virus (AAV) vector expressing Rev-erbα single-guide RNA (sgRNA) into the DR region of SERT-cre;Rosa26-LSL-Cas9animals. Likewise, we generated Pet-1 cKO mice to compare the effects of the PET-1 nuclear activator on circadian depression-like behaviors. Rev-erbα cKO and Pet-1 cKO mice showed significant loss of REV-ERBα and TPH2 positive neurons, respectively, among the AAV-infected SERT positive neurons in DR, compared to LacZ-injected controls. The CRISPR/Cas9 gene KO efficiency for Rev-erbα cKO (91.71% ± 1.990%)16 or Pet-1 cKO (97.44% ± 2.564%) was assessed using immunohistochemistry (Fig. 3a; t = 46.09, df = 4, P < 0.0001 for Rev-erbα cKO; t = 38.00, df = 4, P < 0.0001 for Pet-1 cKO). However, there are no suitable anti-PET-1 antibodies for testing the efficiency of Pet-1 sgRNA gene KO. Therefore, we immunolabeled TPH2 instead of PET-112 to assess the KO efficiency of Pet-1 cKO. We also quantified the DR Tph2 mRNA (F(2, 12) = 8.131, P = 0.0059, interaction) and 5-HT levels (F(2, 12) = 7.384, P = 0.0081, interaction) with Rev-erbα cKO, Pet-1 cKO, and LacZ mice at dawn and dusk to confirm the functional effectiveness of each gene ablation at the molecular level. Rev-erbα cKO showed a significant increase in Tph2 mRNA and 5-HT levels at dusk, whereas Pet-1 cKO exhibited a significant decrease in Tph2 mRNA and 5-HT levels at dawn (Fig. 3b, c). Moreover, the DR ChIP assay with Rev-erbα cKO mice showed significant reductions in REV-ERBα binding with RREs on the Tph2 promoter at dawn and dusk (F(1, 8) = 13.61, P = 0.0061, interaction for REV-ERBα; F(1, 8) = 0.07328, P = 0.7935 interaction for PET-1). Interestingly, the decreased REV-ERBα binding correlated with increased PET-1 binding to the Tph2 promoter region (Fig. 3d). These results confirmed that REV-ERBα directly binds to the Tph2 promoter region and functions as a transcription repressor for Tph2 expression in vitro and in vivo.
a Schematic representation of the designated AAV-sgRNA injection into the DR region of SERT-cre;Rosa26-LSL-Cas9 expressing mice for the generation of 5-HTergic Rev-erbα cKO, Pet-1 cKO, and LacZ control models. The CRISPR/Cas9 KO efficiency for Rev-erbα cKO or Pet-1 cKO was measured (lower left; unpaired Student t test; ****P < 0.0001). Representative confocal images of the REV-ERBα expression in the LacZ and Rev-erbα cKO models (upper right) and TPH2 expression in LacZ and Pet-1 cKO models (lower right). Tph2 mRNA (b) and 5-HT (c) levels were quantified in Rev-erbα cKO, Pet-1 cKO, and LacZ expressing SERT-cre; Rosa26-LSL-Cas9 mice at CT00 vs. CT12 (n = 3, duplicated). d DR ChIP-qPCR analyses with REV-ERBα (left) or PET-1 (right) antibody for the Tph2 promoter region were performed with DR chromatin of Rev-erbα cKO and LacZ at CT00 and CT12. Reduced binding of REV-ERBα and increased PET-1 binding on the Tph2 promoter region were detected in the Rev-erbα cKO DR chromatin (each chromatin sample was pooled from four mice; triplicate samples). Data are presented as mean ± SEM, and the significance was assessed by two-way ANOVA followed by Tukey’s post hoc test; *P < 0.05, **P < 0.01, ***P < 0.001, and ns = not significant.
To assess the physiological importance of DR 5-HTergic REV-ERBα in mood regulation, we examined the circadian depression-like phenotypes in Rev-erbα cKO and Pet-1 cKO mice (Fig. 4a). Both DR 5-HTergic Rev-erbα cKO and Pet-1 cKO mice showed a loss of circadian variations of immobility in the forced swim test (FST; F(2, 44) = 6.348, P = 0.0038, interaction) and tail suspension test (TST; F(2, 44) = 13.91, P < 0.0001, interaction) compared to the LacZ-injected controls. Notably, Rev-erbα cKO mice (Fig. 4b, c), which exhibited increased 5-HT levels, particularly at dusk, displayed reduced depression-like phenotypes, whereas the Pet-1 cKO mice, with significantly decreased 5-HT levels, exhibited increased depression-like behaviors throughout the day (Fig. 3c).
a Experimental design for generating the Rev-erbα cKO, Pet-1 cKO, and LacZ models followed by the designated behavioral assays. Depression-like phenotypes were assessed with Rev-erbα cKO and Pet-1 cKO models and compared with the LacZ controls. Immobility in FST (b) and TST (c) was reduced in Rev-erbα cKO at dusk (CT10-13), whereas Pet-1 cKO showed increased immobility at dawn (CT22-01) (n = 8 for control and Rev-erbα cKO; n = 9 for Pet-1 cKO). Data are presented as mean ± SEM, and significance was assessed by two-way ANOVA followed by Tukey’s post hoc test; *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, and ns = not significant.
Pharmacological manipulation of DR REV-ERBα altered the circadian depression-like phenotypes
Considering that REV-ERBα functions as a transcriptional repressor of Tph2 expression in the DR 5-HTergic system, we investigated whether the regulation of DR 5-HTergic REV-ERBα could alter the circadian depression-like behaviors in mice. To further investigate the effects of DR REV-ERBα on circadian depression-like behaviors, we microinjected the REV-ERBα antagonist, SR8278 (16 μg/mouse), or REV-ERBα agonist, GSK4112 (32 ng/mouse), into the DR, 3-h prior to the behavioral tests at dawn and dusk (Fig. 5a), as previously described. SR8278 microinjection into the DR increased 5-HT levels, whereas GSK4112 reduced 5-HT levels compared to vehicle injection at ZT04 (Fig. 5b; F(2, 6) = 29.27, P = 0.0008). Moreover, local microinjection of SR8278 or GSK4112 altered circadian depression-like phenotypes. The inhibition of REV-ERBα in the DR by SR8278 decreased immobility in both the FST (F(2, 40) = 1.507, P = 0.2339, interaction) and TST (F(2, 40) = 4.268, P = 0.0209, interaction), i.e., promoted active coping behavior, particularly at dusk. GSK4112 administration into the DR significantly induced a significant depression-like phenotype at dawn compared with that of the vehicle-injected groups (Fig. 5c, d). These pharmacological manipulations of the DR REV-ERBα and their behaviors support the role of REV-ERBα as the key transcriptional factor for circadian depression-like behaviors through the modulation of the 5-HTergic system.
a Experimental scheme for the behavioral tests for local microinjection of SR8278 (Rev-erbα antagonist), GSK4112 (Rev-erbα agonist), or vehicle (DMSO) into DR. SR8278, GSK4112, or vehicle was administered 3-h prior to the depression-like behavioral tests. b Quantification of 5-HT after local microinjection of either SR8278, GSK4112, or vehicle in DR at ZT04. Each animal was administered the selected drugs for three consecutive days before sampling. Data are presented as mean ± SEM, and significance was assessed by one-way ANOVA followed by Dunnett’s post-hoc test; *P < 0.05, **P < 0.01. Depression-like phenotypes were measured after SR8278, GSK4112, or vehicle microinjection into the DR. Immobility was reduced in FST (c) and TST (d) after SR8278 microinjection at dusk, whereas GSK4112 increased the immobility at dawn (n = 8 for controls; n = 6 for SR8278; n = 9 for GSK4112). Data are presented as mean ± SEM. Significance was assessed by two-way ANOVA followed by Tukey’s post hoc test; *P < 0.05, **P < 0.01, ****P < 0.0001, and ns = not significant.
In summary, the CRISPR/Cas9 gene-editing system allowed the examination of the gene ablation of REV-ERBα in DR 5-HTergic neurons with respect to molecular, physiological, and behavioral alterations. These findings support the notion that transcriptional repression of REV-ERBα on Tph2 expression is crucial for generating circadian oscillation within the DR 5-HTergic system, thereby emphasizing the importance of temporal regulations of Tph2 and, consequently, mood regulations via the DR 5-HTergic system.
Discussion
Central serotonin dysregulation causes mental instability and is life-threatening in severe conditions24,25. Patients with MDD often suffer from mood instability accompanied by disrupted circadian physiology manifesting as morning blues, and sleep disturbances26. However, the potential role of circadian biology in mood regulation through the serotonin system remains unclear. Here, we provide compelling evidence that the circadian timing system regulates the rhythmic serotonin levels; notably, the nuclear receptor REV-ERBα transcriptionally represses Tph2 expression in a manner functionally antagonistic to the nuclear activator PET-1, thereby driving the rhythmic oscillation of 5-HT biosynthesis.
MDD is primarily treated by regulating the central 5-HT levels to alleviate the disrupted mood states, particularly the depressive symptoms. Selective serotonin reuptake inhibitors (SSRIs; for instance, fluoxetine, brand name Prozac) were first introduced several decades ago, and have since become the most prescribed drugs for treating depression worldwide27. However, SSRIs have delayed antidepressant effects with undesirable side effects, including an increased risk of suicidal thoughts, serotonin syndrome, and sleep disturbances28. Recently, the non-SSRI antidepressant ketamine, an open channel blocker of ionotropic glutamatergic N-methyl-D-aspartate (NMDA) receptors, has been used at low sub-anesthetic dose. Ketamine is a fast-acting and effective antidepressant for patients with MDD, including those with treatment-resistant depression (resistant to regular antidepressants)29,30. However, its use is limited by a narrow treatment window to the profound psychoactive effects associated with repeated administration31,32. Lastly, sleep deprivation also has an anti-depressant effect through an increase in the serotonin 2A receptor (5-HT2A) levels, especially in the hippocampus and frontal cortex33,34. However, the exact mechanism by which sleep depreviation can alter central 5-HT levels is not fully understood.
Unlike SSRIs or ketmaine, targeting 5-HTergic REV-ERBα could be more rapid and effective in alleviating circadian depression-like phenotypes35,36. Although REV-ERBα is a key regulator of circadian clock machinery regulator, most studies have largely focused on its pivotal non-circadian roles in different physiological processes such as mood regulation, metabolism, and neuroinflammation37,38,39. REV-ERBα rhythmically represses the expression of TH (the rate-limiting enzyme of the DA system)6,40 by competing with the nuclear receptor-related 1 (NURR1) transcription factor. Thus, REV-ERBα targets the rate-limiting enzymes of both the monoamine systems — Tph2 in the 5-HTergic system, and Th in the DAergic system, which are critical for rhythmic 5-HT and DA biosynthesis. Our findings demonstrate that REV-ERBα serves as a multifunctional node for mood regulation in different neural populations, including 5-HT and DA neurons, suggesting a new potential therapeutic target for affective disorders.
Transcriptional regulations of the 5-HTergic system in the CNS mainly involves several developmental genes, including NK2 homeobox 2 (Nxk2.2), LIM homeobox transcription factor 1 beta (Lmx1b), Pet-1, and GATA binding protein 3 (GATA3), during the developmental stages of 5-HT neurons11. The identification of the central Tph2 in mammals41,42 has facilitated studies on the regulatory mechanisms of the central 5-HTergic system11,43,44. Recent advances in genetic manipulation using animal models have boosted studies on transcriptional regulators of the adult 5-HTergic system12,45,46. These transcriptional regulations led to rhythmic Tph2 expression and 5-HT biosynthesis in the DR 5-HTergic system. Our recent study demonstrated that the ablation of REV-ERBα specifically in DR 5-HTergic neurons resulted in a significant increase in the activity of DR 5-HTergic neurons and elevated 5-HT release throughout the day. Importantly, we found that the DR to nucleus accumbens (NAc) 5-HTergic circuit is crucial for regulating social preference behavior. The increase in 5-HT release into the NAc was functionally associated with the disruption of social preference in an animal model, without affecting social recognition16. Pharmacological manipulation of REV-ERBα activity, using agonists and antagonists, was able to alter the circadian depression-like phenotypes and partially restore the deficits in social preference, further highlighting the importance of REV-ERBα in regulating 5-HT homeostasis and its impact on depression-like and social interaction behaviors. Ablation of the DR 5-HTergic Rev-erbα resulted not only in an increase in Tph2 mRNA and 5-HT levels (Fig. 3b, c), but also enhanced the 5-HTergic neural activity and 5-HT release in DR16, especially at dusk. These suggest that loss in REV-ERBα repression altered Tph2 expression, which correlates with the highest binding affinities at CT12 (Fig. 2a), and reduced depression-like phenotype at dusk. We observed that there are peak time differences in Tph2 mRNA (CT20) and the 5-HT (CT04) levels, which led us to speculate the following: (i) the time delays in TPH2 protein synthesis is due to plausible post-transcriptional/translational modifications47, (ii) the metabolizing rate of 5-HT by monoamine oxidase48,49 potentially alters the accumulation of 5-HT in DR, shifting the peak time of 5-HT, and (iii) other circadian regulators may be involved in shaping the rhythmicity of Tph2 expression. In addition to REV-ERBα’s repressive function on Tph2 expression, circadian regulation of the 5-HTergic system was also supported by genome-wide mapping of chromatin landscapes with ATAC-seq, showing time-of-the-day dependent rhythmic variations in Tph2 gene loci and RRE and PRE motif changes. We observed that endogenous REV-ERBα binding to Tph2 promoter showed the circadian variations, while PET-1 binding remained constant throughout the day. In vitro ChIP assay also showed that in a same molar ratio of overexpressed REV-ERBα and PET-1, REV-ERBα binding to Tph2 promoter is more favorable than that of PET-1. Moreover, DR 5-HTergic REV-ERBα ablation showed the increase in PET-1 binding to Tph2 promoter region at dawn and at dusk. Overall, these findings show that the circadian oscillation of Tph2 and 5-HT levels in DR are mainly derived by the repressive function of DR 5-HTergic REV-ERBα, the circadian nuclear receptor33.
In this study, we observed that 5-HT-deficient Pet-1 cKO mice exhibited a prominent increase in depression-like behaviors at dawn, whereas Rev-erbα cKO mice exhibited a reduction in depression-like phenotypes at dusk. These results may explain the controversial reports on Tph2 KO or cKO50,51 and Pet-1 cKO52, which showed a lack of depression-like phenotypes compared to their controls. This underscores the importance of factoring in the specific time-of-day, in analyzing the 5-HT dependent depression-like phenotypes in mice. Further, a recent study on Rev-erbα KO mice53 demonstrated that REV-ERBα regulates the 5-HTergic system and mood. However, in contrast to our findings, the Rev-erbα KO mice in this previous study, exhibited decreased depression-like phenotype with a decrease in Tph2 mRNA and 5-HT levels in raphe nuclei compared to control groups. These discrepancies in the molecular findings (between our results and those of the previous study) may have resulted from differences in the genetic models and/or experimental conditions used in both studies. Studying global REV-ERBα KO animal model can provide valuable insights into the developmental alterations associated with the chronic ablation of this transcriptional regulator across all brain cell types. This model allows researchers to investigate the dysregulation of REV-ERBα-related gene populations, which may have far-reaching consequences for various developmental processes. For instance, the observed defects in neurogenesis within the prefrontal cortex (PFC) of REV-ERBα KO mice may not be solely attributable to the loss of REV-ERBα in 5-HTergic neurons. Instead, the PFC-specific neurogenesis defects could be a result of the loss of REV-ERBα function directly within the PFC itself, rather than just in the 5-HT neuronal population. The global loss of REV-ERBα can have widespread impacts on gene regulation and cellular function across different brain regions and cell types, which may contribute to the observed phenotypes. However, further investigations are required to understand these differences.
In conclusion, our comprehensive study demonstrates that the 5-HTergic system is subject to circadian regulation, and the circadian nuclear receptor REV-ERBα plays a key role in modulating the circadian oscillation of 5-HT expression. Our findings were obtained through a multimodal analysis, encompassing molecular, genomics, pharmacological, and behavioral analyses. Our study highlights the local connection between circadian rhythms and 5-HT regulation, contributing to a deeper understanding of the molecular basis of mood regulation and its potential implications in neuropsychiatric disorders.
Methods
Cell culture
The PC12 cell line was obtained from ATCC (Manassas, VA), and the culture materials were obtained from Thermo Fisher Scientific (Waltham, MA). PC12 cells were maintained in an RPMI-1640 medium (Gibco, Waltham, MA) supplemented with 10% fetal bovine serum (Gibco), 5% horse serum (Gibco), and 100 U/mL penicillin/streptomycin (Gibco) in culture dishes coated with poly-L-lysine (PLL, Sigma-Aldrich, Burlington, MO). Upon neural differentiation, PC12 cells were treated with OPTI-Minimum Essential Medium Eagle (OPTI-MEM, Gibco) supplemented with 0.5% fetal bovine serum and 50 ng/μL nerve growth factor (Sigma-Aldrich) for 4 days54.
Animals
Male mice (8–15 weeks of age) used in this study were maintained on a C57BL/6 J background and housed in a temperature-controlled (23 °C–25 °C) environment under a 12-h light/dark (LD) photoperiod (lights on at 7:00) with ad libitum access to food and water. For the CT experiments, mice were kept in constant dark (DD) for a minimum of 3 days; subsequently, the assigned behavioral tests were performed at specified time periods (CT22-01 vs. CT10-13) under dim red light40. For ZT experiments, mice were kept under a 12:12 LD cycle, and behavioral tests were performed at ZT01-03 vs. ZT10-12. Before sacrificing the mice for tissue sampling, avertin (300 mg/kg) was injected intraperitoneally. All animal procedures were approved by the Institutional Animal Care and Use Committee of the DGIST (DGIST-IACUC-21070804) and conformed to the relevant regulatory standards.
Plasmid constructs
Tph2 promoter assay
The 2.7-kb mouse Tph2 promoter (–2758 to –1) genomic sequence (Gene ID: 216343), 1.9-kb human Tph2 promoter (-1915 to +72) genomic sequence (Gene ID: 121278)19 were obtained from the NCBI database55, synthesized and acquired with the pUC-57AMP vector (Bionics, Seoul, Republic of Korea). The synthesized mTph2 promoter region (2.7-kb) was digested with MluI and XmaI and cloned in the pGL3-basic vector (Promega, Madison, WI). For WT and site-directed mutated 330-bp mTph2 promoter (-780 to -450) luciferase reporter vectors, the target Tph2 promoter region was amplified using PCR and transferred into the pGL3-basic vector (Promega) for the luciferase reporter assay. SpectraMax Luminometer (Molecular Devices, LLC, San Jose, CA) was used for the measurement of luciferase activities. The primer designs for site-directed mutagenesis are shown in Supplementary table 1.
sgRNAs
sgRNAs targeting the genes of interest were designed and selected using CHOPCHOP56. The synthesized DNA oligomers with PAM adaptor sequences were annealed and cloned into the sgRNA expression vector, pAAV-U6-sgRNA-hSyn-mCherry-KASH, as previously described57. The sgRNA sequences were designed as follows: sgRev-erbα, 5′-GTT GCG ATT GAT GCG AAC GAT GG-3′ (chr 11: 98,771,255, strand: +); and sgPet-1 (Fev), 5′- CCG CGC CAC CTC GTC CGG GTC GG-3′ (chr 1: 74,882,535, strand: +).
Transfection and promoter assay
PC12 cells were cultured and seeded in the PLL-coated 24-well plate at a density of 1.5 ×105 per well in a neural differentiation medium; cloned pGL3 luciferase reporter (300 ng) and cDNA-containing expression (800 ng) vectors were co-transfected with 50 ng thymidine kinase promoter-driven renilla luciferase (pRL-TK) vector using the Lipofectamine 3000 Transfection Reagent (Invitrogen, Waltham, MA) in a PLL-coated 24-well culture plate. To maintain the total DNA used for each transfection, a pcDNA3.1 vector was added as required. After 48 h of transfection, half of the neural differentiation medium was replaced with a fresh medium to maintain culture conditions. After 96 h of initial transfection, the luciferase activities were measured using the Dual-Glo luciferase assay system (Promega), and the renilla luciferase activities were used to normalize the firefly luciferase activities.
Chromatin immunoprecipitation assay
The ChIP assay was performed as described previously58 with some modifications. For the in vitro ChIP assay, PC12 cells were cultured in a PLL-coated 60-mm2 culture dish containing a neural differentiation medium. CMV promoter-driven expression vectors containing EGFP-REV or HA-PET-1 (total 8 μg) and pGL-Tph2 promoter vector (1 μg) were co-transfected into PC12 cells using Lipofectamine 3000 (Fig. 1g). pcDNA3.1 was co-transfected as required. After 72 h of transfection, cells were treated with pre-chilled DPBS (Gibco) and stored at −80 °C until further processing. For the in vivo ChIP assay, a fresh mouse brain was dissected on an ice-chilled brain matrix, and the DR tissue was acquired from 1-mm-thick brain sections using a 1.5-mm-diameter micropunch (samples from four mice were pooled for each ChIP assay) targeting the bregma between -4.0 to -5.0 mm of Allen mouse brain atlas. Cells or DR tissues were crosslinked for 10 min at room temperature (23 °C–25 °C) with an 11% formaldehyde solution (Cell Signaling Technology, Danvers, MA). Chromatin was sheared with an M220 focused ultrasonicator (Covaris, Bengaluru, India) and precleared with Dynabeads protein A magnetic beads (Invitrogen) for 1 h at 4 °C with agitation. Chromatin was immunoprecipitated with 1.0–1.2 μg of anti-GFP (Santa Cruz Biotechnology, Dallas, TX, sc-9996), anti-HA (Santa Cruz Biotechnology, sc-7392), anti-REV-ERBα (Invitrogen, PA5-29865), anti-PET-1 (Novus Biologicals, Centennial, CO, NBP2-55967), anti-BMAL1 (Cell signaling Technology, 14020), or anti-histone H3 (Cell Signaling Technology, 4620). As a negative control, chromatin was incubated with 5 μL normal rabbit IgG (Cell Signaling Technology, 2729). Immune complexes were then collected using Dynabeads protein A magnetic beads and de-crosslinked by overnight incubation at 65 °C in 5-M NaCl, RNase A (Invitrogen), and Proteinase K (Invitrogen). The primer sets used for real-time qPCR targeting mTph2 promoter region (-718 to -468) were as follows12: mTph2p ChIP forward, 5′-TTG GAA AAG TAC AAA TAT AAT CTT-3′, and mTph2p ChIP reverse, 5′-GCT TCC AAA ACC CAT GGT GTT TCC-3′ and hTph2 promoter region, amplicon 1 (-296 to -177) for hTph2p ChIP (1) forward, 5’-GCT TAA AGA CTG AGG TTA ACT TGC-3’, and hTph2p ChIP (1) reverse, 5’-GGG GTG GTG GAG AAC AAT ACA T-3’; for amplicon 2 (-1256 to -1037) for hTph2p ChIP (2) forward, 5’-CAT TCT TCA GGA GAT TGG TAA CA-3’, and hTph2p ChIP (2) reverse, 5’-CGA GAT CTG TGC GTT ATG TGG TGA-3’.
HPLC-ECD
Sample preparation
The DR regions were dissected (2-mm thickness) using a brain matrix, and samples were stored at −80 °C until further processing.
5-HT measurements
Tissues were washed with pre-chilled phosphate-buffered saline (PBS; Gibco) and homogenized using a sonicator with pulses at 12 V in 0.3-N perchloric acid solution. The cell debris was then pelleted via centrifugation at 14,000 × g for 20 min at 4 °C using a tabletop refrigerated centrifuge (Eppendorf, Hamburg, Germany). The 5-HT contents were measured with high-performance liquid chromatography coupled with electrochemical detection (HPLC-ECD, HTEC-510; Eicom, Tokyo, Japan) using column SC-5ODS (2.1ɸ); 20 µL of supernatant was injected into HPLC-ECD. The mobile phase was prepared using 100-mM citrate-acetate buffer (pH 3.9), methanol (83:17, v/v), 140 mg/L Sodium 1-octane sulfonate, and 5 mg/L EDTA 2Na, according to the manufacturer’s instructions. The reagents were purchased from Sigma-Aldrich and were of HPLC-grade quality.
RNA isolation and real-time qPCR
RNA samples were purified using TRIzol reagent (Invitrogen) according to the manufacturer’s protocol. For RT-PCR, 1 µg of each RNA sample was reverse-transcribed using the PrimeScript RT reagent (Takara Bio, Kyoto, Japan). cDNA aliquots were then subjected to quantitative real-time PCR in the presence of SYBR Green I (Enzynomics, Daejeon, Republic of Korea). The gene expression levels were normalized to that of the gene encoding TATA-box binding protein (Tbp). The primer sequences used for qRT-PCR are listed in Supplementary Table 2.
Production of AAV
All recombinant AAV vectors used in this study were generated in-house as described previously16 with modifications. After the AAV production, the titer was quantified using real-time qPCR. The AAV vectors were injected into 8- to 10-week-old mice and used with the following titers and volumes: AAV2/PHP.eB-U6-Rev-erbα-hSyn-mCherry, 1.0 × 1013 GC/mL; AAV2/PHP.eB-U6-sgPet-1-hSyn-mCherry, 1.0 × 1013 GC/mL; DR, 800 nL; DR, 800 nL; and AAV2/PHP.eB-hU6-sgLacZ1-hU6-sgLacZ2-hSyn-mCherry.
Surgical procedures
Mice were deeply anesthetized with pentobarbital sodium (50 mg/kg, intraperitoneal injection) and placed on a stereotaxic apparatus (Stoelting). The coordinates were as follows: DR (lambda AP, +1.5 mm ML, –3.0 mm DV, angled 20°; for unilateral AAV injection and cannula implantation).
Drug preparation and application
Local injection of SR8278 or GSK4112
The Rev-erbα antagonist SR8278 (Tocris Bioscience, Bristol, UK) or Rev-erbα agonist GSK4112 (Sigma-Aldrich) was dissolved in DMSO (Sigma-Aldrich) to a concentration of 20 μg/μL for SR8278, as previously described6 and 100 μM for GSK4112. SR8278 (16 μg/mouse), GSK4112 (32 ng/mouse), or vehicle (DMSO) was directly microinjected using a 26-gauge cannula (PlasticOne, Boerne, TX) into the DR with a Hamilton syringe at a rate of 0.1 μL/min. SR8278, GSK4112, or DMSO was administered 3 h prior to tissue sampling or behavioral tests.
Behavioral protocols
All behavioral tests were performed under DD conditions (CT) as described previously40.
Forced swim test (FST)
Mice were placed in a 10-cm diameter cylinder filled with warm water (21 °C–25 °C) to assess the depression-like phenotypes. The mice were monitored for 6 min, and immobility was scored using the last 4 min of the video recording.
Tail suspension test (TST)
Mice were suspended in a white box using adhesive tape and monitored for 6 min; the immobility time was scored during the last 4 min of the test period. Immobility was defined as the lack of movement or struggle while hanging.
Immunohistochemistry
Mice were perfused with freshly made 4% paraformaldehyde in PBS. The brain samples were post-fixed in the same fixative overnight at 4 °C. Tissues were then cryoprotected in 20% sucrose and sectioned (50-µm thickness) using a cryostat (Leica). The sections were incubated with a homemade labeling solution at 37 °C for 30 min and incubated with primary antibodies against TPH2 (Abcam, Cambridge, UK; ab121013) overnight at 25 °C. After several washes with PBS, anti-goat Alexa488-labeled antibody (Invitrogen) was added for 3 h. Subsequently, the sections were washed, mounted, and observed under a confocal microscope (Zeiss LSM700 and 800).
ATAC-seq
Sample preparation for ATAC-seq
Rev-erbα cKO and WT mice aged 12 weeks were kept under a 12-h LD photoperiod. The DR regions were dissected into 1-mm thickness using an ice-chilled brain matrix and sampled using a 1.5-mm micropunch in pre-chilled Hanks’ Balanced Salt Solution (Gibco), as previously described40. The DR samples were collected from five mice per group at ZT00 and ZT12. The tissues were then washed with 1 mL fresh neurobasal medium (Gibco) supplemented with B27 (Gibco) and 25-mM HEPES (Gibco). The samples were incubated in a dissociation buffer containing 1 mg/mL papain (Worthington Biochemical Corporation, Lakewood, NJ), B27, and 25-mM HEPES diluted in a neurobasal medium for 30 min at 37 °C with gentle agitation. After incubation, gentle trituration was performed by pipetting 25 times with a 1-mL pipette. The transposition reaction, purification, and library preparation procedures were performed as described previously59. The libraries were quantified using real-time qPCR and sequenced on a single lane of Hi-seq 2500 (Illumina) with 100-bp paired-end reads (Macrogen).
Data processing for ATAC-seq
The sequencing files were first checked for quality control using FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc). Trimmomatic60 was used to remove the adapter sequences before mapping with Bowtie2 with the Mus musculus reference genome (mm10) parameters “--very sensitive --dovetail”61. Further, the unmapped and mapped reads were sorted, and the mapping quality below 10 was eliminated with SAMtools version 1.1262. To remove the PCR duplications during library preparation, the sorted reads were subjected to Picard deduplication (http://broadinstitute.github.io/picard/). Before further analysis, the final reads were checked again with FastQC, and we proceeded with further analysis when the quality control results were acceptable. The ATAC-seq peaks were called using HOMER with an FDR of 0.05 as the threshold. The called peaks were visualized using the UCSC Genome Browser. For annotations and motif analysis from the called peaks, their genomic regions and REV-ERBα40 and PET-163 motifs were processed with annotatePeaks using HOMER. For post-alignment quality control, ATACseqQC was performed to assess the fragment sizes of ATAC-seq libraries64, and chromatin accessibility from transcription start sites was visualized using deepTools (version 3.5.1)65.
Quantification and statistical analysis
Statistical analyses were performed using Prism9 (GraphPad, La Jolla, CA). We performed unpaired Student’s t test, one-way analysis of variance (ANOVA) followed by Dunnett’s post-hoc test, two-way ANOVA followed by Tukey’s post-hoc test. Data are presented as mean ± standard error of the mean. Significance was defined as *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, and “ns” indicated not significant. No statistical approach was used to determine the sample size. To minimize any observer bias in data analysis, the observers were randomly assigned with video recordings, and a blind data analysis was performed.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request. Key plasmids generated from this work are available from Addgene: pAAV-U6-sgRev-erba-hSyn-mCherry-KASH (Addgene ID:223226) and pAAV-U6-sgPet-1-hSyn-mCherry-KASH (Addgene ID:223227), mTph2p-Luc (Addgene ID: 223525), hTph2p-Luc (Addgene ID: 223526) and mTph2p-330bp-Luc WT (Addgene ID: 223663), mTph2p-330bp-Luc mut1 (Addgene ID: 223664), mTph2p-330bp-Luc mut2 (Addgene ID: 223665), mTph2p-330bp-Luc mut3 (Addgene ID: 223666), and mTph2p-330bp-Luc mut4 (Addgene ID: 223667). The raw dataset for ATAC-sequencing are available on Genome Sequence Archive, with accession code: CRA017728 and processed dataset for the ATAC-sequencing are uploaded on Figshare (https://doi.org/10.6084/m9.figshare.21806667.v1)66.
References
Takahashi, J. S. Transcriptional architecture of the mammalian circadian clock. Nat. Rev. Genet. 18, 164–179 (2017).
Mohawk, J. A., Green, C. B. & Takahashi, J. S. Central and peripheral circadian clocks in mammals. Annu. Rev. Neurosci. 35, 445–462 (2012).
Dibner, C., Schibler, U. & Albrecht, U. The mammalian circadian timing system: organization and coordination of central and peripheral clocks. Annu. Rev. Physiol. 72, 517–549 (2010).
Welz, P. S. et al. BMAL1-driven tissue clocks respond independently to light to maintain homeostasis. Cell 177, 1436–1447 (2019).
Koronowski, K. B. et al. Defining the independence of the liver circadian clock. Cell 177, 1448–1462 (2019).
Chung, S. et al. Impact of circadian nuclear receptor REV-ERBα on midbrain dopamine production and mood regulation. Cell 157, 858–868 (2014).
Landgraf, D. et al. Genetic disruption of circadian rhythms in the suprachiasmatic nucleus causes helplessness, behavioral despair, and anxiety-like behavior in mice. Biol. Psychiatry 80, 827–835 (2016).
Mukherjee, S. et al. Knockdown of clock in the ventral tegmental area through RNA interference results in a mixed state of mania and depression-like behavior. Biol. Psychiatry 68, 503–511 (2010).
Martini, T. et al. Deletion of the clock gene Period2 (Per2) in glial cells alters mood-related behavior in mice. Sci. Rep. 11, 12242 (2021).
Roybal, K. et al. Mania-like behavior induced by disruption of CLOCK. Proc. Natl. Acad. Sci. USA 104, 6406–6411 (2007).
Deneris, E. S. & Wyler, S. C. Serotonergic transcriptional networks and potential importance to mental health. Nat. Neurosci. 15, 519–527 (2012).
Liu, C. et al. Pet-1 is required across different stages of life to regulate serotonergic function. Nat. Neurosci. 13, 1190–1198 (2010).
Ren, J. et al. Single-cell transcriptomes and whole-brain projections of serotonin neurons in the mouse dorsal and median raphe nuclei. eLife 8, e49424 (2019).
Okaty, B. W., Commons, K. G. & Dymecki, S. M. Embracing diversity in the 5-HT neuronal system. Nat. Rev. Neurosci. 20, 397–424 (2019).
Miyazaki, K. W. et al. Optogenetic activation of dorsal raphe serotonin neurons enhances patience for future rewards. Curr. Biol. 24, 2033–2040 (2014).
Jang, S. et al. Impact of the circadian nuclear receptor REV-ERBα in dorsal raphe 5-HT neurons on social interaction behavior, especially social preference. Exp. Mol. Med. 55, 1806–1819 (2023).
Bacqué-Cazenave, J. et al. Serotonin in animal cognition and behavior. Int. J. Mol. Sci. 21, 1649 (2020).
Hensler, J. Serotonin in mood and emotion, in Handbook of the behavioral neurobiology of serotonin, Edn. 1st. (eds. Mueller C. & B. Jacobs) 367-378 (Academic Press, New York; 2010).
Zhang, X. et al. Loss-of-function mutation in tryptophan hydroxylase-2 identified in unipolar major depression. Neuron 45, 11–16 (2005).
Daut, R. A. & Fonken, L. K. Circadian regulation of depression: A role for serotonin. Front. Neuroendocrinol. 54, 100746 (2019).
Ciarleglio, C. M., Resuehr, H. E. & McMahon, D. G. Interactions of the serotonin and circadian systems: nature and nurture in rhythms and blues. Neuroscience 197, 8–16 (2011).
Malek, Z. S., Dardente, H., Pevet, P. & Raison, S. Tissue-specific expression of tryptophan hydroxylase mRNAs in the rat midbrain: anatomical evidence and daily profiles. Eur. J. Neurosci. 22, 895–901 (2005).
Ueda, H. R. et al. System-level identification of transcriptional circuits underlying mammalian circadian clocks. Nat. Genet. 37, 187–192 (2005).
Smith, K. A., Fairburn, C. G. & Cowen, P. J. Relapse of depression after rapid depletion of tryptophan. Lancet 349, 915–919 (1997).
Boyer, E. W. & Shannon, M. The serotonin syndrome. N. Engl. J. Med. 352, 1112–1120 (2005).
Antypa, N. et al. Associations between chronotypes and psychological vulnerability factors of depression. Chronobiol. Int. 34, 1125–1135 (2017).
Wong, D. T., Horng, J. S., Bymaster, F. P., Hauser, K. L. & Molloy, B. B. A selective inhibitor of serotonin uptake: Lilly 110140, 3-(p-trifluoromethylphenoxy)-N-methyl-3-phenylpropylamine. Life Sci. 15, 471–479 (1974).
Mann, J. J. The medical management of depression. N. Engl. J. Med. 353, 1819–1834 (2005).
Matveychuk, D. et al. Ketamine as an antidepressant: Overview of its mechanisms of action and potential predictive biomarkers. Ther. Adv. Psychopharmacol. 10, https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7225830/ (2020).
Krystal, J. H., Kavalali, E. T. & Monteggia, L. M. Ketamine and rapid antidepressant action: new treatments and novel synaptic signaling mechanisms. Neuropsychopharmacology 49, 41–50 (2024).
Nichols, D. E. Psychedelics. Pharmacol. Rev. 68, 264–355 (2016).
Miller, A. H. & Raison, C. L. Psychedelics and ketamine are a symptom of psychiatry’s woes, not a cure. Mol. Psychiatry 28, 3167–3168 (2023).
Gardner, J. P., Fornal, C. A. & Jacobs, B. L. Effects of sleep deprivation on serotonergic neuronal activity in the dorsal raphe nucleus of the freely moving cat. Neuropsychopharmacology 17, 72–81 (1997).
Zhao, X. et al. Acute sleep deprivation upregulates serotonin 2A receptors in the frontal cortex of mice via the immediate early gene Egr3. Mol. Psychiatry 27, 1599–1610 (2022).
Cederroth, C. R. et al. Medicine in the fourth dimension. Cell Metab. 30, 238–250 (2019).
Allada, R. & Bass, J. Circadian mechanisms in medicine. N. Engl. J. Med. 384, 550–561 (2021).
Zhang, Y. et al. Discrete functions of nuclear receptor Rev-erbα couple metabolism to the clock. Science 348, 1488–1492 (2015).
Banerjee, S. et al. Pharmacological targeting of the mammalian clock regulates sleep architecture and emotional behaviour. Nat. Commun. 5, 5759 (2014).
Griffin, P. et al. Circadian clock protein Rev-erbα regulates neuroinflammation. Proc. Natl. Acad. Sci. USA 116, 5102–5107 (2019).
Kim, J. et al. Pharmacological rescue with SR8278, a circadian nuclear receptor REV-ERBα antagonist as a therapy for mood disorders in Parkinson’s disease. Neurotherapeutics 19, 592–607 (2022).
Walther, D. J. & Bader, M. A unique central tryptophan hydroxylase isoform. Biochem. Pharmacol. 66, 1673–1680 (2003).
Zhang, X., Beaulieu, J. M., Sotnikova, T. D., Gainetdinov, R. R. & Caron, M. G. Tryptophan hydroxylase-2 controls brain serotonin synthesis. Science 305, 217 (2004).
Patel, P. D. et al. Regulation of tryptophan hydroxylase-2 gene expression by a bipartite RE-1 silencer of transcription/neuron restrictive silencing factor (REST/NRSF) binding motif. J. Biol. Chem. 282, 26717–26724 (2007).
Hiroi, R. & Handa, R. J. Estrogen receptor-β regulates human tryptophan hydroxylase-2 through an estrogen response element in the 5′ untranslated region. J. Neurochem. 127, 487–495 (2013).
Whitney, M. S. et al. Adult brain serotonin deficiency causes hyperactivity, circadian disruption, and elimination of siestas. J. Neurosci. 36, 9828–9842 (2016).
Kitt, M. M. et al. An adult-stage transcriptional program for survival of serotonergic connectivity. Cell Rep. 39, 110711 (2022).
Valdés-Fuentes, M. et al. Effect of daytime-restricted feeding in the daily variations of liver metabolism and blood transport of serotonin in rat. Physiol. Rep. 3, e12389 (2015).
Menon, J. M. L. et al. Brain microdialysate monoamines in relation to circadian rhythms, sleep, and sleep deprivation - a systematic review, network meta-analysis, and new primary data. J. Circadian Rhythms 17, 1 (2019).
Hampp, G. et al. Regulation of monoamine oxidase A by circadian-clock components Implies clock influence on mood. Curr. Biol. 18, 678–683 (2008).
Angoa-Pérez, M. et al. Mice genetically depleted of brain serotonin do not display a depression-like behavioral phenotype. ACS Chem. Neurosci. 5, 908–919 (2014).
Jia, Y.-F. et al. Abnormal anxiety- and depression-like behaviors in mice lacking both central serotonergic neurons and pancreatic islet cells. Front. Behav. Neurosci. 8, 325 (2014).
Lyon, K. A. et al. Sex-specific role for dopamine receptor D2 in dorsal raphe serotonergic neuron modulation of defensive acoustic startle and dominance behavior. eNeuro 7, 1–23 (2020).
Otsuka, T. et al. Deficiency of the circadian clock gene Rev-erbα induces mood disorder-like behaviours and dysregulation of the serotonergic system in mice. Physiol. Behav. 256, 113960 (2022).
Hu, R. et al. A novel method of neural differentiation of PC12 cells by using Opti-MEM as a basic induction medium. Int. J. Mol. Med. 41, 195–201 (2018).
Sayers, E. W. et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 49, D10–D17 (2021).
Montague, T. G., Cruz, J. M., Gagnon, J. A., Church, G. M. & Valen, E. CHOPCHOP: a CRISPR/Cas9 and TALEN web tool for genome editing. Nucleic Acids Res. 42, W401–W407 (2014).
Kim, B. et al. Multiplexed CRISPR-Cas9 system in a single adeno-associated virus to simultaneously knock out redundant clock genes. Sci. Rep. 11, 2575 (2021).
Park, S. H. et al. Type I interferons and the cytokine TNF cooperatively reprogram the macrophage epigenome to promote inflammatory activation. Nat. Immunol. 18, 1104–1116 (2017).
Buenrostro, J. D., Wu, B., Chang, H. Y. & Greenleaf, W. J. ATAC-seq: A method for assaying chromatin accessibility genome-wide. Curr. Protoc. Mol. Biol. 109, 21.29.21–21.29.29 (2015).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Li, H. et al. The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Sandelin, A., Alkema, W., Engström, P., Wasserman, W. W. & Lenhard, B. JASPAR: An open-access database for eukaryotic transcription factor binding profiles. Nucleic Acids Res. 32, D91–D94 (2004).
Ou, J. et al. ATACseqQC: a Bioconductor package for post-alignment quality assessment of ATAC-seq data. BMC Genomics 19, 169 (2018).
Ramirez, F., Dundar, F., Diehl, S., Gruning, B. A. & Manke, T. deepTools: A flexible platform for exploring deep-sequencing data. Nucleic Acids Res. 42, W187–W191 (2014).
Park, I. et al. Role of the circadian nuclear receptor REV-ERBα in dorsal raphe serotonin synthesis in mood regulation [Data set]. Figshare https://doi.org/10.6084/m9.figshare.21806667.v1 (2023).
Acknowledgements
This work was supported by the National Research Foundation (NRF) of Korea (2021R1A2C1004803) awarded to K.K., a grant from the basic science research program through the NSF funded by the Ministry of Education (2020R1A6A1A03040516) to CM, grants from NRF funded by the Ministry of Science and ICT (2019M3C1B8090845 and 2018M3C7A1022310) to H.K.C., a grant from Korea Brain Research Institute (22-BR-02-01) to Y.C. and a grant from the Bio & Medical Technology Development Program of the NRF funded by the Ministry of Science and ICT (2017M3A9G8084463) to J.-W.C.
Author information
Authors and Affiliations
Contributions
K.K. supervised the project. I.P., G.H.S., and K.K. designed the experiments and wrote the manuscript with contributions from all coauthors. I.P. and M.C. conducted most of the experiments and analyzed the data. S.J. assisted in behavioral data analyses. J.K1. acquired the preliminary results with optogenetic experiment. Y.C. conducted a single-cell RNA sequencing analysis for preliminary study and profiled candidate circadian transcription factors in 5-HTergic neurons. J.K3., D.K., Y.C., S.-W.Y., C.M., and H.K.C. shared their comments and reviewed the manuscript. D.G. helped develop the hypothesis of this work. J.-W.C., C.M., and H.K.C. contributed experimental reagents and analytic tools.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Biology thanks the anonymous reviewers for their contribution to the peer review of this work. Primary Handling Editors: Christoph Anacker and Joao Valente.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Park, I., Choi, M., Kim, J. et al. Role of the circadian nuclear receptor REV-ERBα in dorsal raphe serotonin synthesis in mood regulation. Commun Biol 7, 998 (2024). https://doi.org/10.1038/s42003-024-06647-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s42003-024-06647-y
- Springer Nature Limited