Abstract
Replicative senescence is triggered when telomeres reach critically short length and activate permanent DNA damage checkpoint-dependent cell cycle arrest. Mitochondrial dysfunction and increase in oxidative stress are both features of replicative senescence in mammalian cells. However, how reactive oxygen species levels are controlled during senescence is elusive. Here, we show that reactive oxygen species levels increase in the telomerase-negative cells of Saccharomyces cerevisiae during replicative senescence, and that this coincides with the activation of Hog1, a mammalian p38 MAPK ortholog. Hog1 counteracts increased ROS levels during replicative senescence. While Hog1 deletion accelerates replicative senescence, we found this could stem from a reduced cell viability prior to telomerase inactivation. ROS levels also increase upon telomerase inactivation when Mec1, the yeast ortholog of ATR, is mutated, suggesting that oxidative stress is not simply a consequence of DNA damage checkpoint activation in budding yeast. We speculate that oxidative stress is a conserved hallmark of telomerase-negative eukaryote cells, and that its sources and consequences can be dissected in S. cerevisiae.
Similar content being viewed by others
Introduction
Telomeres are essential structures found at the ends of linear eukaryotic chromosomes, consisting of DNA sequences, proteins, and long non-coding RNA (LncRNA) telomeric transcripts1. Telomeres crucially safeguard chromosome integrity by protecting against degradation and fusion events2. However, due to the “DNA end-replication problem”, telomeres gradually shorten with each cell cycle. Telomerase, a specialized reverse transcriptase, counteracts telomere shortening by adding repetitive telomeric sequences to chromosome ends. In human somatic cells, the telomere-protective functions become compromised when the telomeres shorten with cell divisions due to telomerase inactivation coupled with the “DNA end-replication problem”. When telomere lengths become critically short, they activate an irreversible DNA damage checkpoint-dependent cell cycle arrest, known as replicative senescence3,4. The unicellular eukaryote, Saccharomyces cerevisiae, relies on telomerase for its long-term viability5, but similar to human somatic cells, telomerase inactivation in budding yeast also leads to replicative senescence. When budding yeast cells divide in the absence of telomerase, they cease proliferation and enter a metabolically active state, arresting in the G2/M phase of the cell cycle6,7. This cell cycle arrest in S. cerevisiae, which is akin to mammalian cells, relies on the activation of the DNA damage checkpoint kinases, Mec1 and Tel1 (the yeast orthologs of ATR and ATM, respectively), in addition to Rad53 phosphorylation8,9,10. Remarkably, not only is the triggering of replicative senescence in response to short telomeres evolutionarily conserved, but also many other essential telomeric functions and maintenance mechanisms11,12.
Studying replicative senescence is challenging due to the inherent heterogeneity resulting from intracellular differences in telomere lengths and the immense intercellular variations13. Intriguingly, data collected from various organisms indicate that mitochondrial defects, oxidative stress, and chronic inflammation can accelerate telomere shortening and dysfunction14,15,16,17. These factors are potential sources of cell-to-cell variation and contribute to genome instability during replicative senescence18. Notably, senescent human fibroblasts exhibit modifications in mitochondrial structure and function, accompanied by elevated reactive oxygen species (ROS) levels and oxidative damage19,20,21,22,23,24. Similar metabolic alterations have also been observed in budding yeast during replicative senescence. A previous study revealed that the absence of telomerase resulted in increased mitochondrial mass, and a transcriptomic analysis indicated that energy production genes were up-regulated and stress response genes were induced25. However, data regarding ROS level alterations and their regulation during senescence in budding yeast is currently lacking.
P38, a member of the mitogen-activated protein kinase (MAPK) family, is critical for various cellular processes, including cellular senescence and oxidative stress responses26. In budding yeast, the MAPK Hog1, the ortholog of mammalian p38, is crucial for the defence against many stressors, including osmotic27 and oxidative stress28. The canonical pathway of Hog1 activation involves two branches, the Sho1 and Sln1 branches, which converge to activate the MAPK kinase (MAPKK), Pbs229. Pbs2 then interacts with and phosphorylates Hog1 at the conserved residues, Thr174 and Tyr176, leading to its activation. Hog1 is a multifunctional protein with important functions in both the cytoplasm and nucleus, and it is vital for stress adaptation30. Its roles encompass regulating gene expression by activating transcription factors, participating in gene initiation and elongation, regulating the cell cycle, and contributing to various steps in mRNA metabolism. Notably, Hog1 is activated in response to H2O2 stress28 and regulates antioxidant genes by activating the transcription factors, Msn2/Msn431 and Sko132. In the absence of Hog1, cells become more sensitive to H2O2, which was previously shown to correlate with reduced expression of the TSA2 gene31. Conversely, sustained Hog1 activation can lead to cell death, which has been linked to alterations in mitochondrial respiration and increases in ROS levels33. Hog1 counters this ROS increase by inducing PNC1 and activating Sir2. Multiple studies have demonstrated that uncontrolled Hog1 activation disrupts mitochondrial function and elevates ROS levels, underscoring the critical importance of regulating Hog1 activation34,35. Furthermore, Hog1 has been implicated in autophagic processes and is required for mitophagy, the selective process of mitochondria degradation36,37,38. Interestingly, Hog1 also positively regulates the localization of the Sir complex to telomeres following osmotic stress and the silencing of telomeric regions39.
Here, we report that ROS levels increase during replicative senescence in budding yeast. During this process, Hog1 is activated by Pbs2 and plays a role in ROS detoxification. This countering of ROS increase occurs independently from the actions of Mec1. We also find that autophagy does not participate in replicative senescence in budding yeast. However, Hog1 participates in maintaining telomere length homeostasis and affects cell viability. Our results thus suggest that Hog1 serves as a link between telomeres and ROS metabolism.
Results
ROS levels increase during replicative senescence in budding yeast
Human senescent cells exhibit increased ROS levels during replicative senescence, however, data relating to S. cerevisiae senescent cells is lacking19,20. Budding yeast telomerase is constitutively active but experimentally inactivating it triggers replicative senescence8. We used a validated TetO2-TLC1 system, where TLC1, which encodes the telomerase RNA template, is controlled by a repressible promotor by doxycycline, enabling the conditional shut-off of telomerase10,40,41. To measure ROS levels, we used DCF, a molecule that can be directly oxidized by ROS and produce fluorescence in quantities reflecting ROS levels, which we can quantify by flow cytometry. Replicative senescence was observed in the PTetO2-TLC1 strain starting from day three of culture with doxycycline, as the cell proliferation capacity gradually decreased, eventually reaching a crisis point by day 5 (Fig. 1a). We also observed a simultaneous increase in ROS levels in the absence of telomerase all along the experiment in presence of doxycycline with a significant increase as compare to the untreated condition starting from the day 5 (Fig. 1b). These results were recapitulated in strains where TLC1 was deleted; in these strains, we also found that ROS levels declined as cultures recovered their initial proliferation capacity following the emergence of post-senescence survivors (Supplementary Fig. 1). As previously outlined, during prolonged culturing of senescent cells, rare events, estimated to occur at a frequency of approximately 2 × 10−5 cells, enables cells to circumvent senescence and resume cellular divisions42,43. These dividing cells, referred to as post-survivors, have the ability to replenish the culture and sustain their telomere length independently of telomerase activity. Instead, they rely on recombination mechanisms, known as the alternative lengthening of telomeres mechanism, which are conserved in yeast and mammals. These data indicate that similar to mammalian models, budding yeast telomerase-negative senescent cultures exhibit increased ROS levels.
Each consecutive day, cells with the genotype indicated were diluted in media either with or without doxycycline (Dox) to enable conditional shut-off of telomerase, and grown for 24 h. Cell density at OD600nm (a) and ROS levels normalized to the strain without doxycycline (b) are plotted as mean ± SD of three independent clones. P values were calculated by two-tailed Student’s t test (* < 0.05; ** < 0.01) and only significant differences have been represented.
Hog1 is activated during replicative senescence in a Pbs2-dependent manner and counteracts increase in ROS levels
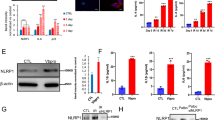
We next investigated how ROS are regulated during replicative senescence. We focused on the multifunctional MAPK, Hog1, which is activated by oxidative stress28,30. To determine whether Hog1 activation occurred during replicative senescence, we prepared protein extracts from senescent cultures and used a specific antibody to detect phosphorylated forms of Hog1. We observed that Hog1 was phosphorylated during replicative senescence from day three, concomitant with an increase in ROS levels (Fig. 2d or Supplementary Fig. 2b and Fig. 2b, respectively). The MAPKK Pbs2 precedes Hog1 in its canonical pathway and is the only Kinase capable of phosphorylating Hog129. Therefore, the Western blot presented in Fig. 2d, showing that the phosphorylated form of Hog1 is no longer detectable in the PBS2-deleted mutant, confirms the deletion. We also observed that HOG1 or PBS2 deletion (PTetO2-TLC1 hog1∆ or PTetO2-TLC1 pbs2∆) (Fig. 2c and Supplementary Fig. 2a or Fig. 2d, respectively) resulted in a similar premature loss of cell viability, when telomerase was inactivated (Fig. 2a). This suggests that the Hog1 pathway may play a role in senescent cells. While the HOG1 pathway activates transcription factors of antioxidant genes to reduce ROS levels33, excessive Hog1 activity can increase ROS levels by disrupting mitochondrial respiration33,34. To understand which of these Hog1 functions was involved in replicative senescence, we measured ROS levels in the absence or presence of Hog1 during senescence. Our results showed that the increases in ROS levels started earlier and reached higher levels in PTetO2-TLC1 hog1∆ compared to PTetO2-TLC1 strains (Fig. 2b). We also observed that HOG1 or PBS2 deletion showed a similar increase in the level of ROS in the course of senescence (Supplementary Fig. 3), confirming the epistatic interaction between the two genes. This suggests that increases in ROS levels trigger an oxidative stress response that activates via Pbs2 the Hog1 pathway, which is required for ROS detoxification in telomerase-inactivated cells.
Cells with the genotypes indicated were treated (D1 to D5) or not (D0) with doxycycline as described in Fig. 1. Cell density at OD600nm (a) and ROS levels normalized to the PTetO2-TLC1 strain without doxycycline (b) are plotted as mean ± SD of three independent clones. P values were calculated by two-tailed Student’s t test (* < 0.05; ** < 0.01; *** < 0.001) and only significant differences between the two strains used have been represented. c, d Protein extracts, from experiment presented in (a), analysed by Western blot using an antibody against phosphorylated forms of Hog1’s human ortholog p38 (Hog1-P), total Hog1 (Hog1-T) or against Pgk1.
HOG1 deletion affects telomere length homeostasis and cell viability
Given the direct relationship between replicative senescence and telomere shortening, we investigated the potential influence of Hog1 on telomere length homeostasis prior to telomerase inactivation, and the rate of telomere shortening in the absence of telomerase. We thus performed telomere-PCR on DNA samples from PTetO2-TLC1 and PTetO2-TLC1 hog1∆ strains to determine telomere length. Our results showed that HOG1 deletion resulted in slightly shorter telomeres of ~30 bp prior to telomerase inactivation (Supplementary Fig. 5a, b). However, no significant differences in telomere shortening rates were observed in the absence of telomerase in either the PTetO2-TLC1 or PTetO2-TLC1 hog1Δ strains; shortening rates were measured to be approximately 2.5 bp/cell population doubling (Fig. 3a, b, Supplementary Fig. 4), similar to previously published results40,44. Therefore, one plausible hypothesis is that Hog1 contributes to the maintenance of telomere length homeostasis. As telomere length homeostasis results from the balance between telomere lengthening by telomerase and telomere shortening due to “DNA end replication problem”, we suggest that Hog1 might promote telomerase recruitment or activity.
a Cells of the genotypes indicated were pre-cultured overnight in doxycycline-containing media to enable telomerase shut-off. Cells were then diluted and grown in the same media for the indicated population doublings (PD). Representative telomere-PCR of Y’ telomeres from genomic DNA extracts are shown. b Quantification of telomere shortening was measured between PD 1 to 5 and plotted as mean ± SD. c, d Microfluidics results of independent lineages with the genotypes indicated. Cells were introduced into the microfluidics microcavities and cultured in SD without doxycycline. Each horizontal line represents the consecutive cell cycles (generations) of a single lineage, and each segment corresponds to one cell cycle. An ellipsis (…) at the end of the lineage line indicates that the cell was living after the experiment, whereas an X indicates cell death. Cell cycle duration is indicated by the colored bar. Scripts used to generate Fig. 3c, d are available: https://doi.org/10.5281/zenodo.11634847.
To test if the accelerated senescence we observed in the hog1∆ strains, when telomerase was inactivated, is due to shorter telomere lengths prior to telomerase inactivation, we evaluated the senescence curve of the TLC1 HOG1 (the wild type condition), TLC1 hog1∆, PTetO2-TLC1 and PTetO2-TLC1 hog1∆ haploids cells from tetrad dissection. We can observe that the change from the endogenous TLC1 promoter to the PTetO2 promoter induces a shortening of telomere length, as previously characterized10 (Supplementary Fig. 5c, d). Moreover, significant clonal variability in telomere length was previously reported45. We find this variability among the majority of clones exhibiting the same genotype (Supplementary Fig. 5c, d). In this context, the small but reproducible decrease in telomere size, significantly observed in three independent PTetO2-TLC1 hog1∆ transformants compared to PTetO2-TLC1 (Supplementary Fig. 5a, b), is no longer observed in a context of tetrad dissection (Supplementary Fig. 5c, d). Yet, the proliferation of PTetO2-TLC1 hog1∆ spores in the presence of doxycycline, consistently show accelerated senescence compared to PTetO2-TLC1 sister spores (Supplementary Fig. 5e). Based on these results, we can conclude that the acceleration of senescence in the absence of Hog1 is independent of the initial shorter telomere length observed in this strain. As expected, the growth curves of the HOG1 and the hog1∆ cells, where TLC1 has is endogenous promoter, are not affected over five days with doxycycline (Supplementary Fig. 5e). Altogether, these data help us separate the slight decrease in telomere size observed in PTetO2-TLC1 hog1∆ before telomerase inactivation from the accelerated senescence observed in the same strain after telomerase inactivation.
Another hypothesis to account for the acceleration of senescence in PTetO2-TLC1 hog1∆ cells is that the deletion of HOG1 might influence cellular mortality. To test this, we employed a microfluidics system, which allows consecutive cell cycles from individual cell lineages (herein referred to as lineages) to be tracked, thereby enabling more precise quantification of cell proliferation46. In the presence of telomerase, PTetO2-TLC1 cells grew indefinitely with a spontaneous mortality rate of ~0.38% (Fig. 3c). In contrast, the absence of HOG1 caused the mortality rate to increase to ~5.8% even in the presence of telomerase (Fig. 3d). This has not previously been observed with cell population growth (liquid or solid). This data indicates that HOG1 loss alone causes some cell death, which, when combined with telomerase inactivation, could have contributed to the accelerated senescence we observed.
Collectively, these findings suggest that the apparent acceleration of senescence we observed in the absence of Hog1 could be attributable to a marked increase in intrinsic mortality rates. However, this does not preclude a significant role for Hog1 in detoxifying ROS during replicative senescence, particularly as PTetO2-TLC1 hog1∆ strains exhibit much higher ROS levels throughout replicative senescence compared to PTetO2-TLC1 strains (Fig. 2b and Supplementary Fig. 3). To understand the impact of higher basis level ROS on the viability of PTetO2-TLC1 hog1∆ cells, we evaluated the cell proliferation of PTetO2-TLC1 hog1∆ cells versus PTetO2-TLC1 treated with the antioxidant N-acetyl-L-cysteine (NAC) 10 mM over 10 generations in the exponential phase. The NAC treatment significantly improved proliferation and ROS levels in both strains, however PTetO2-TLC1 hog1∆ showed the most improvement (Supplementary Fig. 6a, b, respectively). Interestingly, we observed that PTetO2-TLC1 hog1∆ cells treated with 10 mM NAC during exponential phase exhibited a partial rescue of telomere length. However, the telomere length of PTetO2-TLC1 cells remained unaffected (Supplementary Fig. 6c, d). This data underscores the sensitivity of telomeric sequences to ROS compared to the rest of the genome47, emphasizing the importance of Hog1 in maintaining ROS balance under basal condition.
Autophagic processes do not modify the onset of replicative senescence
Autophagy is a process that involves self-eating and bulk degradation, where organelles and their components are delivered to vacuoles to be degraded and recycled48. Autophagy can be selective, when specific cargo, such as damaged organelles, are degraded49. The specific degradation of dysfunctional mitochondria is known as mitophagy. Hog1 plays a role in autophagy under certain conditions50,51,52 and is considered to be a mitophagy activator36,37,53. We therefore investigated whether autophagic processes were essential during replicative senescence. We deleted the ATG8 and ATG32 genes, which encode two proteins essential for bulk autophagy and mitophagy in budding yeast, respectively49. These mutants PTetO2-TLC1 atg8∆ and PTetO2-TLC1 atg32∆ displayed a blockage of bulk autophagy and mitophagy respectively, confirmed by an assay based on Rosella, a fluorescence-based pH biosensor54 (Supplementary Fig. 7).
We then investigated whether blocking these processes in wild-type cells would affect senescence dynamics. Liquid senescence assays revealed that senescence remained unchanged in the absence of autophagy or mitophagy, suggesting that these processes were not essential for the viability of telomerase-negative budding yeast cells (Fig. 4a). Similarly, following telomerase inactivation, the senescence profiles remained unchanged when either ATG8 or ATG32 were deleted in a hog1∆ background (Fig. 4a). We concluded that Hog1 activity occurs independently from autophagic processes, which do not alter the onset of replicative senescence in budding yeast.
a, b Cells with the genotypes indicated were treated as described in Fig. 1. Cell density at OD600nm from the strains indicated are plotted as mean ± SD of three independent clones. c Results derived from the senescence experiment depicted in Fig. 4b. Protein extracts analysed by Western blot using an antibody against Rad53. The mobility shift of the band indicates Rad53 phosphorylation (the upper arrow). d ROS levels normalized to the PTetO2-TLC1 strain without doxycycline derived from the senescence experiment depicted in Fig. 4b using the same legend are plotted as mean ± SD of three independent clones. P values were calculated by two-tailed Student’s t test (* < 0.05; ** < 0.01; *** < 0.001) and only significant differences between the triple mutant and the double mutants have been represented.
Hog1 acts in a Mec1-independent manner to regulate ROS levels during replicative senescence
We hypothesized that ROS level increases could result from cells being in a senescent state. Mec1 is a pivotal kinase in budding yeast, essential for the DNA damage checkpoint and the onset of replicative senescence55. Mec1 and the DNA damage checkpoint pathway are also known to protect cells against oxidative stress56. Hog1 and Mec1 are both necessary to combat the oxidative stress induced by hydrogen peroxide, but act independently28. A hypomorph mutant of MEC1, mec1-21, contains a G to A substitution at position 2644, outside the kinase domain57. The mec1-21 mutant displays lower dNTP levels and shorter telomeres (~50 bp) compared to wild type strains57,58. The mec1-21 mutant retains essential functions but is defective for the S phase checkpoint and Rad53 activation following UV and HU exposure59,60. We thus used the mec1-21 mutant to investigate whether the actions of Hog1 against ROS were Mec1-dependent during replicative senescence. Even prior to telomerase inactivation (D0), we observed a slight decrease in proliferation in the single PTetO2-TLC1 mec1-21 strain indicating an increase in the proportion of deceased cells (Fig. 4b). This trend is further pronounced in the double mutant PTetO2-TLC1 mec1-21 hog1. This observation remains consistent for the ROS levels observed in Fig. 4d at D0. Therefore, it is well possible that ROS levels follow mortality. Then we inactivated telomerase and measured cell growth over time in the PTetO2-TLC1, PTetO2-TLC1 hog1∆, PTetO2-TLC1 mec1-21, and triple mutant strains. When compared to MEC1 cells, we observed that mec1-21 accelerated the loss of viability under telomerase-negative conditions (Fig. 4b), consistent with the initial shorter telomeres (Supplementary Fig. 8a, b). We also verified that Rad53 phosphorylation was impaired in the mec1-21 strains (Fig. 4c and Supplementary Fig. 9). Yet, under these conditions, where the DNA damage checkpoint was disabled, telomerase inactivation resulted in more pronounced increase in ROS levels. This indicates that Mec1 also participates in ROS detoxification in the absence of telomerase (Fig. 4d). In addition, while the triple mutant, PTetO2-TLC1 hog1∆ mec1-21, displayed a much lower proliferation capacity, it showed an even more pronounced increase in ROS compared to the respective single mutants (Fig. 4d). As we showed in Supplementary Fig. 6a, b, The PTetO2-TLC1 hog1∆ cells, treated with NAC 10 mM during the exponential phase, exhibit an increase in cell viability coupled with a decrease of ROS levels. These results are consistent with a model where Hog1 and Mec1 are both involved in ROS detoxification during replicative senescence but act in independent pathways.
Discussion
Here, we have shown that increased ROS levels are a feature of replicative senescence in budding yeast. Hog1, one of the five MAPKs of S. cerevisiae, is activated by Pbs2 during replicative senescence and counteracts increases in ROS levels, likely independently of Mec1. In addition, Hog1 regulates telomere length homeostasis, and its deletion results in a marked increase in cell mortality rates. Our findings also indicate that autophagic processes are not essential in the context of replicative senescence in budding yeast.
Previous studies have shown that Hog1 is activated in response to exogenous acute stresses, such as H2O2 exposure, where it is essential for triggering antioxidant genes and maintaining cell viability28,31. Given our findings that ROS levels increase during replicative senescence, it is plausible that oxidative stress directly triggers Hog1 pathway activation. Replicative senescence is an endogenous process resulting from telomerase inhibition, that leads to numerous cellular modifications at both genomic and metabolic levels. Consequently, other modifications may also contribute to Hog1 activation. Notably, a previous study proposed that Hog1 activation in response to H2O2 stress primarily occurs through the Sln1-Ypd1-Ssk1-Ssk2-Pbs2 pathway, with Ssk2 acting as the MAPKKK that specifically activates Hog1 in response to oxidative stress, but not Sho1 branch61. Therefore, determining the Hog1 pathway upstream of Pbs2 might help clarify the origin of Hog1 pathway activation in the absence of telomerase.
Microfluidics analysis, where cells grow individually, showed that HOG1 deletion in the presence of telomerase increased cell mortality rates by ~15 fold. This increase may have gone undetected in other studies where cells were grown in populations as colonies or liquid cultures due to competition and selection of the fittest cells. Similar mortality rates have been described for other mutants considered “viable”, underscoring the high sensitivity of the microfluidics method46. We speculate that the Hog1 pathway might be important in response to certain intrinsic stresses, and that it becomes essential in rare situations. Accordingly, a potential role for Hog1 under normal stress-free cellular conditions, unrelated to telomeres, has been described62.
We have shown that Hog1 participates in telomere length homeostasis in budding yeast. This could be due to Hog1 acting to positively regulate telomere transcriptional silencing through the localization of the Sir complex following osmotic stress39. It could also be that the absence of Hog1 disrupts subtelomeric heterochromatin, which would alter telomere length homeostasis.
In conclusion, this study has shown that the metabolic alterations observed in human senescent cells are conserved in budding yeast. These alterations involve a conserved MAPK Hog1/p38 pathway, although the outcome might differ in different species. As most basic functions in telomere biology are conserved in eukaryotes, determining the mechanistic link between telomere shortening and increases in ROS levels in budding yeast will be essential to clarify how telomeres have evolved in the context of eukaryotic evolution.
Material and methods
Yeast strains
All yeast strains used in this study were derived from a W303 background corrected for RAD5 and ADE2 (Table 1). Gene deletions were constructed as previously described63. Mec1-21 point mutation was constructed using Crispr-Cas9, as previously described64. Strains expressing Rosella constructs from plasmids were constructed as previously described54. Primers used are listed in Table 2.
Liquid senescence experiments
Strains were grown at 30 °C in liquid-rich media (YPD). Cell suspensions were diluted to 0.001 OD600nm with a final concentration of 30 µg/mL of doxycycline (Sigma-Aldrich #D9891), and the OD600nm was measured after 24 h. Cultures were similarly diluted for several days and daily samples were taken for analysis.
ROS detection
Yeast cultures with an OD600nm of 0.4 were incubated at 30 °C in darkness for one hour in 500 μL of sterile 1X PBS, containing DCF-DA (2’,7’-Dichlorofluorescin diacetate) (Sigma-Aldrich #D6883) at a final concentration of 10 μM. Samples were washed and then resuspended in 500 μL of 1X PBS. Fluorescence was then analysed by flow cytometry using the settings, 488 (λex)/533 (λem) in Accuri C6 or MACSQuant Analyzer 10. The mean intensity values were then plotted. The mean values of PTetO2-TLC1 at day 0 without doxycycline were subtracted.
DNA extraction
Cells with an OD600nm of 5 were centrifuged for 4 min at 2000 g and washed in 500 μL of sterile distilled water. After centrifugation, 200 μL of lysis buffer (Triton 100 X - 10% SDS sodium dodecyl sulfate - 5 M NaCl – 0.5 M EDTA ethylenediaminetetraacetic acid - 1 M Tris - H2O), 200 μL of 0.45 μm acid-washed glass beads (Sigma-Aldrich #G8772), and 200 μL of phenol: chloroform: isoamyl alcohol solution (25:24:1, Sigma-Aldrich #77617) were added to the cell pellet. The tubes were vortexed for 15 min at 4 °C, followed by the addition of 200 μL of TE (Tris/EDTA) at pH 8. After five minutes of centrifugation at maximum speed, the aqueous phase was transferred to a new tube containing the same volume of isopropanol. After mixing by inversion, the samples were centrifuged for 1 min at maximum speed. The resulting DNA pellet was washed in 500 μL of ethanol. Finally, after centrifuging and drying in a speed vacuum (3 min at 40 °C), the DNA pellet was resuspended in 50 μL of TE and 0.1 μL of RNase A (100 mg/ml) and incubated for 30 min at 37 °C. The quality and integrity of the DNA were checked by agarose gel electrophoresis (1% agarose, 0.5X TBE). The quantity was evaluated using Qubit2 (Thermofisher). Samples were stored at −20 °C.
SDS-PAGE and western blot
Cells with an OD600nm of 5 were collected by centrifugation. The pellet was lysed in 0.2 M of sodium hydroxide (NaOH) for 10 min on ice. After adding trichloroacetic acid (TCA) at a final concentration of 0.5%, the samples were incubated again on ice for 10 min. After centrifuging for 10 min at maximum speed at 4 °C, the pellet was resuspended in 50 μL of Laemmli 4X buffer and denatured at 95 °C for five minutes. Protein samples were electrophoresed on a 10% denaturing gel (or 7.5% for Rad53 detection) of 37.5:1 Acrylamide: Bis-acrylamide (Sigma-Aldrich #A3699). Proteins were then transferred to a nitrocellulose membrane (Amersham Protran 0.45 NC, GE HealthCare) and stained with Ponceau red. The following antibodies were used: Cell Signaling, #9211 to detect Phospho-Hog1, Santa Cruz, #sc-165978 to detect total Hog1, Abcam, #ab104252 to detect both unphosphorylated and phosphorylated forms of Rad53, and the horseradish peroxidase-coupled secondary antibody (HRP) (Sigma, #A9044 and #A9169). The signals were revealed using an electrochemiluminescence reagent (ClarityWestern ECL, Biorad) and recorded using the ChemiDoc Imaging System (Biorad).
Telomere-PCR
This method was adapted from ref. 65. In brief, 40 ng of genomic DNA was denatured at 98 °C for 5 min before the tailing reaction in 20 μL of New England Biolabs Buffer 4 (1X), 100 μM of dCTP, and 1U of terminal transferase (New England Biolabs #M0315L). The reaction was incubated at 37 °C for five minutes, followed by 5 min at 94 °C, and then maintained at 4 °C. For the PCR reactions, 5 μL of the polyC-tailed products were used with 1X Taq Mg-free Buffer (New England Biolabs), 500 nM of each primer (Table 2), 200 μM of dNTPs, and 1 U of Taq Standard Polymerase (New England Biolabs #M0273) in a final volume of 30 μL. The following PCR program was used: 3 min at 94 °C, followed by 45 cycles of 20 s at 94 °C, 15 s at 57 °C, 20 s at 72 °C, and finally, five minutes at 72 °C. The TeloPCR products were then loaded onto a large 2% agarose gel with 1X TBE buffer and 100 ng/ml of ethidium bromide (BET). A 50 bp molecular weight marker was also loaded (New England Biolabs #N3236). Electrophoresis was then performed at 50 V for 15 h. Visualization and analysis were performed using the ImageLab® software (Biorad).
Microfluidics analysis
Microfluidics analyses were performed as previously described46.
Statistics and reproducibility
All data were analysed with excel and are presented as means ± SD. Comparisons between two groups were performed using a two-tailed Student’s t test. A P < 0.05 was considered to be statistically significant. To ensure reproducibility, a minimum of three replicates were conducted with three independent clones. The figure legend details sample sizes and specifies the replicates.
Data availability
Code availability
Scripts used with Matlab 2014a to create graphs from microfluidic analysis results are available in Zenodo, a DOI-minting repository. https://doi.org/10.5281/zenodo.11634847.
References
Shay, J. W. & Wright, W. E. Telomeres and telomerase: three decades of progress. Nat. Rev. Genet. 20, 299–309 (2019).
Jain, D. & Cooper, J. P. Telomeric strategies: means to an end. Annu. Rev. Genet. 44, 243–269 (2010).
d’Adda, D. F. F. et al. A DNA damage checkpoint response in telomere-initiated senescence. Nature 426, 194–198 (2003).
Campisi, J. & d’Adda, D. F. F. Cellular senescence: when bad things happen to good cells. Nat. Rev. Mol. Cell Biol. 8, 729–740 (2007).
Lundblad, V. & Szostak, J. W. A mutant with a defect in telomere elongation leads to senescence in yeast. Cell 57, 633–643 (1989).
Enomoto, S., Glowczewski, L. & Berman, J. MEC3, MEC1, and DDC2 are essential components of a telomere checkpoint pathway required for cell cycle arrest during senescence in Saccharomyces cerevisiae. Mol. Biol. Cell 13, 2626–2638 (2002).
Ijpma, A. S. & Greider, C. W. Short telomeres induce a DNA damage response in Saccharomyces cerevisiae. Mol. Biol. Cell 14, 987–1001 (2003).
Teixeira, M. T. Saccharomyces cerevisiae as a model to study replicative senescence triggered by telomere shortening. Front. Oncol. 3, 101 (2013).
Abdallah, P. et al. A two-step model for senescence triggered by a single critically short telomere. Nat. Cell Biol. 11, 988–993 (2009).
Khadaroo, B. et al. The DNA damage response at eroded telomeres and tethering to the nuclear pore complex. Nat. Cell Biol. 11, 980–987 (2009).
Kupiec, M. Biology of telomeres: lessons from budding yeast. FEMS Microbiol. Rev. 38, 144–171 (2014).
Wellinger, R. J. & Zakian, V. A. Everything you ever wanted to know about Saccharomyces cerevisiae telomeres: beginning to end. Genetics 191, 1073–1105 (2012).
Xu, Z. & Teixeira, M. T. The many types of heterogeneity in replicative senescence. Yeast 36, 637–648 (2019).
Opresko, P. L. et al. Telomere-binding protein TRF2 binds to and stimulates the Werner and Bloom syndrome helicases. J. Biol. Chem. 277, 41110–41119 (2002).
Fouquerel, E. et al. Targeted and Persistent 8-Oxoguanine base damage at telomeres promotes telomere loss and crisis. Mol. Cell 75, 117–130 e116 (2019).
Ahmed, W. & Lingner, J. PRDX1 and MTH1 cooperate to prevent ROS-mediated inhibition of telomerase. Genes Dev. 32, 658–669 (2018).
Ahmed, W. & Lingner, J. PRDX1 counteracts catastrophic telomeric cleavage events that are triggered by DNA repair activities post oxidative damage. Cell Rep. 33, 108347 (2020).
Passos, J. F. & von Zglinicki, T. Mitochondria, telomeres and cell senescence. Exp. Gerontol. 40, 466–472 (2005).
Passos, J. F. et al. Feedback between p21 and reactive oxygen production is necessary for cell senescence. Mol. Syst. Biol. 6, 347 (2010).
Passos, J. F. et al. Mitochondrial dysfunction accounts for the stochastic heterogeneity in telomere-dependent senescence. PLoS Biol. 5, e110 (2007).
Hutter, E. et al. Senescence-associated changes in respiration and oxidative phosphorylation in primary human fibroblasts. Biochem. J. 380, 919–928 (2004).
Sitte, N., Merker, K., Grune, T. & von, Z. T. Lipofuscin accumulation in proliferating fibroblasts in vitro: an indicator of oxidative stress. Exp. Gerontol. 36, 475–486 (2001).
Sitte, N., Merker, K., Von, Z. T., Davies, K. J. & Grune, T. Protein oxidation and degradation during cellular senescence of human BJ fibroblasts: part II–aging of nondividing cells. FASEB J. 14, 2503–2510 (2000).
Mai, S., Klinkenberg, M., Auburger, G., Bereiter-Hahn, J. & Jendrach, M. Decreased expression of Drp1 and Fis1 mediates mitochondrial elongation in senescent cells and enhances resistance to oxidative stress through PINK1. J. Cell Sci. 123, 917–926 (2010).
Nautiyal, S., DeRisi, J. L. & Blackburn, E. H. The genome-wide expression response to telomerase deletion in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. 99, 9316–9321 (2002).
Martínez-Limón, A., Joaquin, M., Caballero, M., Posas, F. & de Nadal, E. The p38 pathway: from biology to cancer therapy. Int. J. Mol. Sci. 21 https://doi.org/10.3390/ijms21061913 (2020).
Brewster, J. L., de Valoir, T., Dwyer, N. D., Winter, E. & Gustin, M. C. An osmosensing signal transduction pathway in yeast. Science 259, 1760–1763 (1993).
Haghnazari, E. & Heyer, W. D. The Hog1 MAP kinase pathway and the Mec1 DNA damage checkpoint pathway independently control the cellular responses to hydrogen peroxide. DNA Repair 3, 769–776 (2004).
O’Rourke, S. M., Herskowitz, I. & O’Shea, E. K. Yeast go the whole HOG for the hyperosmotic response. Trends Genet. 18, 405–412 (2002).
de Nadal, E. & Posas, F. The HOG pathway and the regulation of osmoadaptive responses in yeast. FEMS Yeast Res. 22 https://doi.org/10.1093/femsyr/foac013 (2022).
Wong, C. M., Ching, Y. P., Zhou, Y., Kung, H. F. & Jin, D. Y. Transcriptional regulation of yeast peroxiredoxin gene TSA2 through Hap1p, Rox1p, and Hap2/3/5p. Free Radic. Biol. Med. 34, 585–597 (2003).
Rep, M. et al. The Saccharomyces cerevisiae Sko1p transcription factor mediates HOG pathway-dependent osmotic regulation of a set of genes encoding enzymes implicated in protection from oxidative damage. Mol. Microbiol. 40, 1067–1083 (2001).
Vendrell, A. & Posas, F. Sir2 plays a key role in cell fate determination upon SAPK activation. Aging 3, 1163–1168 (2011).
Barbosa, A. D. et al. Activation of the Hog1p kinase in Isc1p-deficient yeast cells is associated with mitochondrial dysfunction, oxidative stress sensitivity and premature aging. Mech. Ageing Dev. 133, 317–330 (2012).
Zhou, J., Zhong, Q., Li, G. & Greenberg, M. L. Loss of cardiolipin leads to longevity defects that are alleviated by alterations in stress response signaling. J. Biol. Chem. 284, 18106–18114 (2009).
Mao, K., Wang, K., Zhao, M., Xu, T. & Klionsky, D. J. Two MAPK-signaling pathways are required for mitophagy in Saccharomyces cerevisiae. J. Cell Biol. 193, 755–767 (2011).
Aoki, Y. et al. Phosphorylation of Serine 114 on Atg32 mediates mitophagy. Mol. Biol. Cell 22, 3206–3217 (2011).
Huang, S., Zhang, D., Weng, F. & Wang, Y. Activation of a mitogen-activated protein kinase Hog1 by DNA damaging agent methyl methanesulfonate in yeast. Front. Mol. Biosci. 7, 581095 (2020).
Mazor, Y. & Kupiec, M. Developmentally regulated MAPK pathways modulate heterochromatin in Saccharomyces cerevisiae. Nucleic Acids Res. 37, 4839–4849 (2009).
Soudet, J., Jolivet, P. & Teixeira, M. T. Elucidation of the DNA end-replication problem in Saccharomyces cerevisiae. Mol. Cell 53, 954–964 (2014).
Bah, A., Gilson, E. & Wellinger, R. J. Telomerase is required to protect chromosomes with vertebrate-type T2AG3 3’ ends in Saccharomyces cerevisiae. J. Biol. Chem. 286, 27132–27138 (2011).
Kockler, Z. W., Comeron, J. M. & Malkova, A. A unified alternative telomere-lengthening pathway in yeast survivor cells. Mol. Cell 81, 1816–1829 e1815 (2021).
Lundblad, V. & Blackburn, E. H. An alternative pathway for yeast telomere maintenance rescues est1- senescence. Cell 73, 347–360 (1993).
Marcand, S., Brevet, V. & Gilson, E. Progressive cis-inhibition of telomerase upon telomere elongation. EMBO J. 18, 3509–3519 (1999).
Shampay, J. & Blackburn, E. H. Generation of telomere-length heterogeneity in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA 85, 534–538 (1988).
Xu, Z. et al. Two routes to senescence revealed by real-time analysis of telomerase-negative single lineages. Nat. Commun. 6, 7680 (2015).
Barnes, R. P., Fouquerel, E. & Opresko, P. L. The impact of oxidative DNA damage and stress on telomere homeostasis. Mech. Ageing Dev. 177, 37–45 (2019).
Yin, Z., Pascual, C. & Klionsky, D. J. Autophagy: machinery and regulation. Microb. Cell 3, 588–596 (2016).
Suzuki, K. Selective autophagy in budding yeast. Cell Death Differ. 20, 43–48 (2013).
Bicknell, A. A., Tourtellotte, J. & Niwa, M. Late phase of the endoplasmic reticulum stress response pathway is regulated by Hog1 MAP kinase. J. Biol. Chem. 285, 17545–17555 (2010).
Prick, T., Thumm, M., Köhrer, K., Häussinger, D. & Vom Dahl, S. In yeast, loss of Hog1 leads to osmosensitivity of autophagy. Biochem. J. 394, 153–161 (2006).
Babele, P. K., Thakre, P. K., Kumawat, R. & Tomar, R. S. Zinc oxide nanoparticles induce toxicity by affecting cell wall integrity pathway, mitochondrial function and lipid homeostasis in Saccharomyces cerevisiae. Chemosphere 213, 65–75 (2018).
Shen, Z., Li, Y., Gasparski, A. N., Abeliovich, H. & Greenberg, M. L. Cardiolipin regulates mitophagy through the protein kinase C Pathway. J. Biol. Chem. 292, 2916–2923 (2017).
Rosado, C. J., Mijaljica, D., Hatzinisiriou, I., Prescott, M. & Devenish, R. J. Rosella: a fluorescent pH-biosensor for reporting vacuolar turnover of cytosol and organelles in yeast. Autophagy 4, 205–213 (2008).
Hector, R. E. et al. Mec1p associates with functionally compromised telomeres. Chromosoma 121, 277–290 (2012).
Tsang, C. K., Liu, Y., Thomas, J., Zhang, Y. & Zheng, X. F. Superoxide dismutase 1 acts as a nuclear transcription factor to regulate oxidative stress resistance. Nat. Commun. 5, 3446 (2014).
Ritchie, K. B., Mallory, J. C. & Petes, T. D. Interactions of TLC1 (which encodes the RNA subunit of telomerase), TEL1, and MEC1 in regulating telomere length in the yeast Saccharomyces cerevisiae. Mol. Cell Biol. 19, 6065–6075 (1999).
Fasullo, M., Tsaponina, O., Sun, M. & Chabes, A. Elevated dNTP levels suppress hyper-recombination in Saccharomyces cerevisiae S-phase checkpoint mutants. Nucleic Acids Res. 38, 1195–1203 (2009).
Sun, M. & Fasullo, M. Activation of the budding yeast securin Pds1 but not Rad53 correlates with double-strand break-associated G2/M cell cycle arrest in a mec1 hypomorphic mutant. Cell Cycle 6, 1896–1902 (2007).
Fasullo, M. & Sun, M. The Saccharomyces cerevisiae checkpoint genes RAD9, CHK1 and PDS1 are required for elevated homologous recombination in a mec1 (ATR) hypomorphic mutant. Cell Cycle 7, 2418–2426 (2008).
Lee, Y. M. et al. Dissection of the HOG pathway activated by hydrogen peroxide in Saccharomyces cerevisiae. Environ. Microbiol 19, 584–597 (2017).
Reynolds, T. B., Hopkins, B. D., Lyons, M. R. & Graham, T. R. The high osmolarity glycerol response (HOG) MAP kinase pathway controls localization of a yeast golgi glycosyltransferase. J. Cell Biol. 143, 935–946 (1998).
Longtine, M. S. et al. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14, 953–961 (1998).
Lemos, B. R. et al. CRISPR/Cas9 cleavages in budding yeast reveal templated insertions and strand-specific insertion/deletion profiles. Proc. Natl Acad. Sci. USA 115, E2040–E2047 (2018).
Forstemann, K., Hoss, M. & Lingner, J. Telomerase-dependent repeat divergence at the 3’ ends of yeast telomeres. Nucleic Acids Res. 28, 2690–2694 (2000).
Acknowledgements
We wish to thank Mohcen Benmounah for writing the script that simplified the analysis and graph creation for microfluidic analysis. Thank to Naima Belgareh-Touzé and Oznür Ozturk for sharing reagents and technical advice, the Teixeira lab, and the UMR8226 unit members for technical support and fruitful discussions. We also wish to thank Pascale Jolivet, Prisca Berardi, Yann Lustig, Juan Manuel Peralta, Pol Ubeda, Victoria Rojat and Clara Basto for technical help, and Francesc Posas’ lab, Miguel Godinho Ferreira, and Zhou Xu for fruitful discussions. During the preparation of this work, we occasionally used ChatGPT and DeepL to improve the readability and language of the manuscript. After using this tool, the authors reviewed and edited the content as needed and take full responsibility for the content of the published article. Work in MTT’s laboratory was supported by the CNRS, Sorbonne University, the “Fondation de la Recherche Medicale” (“Equipe FRM EQU202003010428”), by the French National Research Agency (ANR) as part of the “Investissements d’Avenir” Program (LabEx Dynamo ANR-11-LABX-0011-01) and the French National Cancer Institute (INCa_15192). B.Z. is a recipient of fellowship from the «Ministère de l’enseignement supérieur et de la Recherche» (MESR).
Author information
Authors and Affiliations
Contributions
A.B. conceived the study. B.Z. and A.B. performed experiments and analyzed the data. B.Z., M.T.T., and A.B. wrote the manuscript. A.B. and M.T.T. supervised the study. M.T.T. provided funding support. All authors discussed the data, read, and edited the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Biology thanks the anonymous reviewers for their contribution to the peer review of this work. Primary Handling Editors: Valeria Naim and Dario Ummarino. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zeinoun, B., Teixeira, M.T. & Barascu, A. Hog1 acts in a Mec1-independent manner to counteract oxidative stress following telomerase inactivation in Saccharomyces cerevisiae. Commun Biol 7, 761 (2024). https://doi.org/10.1038/s42003-024-06464-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s42003-024-06464-3
- Springer Nature Limited