Abstract
Rhabdomyosarcoma, the most common pediatric sarcoma, has no effective treatment for the pleomorphic subtype. Still, what triggers transformation into this aggressive phenotype remains poorly understood. Here we used Ptch1+/−/ETV7TG/+/− mice with enhanced incidence of rhabdomyosarcoma to generate a model of pleomorphic rhabdomyosarcoma driven by haploinsufficiency of the lysosomal sialidase neuraminidase 1. These tumors share mostly features of embryonal and some of alveolar rhabdomyosarcoma. Mechanistically, we show that the transforming pathway is increased lysosomal exocytosis downstream of reduced neuraminidase 1, exemplified by the redistribution of the lysosomal associated membrane protein 1 at the plasma membrane of tumor and stromal cells. Here we exploit this unique feature for single cell analysis and define heterogeneous populations of exocytic, only partially differentiated cells that force tumors to pleomorphism and promote a fibrotic microenvironment. These data together with the identification of an adipogenic signature shared by human rhabdomyosarcoma, and likely fueling the tumor’s metabolism, make this model of pleomorphic rhabdomyosarcoma ideal for diagnostic and therapeutic studies.
Similar content being viewed by others
Introduction
Genetic, epigenetic, and environmental cues shape molecularly distinct subpopulations of cells in an evolving solid tumor1,2. This intratumoral heterogeneity is a likely prelude to chemotherapy resistance and metastases. Rhabdomyosarcoma (RMS) is the most common pediatric soft tissue sarcoma with poor outcomes for high-risk patients3. Although it usually displays myogenic features, RMS can originate from both myogenic and non-myogenic progenitor cells, which explains, at least in part, its different sites of occurrence (e.g., limbs, chest, head and neck, and retroperitoneum)4. Generally, these tumors are classified into two major histological subtypes: (1) ERMS (embryonal RMS) (~60%) has a mostly favorable outcome, which decreases to only 40% overall survival when metastatic; (2) ARMS (alveolar RMS) (~20%) is the aggressive form associated with early metastatic dissemination and poor prognosis4. More than 80% of ARMS are defined by two chromosome translocations, involving PAX3/7 and FOXO14,5. Both ERMS and the rare pleomorphic RMS subtypes lack these chromosome translocations and develop into genetically more complex tumors, frequently showing inactivation of the p53 pathway and/or another oncogenic driver mutations6,7,8,9,10,11,12. These tumors express both developmentally early myogenic markers (e.g., PAX3/7, MyoD, and Myogenin) and markers of terminal differentiation (e.g., Desmin and SMA)13,14,15. Although several mouse models of ERMS have been reported16,17,18, none addressed the molecular and cellular events that drive RMS into a pleomorphic state, which therefore remains poorly understood.
We have shown earlier that haploinsufficiency of the gene encoding the lysosomal sialidase Neu1 in Arf–/– mice rendered different types of sarcomas more aggressive19. NEU1 hydrolyzes terminal sialic acids from sialylated glycoproteins, changing their biochemical properties and affecting downstream pathways20,21. One of these pathways is lysosomal exocytosis which NEU1 negatively regulates by cleaving the sialic acids of LAMP1. This lysosomal membrane protein is responsible for docking lysosomes at the plasma membrane (PM) of cells, prior to their fusion with the PM22,23,24. Low NEU1 activity increases the number of lysosomes, decorated with a long-lived, sialylated LAMP1, that dock at the PM. This ultimately results in excessive lysosomal exocytosis with deleterious consequences for the integrity of PMs and the extracellular matrix (ECM)19,22,24. The common readouts of this aberrant process in cells are increased activity of lysosomal enzymes (e.g., β-hexosaminidase) extracellularly and relocation of LAMP1 to the PM22,25.
Dysregulated lysosomal exocytosis is the basis of disease pathogenesis in Neu1−/− mice, a model of the lysosomal storage disease sialidosis20,22,26,27. Relevant to these studies is the muscle connective tissue phenotype in Neu1–/– mice that undergo a precancerous transformation, leading to expansion of the tissue via increased proliferation and generalized fibrosis26,27. Neu1 deficient fibroblasts behave as myofibroblasts/mesenchymal cells, being simultaneously proliferative and migratory/invasive. They show increased exocytosis of soluble proteolytic enzymes and exosomes, which together remodel the ECM and propagate the fibrotic disease26. We found that this pathogenic cascade accelerated the occurrence of several types of sarcomas in Neu1+/−/Arf−/− mice and transformed these tumors into an aggressive and chemo-resistant phenotype19. We sought to explore these Neu1-mediated phenomena in a mouse model of RMS.
The Patched1 (Ptch1)+/− mouse, a model of medulloblastoma, also develops ERMS-like tumors at low incidence9,28,29. These tumors are driven by activation of the Shh pathway9,28,30, which is normally kept inactive by binding of the Ptch1 receptor to Smo (smoothened)29,31. Reduced Ptch1 expression frees Smo, which then hyperactivates Gli transcription factors, triggering growth and proliferation. Crossing the Ptch1+/− mice with a transgenic mouse line (ETV7TG+/−) expressing human ETV7 increased the incidence and penetrance of ERMS and hematopoietic malignancy32,33. ETV7, an ETS transcription factor34, promotes the assembly of a novel rapamycin-resistant mTOR complex, mTORC3, by directly binding to mTOR32. ETV7 is differentially expressed in all human RMS, and mTORC3 increases the proliferation of several human ERMS cell lines. Furthermore, it was shown that the higher incidence of RMS in Ptch1+/−/ETV7TG+/− mice is driven by mTORC332.
Here, we show that reducing Neu1 expression in the Ptch1+/−/ETV7TG+/− model changes RMS tumors into a pleomorphic state. These tumors have increased cell heterogeneity, a distinctly fibrotic microenvironment, and share features with both human ERMS and ARMS. We found that deregulated lysosomal exocytosis downstream of low Neu1 is the underlying pathway driving transformation in these tumors, which are also fueled by an adipogenic signature. Thus, Neu1+/−/Ptch1+/−/ETV7TG+/− mice are a model of human pleomorphic RMS and may represent a powerful means for diagnostic and therapeutic studies of these aggressive and incurable tumors.
Results
Neu1 haploinsufficiency promotes the development of poorly differentiated RMS in the Ptch1 +/− /ETV7 TG+/− mouse model
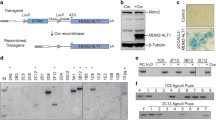
In Neu1+/−/Ptch1+/−/ETV7TG+/− (NPE) mice, low Neu1 RNA and protein expression (Supplementary Fig. 1a, b) was sufficient to increase the incidence of RMS to 62%, as compared to 54% in the Ptch1+/−/ETV7TG+/− (PE) mice and only 8% in the Ptch1+/− mice in 2 years (Fig. 1a). RMS was diagnosed based on morphology and immune reactivity to the myogenic regulatory factors (Mrfs) MyoD and Myogenin, and the muscle-specific type III intermediate filament desmin (Des) (Supplementary Fig. 1c–e)35. The survival rate of both NPE and PE mice was comparable but significantly lower than that of Ptch1+/− mice (Fig. 1b). All RMS tumors in NPE and PE models developed in either the extremities or the trunk, reflecting the location of human ARMS and pleomorphic RMS with poor prognosis and short survival4 (Fig. 1c). Within the cohort of mice with RMS tumors 4/35 (11.4%) NPE and 1/23 (4.3%) PE mice also developed secondary growths (Fig. 1a, c), a finding that may indicate metastatic spread, although without additional genomic analysis we cannot exclude that these tumors are synchronous rather than metachronous. We next established that ETV7 was expressed in both NPE and PE tumors, as compared to Neu1+/−/Ptch1+/− (NP) controls (Fig. 1d). We also confirmed that ETV7 expression in both models supported the assembly of mTORC3, as shown by two-way co-immunoprecipitation of ETV7 and mTOR (Fig. 1e, f and Supplementary Fig. 2a, b). In addition, the PP2Ac and phospho-4E-BP1Thr37/46components of mTORC3, also co-immunoprecipitated with mTOR and ETV7 (Fig. 1f and Supplementary Fig. 2b), further supporting the presence of the complex. These results suggest that the rapid growth of RMS in NPE and PE mice is driven in part by mTORC3 activity, although we cannot exclude the possible contribution of Neu1 heterozygosity, giving these tumors an additional proliferative advantage.
a Percentages of RMS types developed by Ptch1+/−, PE, and NPE mice. b Percent survival of Ptch1+/−, PE, and NPE mice. P calculated with log-rank test for trend. c Location, number and percentage of RMS developed in Ptch1+/−, PE, and NPE mice. d ETV7 mRNA expression in PE and NPE tumors. NP: Neu1+/−/Ptch1+/− tumors were used as control. Mean ± s.d.; One-way ANOVA t-test; n = 7 biologically independent tumors. e Western blot of mTOR and ETV7 in PE, NPE, and NP tumors. f Two-way co-immunoprecipitation of mTOR and ETV7 and co-immunoprecipitation of mTOR and ETV7 with mTORC3 components, PP2Ac and phospho-4E-BP1Thr37/46, in PE and NPE tumors. NP tumors were used as control. g, h Morphological characteristics of PE (g) and NPE (h) tumors by H&E staining. Arrow—large abnormal and multinucleated cancer cells; Arrowhead—undifferentiated small cancer cells; asterisk—cytoplasmic vacuolation consistent with accumulation of neutral lipids. Scale bar: 50 µm. i, j Quantification of the size of nuclei (i) and quantification of number of nuclei (j) in tumor sections of NPE and PE mice. Mean ± s.d.; Student’s (unpaired) t-test; PE: n = 10 and NPE: n = 6 biologically independent tumors.
What distinguished NPE from PE tumors was their histopathological appearance. RMS in the NPE model showed pronounced cellular heterogeneity, associated with the presence of numerous undifferentiated, anaplastic foci, rich in pleomorphic and rhabdoid cells, as compared with the more differentiated morphology of PE RMS (Fig. 1g, h, Supplementary Fig. 1c, d). These pleomorphic foci were populated with intertwining bundles of tumor cells of abnormal size and shape (Fig. 1h arrows), and small undifferentiated tumor cells (Fig. 1h arrowhead). An algorithm designed to quantify nuclear morphometry in tumor H&E sections revealed an increased number of aberrantly large, multinucleated cells with atypical mitotic figures, predominantly in NPE tumors (Fig. 1i). Because of the very large size of most of the nuclei in the NPE tumors, their total number per annotated area was lower than that in PE tumors (Fig. 1j). Instead, other typical RMS features were common to both NPE and PE tumors, including spindle-shaped cells, round rhabdomyoblasts, cross-striated mature skeletal myocytes and myofibers (Fig. 1g, h), as well as expression of MyoD, Myogenin, and Desmin (Supplementary Fig. 1e)35. Together, these results suggest that Neu1 haploinsufficiency superimposed on the spontaneously occurring PE tumors changes the overall morphology and differentiation status of RMS.
Neu1-mediated pleomorphism is associated with neoplastic fibrosis
In general, aggressive/invasive tumors undergo active remodeling of their ECM, which creates a fibrotic microenvironment that facilitates metastatic spread and promotes drug resistance36. To assess the extent of collagen deposition and ECM remodeling, Masson’s trichrome staining identified extensive areas of collagenous connective tissue that were much more prominent in the NPE (blue) than in PE tumors, particularly at invasive fronts and pleomorphic foci (Fig. 2a, b). A similar increase in connective tissue was observed in Masson’s trichrome-stained human tissue microarrays (TMAs) from several RMS patients (ERMS n = 16, ARMS n = 17, and spindle cell/sclerosing RMS n = 3), where 50% of all TMA cores had more than 9% of the total area as collagenous material, with an average of 28.7% in ERMS, 9.7% in ARMS, and 23.3% in spindle cell/sclerosing RMS, although the latter only comprised three samples (Fig. 2c and Supplementary Fig. 3a). In addition, we observed a substantial number of cells that showed intracytoplasmic collagen as well, with an average of 12.8% in ERMS, 8.5% in ARMS, and 13.7% in spindle cell/sclerosing RMS (Supplementary Fig. 3b). Quantification of the staining of the mouse tumors revealed a significant increase of connective tissue in the NPE tumors compared to PE tumors, underscoring their more aggressive nature (Fig. 2d). Similar to what is seen in human RMS tumors, subpopulations of NPE cells also showed quantifiable intracytoplasmic collagen (Fig. 2e and Supplementary Fig. 3b), indicating their cell-autonomous capacity to deposit ECM, like that of myofibroblasts during fibrosis26,36. The latter was further confirmed by the significant upregulation of Col1a2 and Col4a1 expression in NPE versus PE RMS (Fig. 2f, g). These combined observations were not seen in normal muscle tissue of these mice (Supplementary Fig. 3c–e), emphasizing that the connective tissue deposition observed in the NPE RMS tumors was caused by the combination of reduced activity of Neu1 within tumor cells and stroma. Thus, reduced Neu1 expression promotes pleomorphism and transforms the tumor stroma into a desmoplastic, fibrotic state.
a, b Masson’s Trichrome staining of PE RMS (a) and NPE (b) RMS depicting ECM/collagenous material in blue and cell cytoplasm in red. Scale bar: 50 µm. c Representative micrograph of one core of human RMS TMAs (n = 36 biologically independent samples) stained with Masson’s Trichrome. Scale bar: 50 µm, inset scale bar: 10 µm. d Quantification of ECM/collagen deposition in PE and NPE RMS; Mean ± s.d.; Mann–Whitney (unpaired) t-test; PE: n = 9 and NPE: n = 12 biologically independent tumors. e Quantification of tumor intracytoplasmic collagen from PE and NPE RMS; Mean ± s.d.; Mann–Whitney (unpaired) test; PE: n = 10 and NPE: n = 11 biologically independent tumors. f PE and NPE Col1a2 mRNA expression relative to PE. Mean ± s.d.; Welch (unpaired) t-test; PE: n = 6 and NPE: n = 11 biologically independent tumors. g PE and NPE Col4a1 mRNA expression relative to PE. Mean ± s.d.; Welch (unpaired) t-test; PE: n = 6 and NPE: n = 12 biologically independent tumors. h Heatmap of gene expression comparison between murine and human RMS. The nine genes that correlated between murine RMS and ARMS are boxed in orange. Pearson’s correlation was performed on 91 genes analyzed.
Neu1 +/− /Ptch1 +/− /ETV7 TG+/− mice share molecular characteristics of human ARMS and ERMS
We next compared the gene expression profiles of NPE and PE tumors with RNA-seq data from human ERMS and ARMS available from the St Jude Pediatric Cancer Genome Project (PCGP). As shown in the heatmap (Fig. 2h), the pool of human genes that could be analyzed and compared were 91 genes from different platforms (murine microarray and Pediatric Cancer Genome Project (PCGP) RNAseq) that passed the principal component analysis (PCA), quality control, and significant Pearson correlation criteria. This comparative analysis showed no significant differences between human RMS and our NPE and PE tumors. Within this geneset, the highest positive correlation was found between the expression profiles of NPE (r = 0.71) and PE (r = 0.72) tumors with human ERMS that comprise the most genetically heterogeneous subtypes4 (Fig. 2h). Although no significant differences at the transcriptional levels were observed between NPE or PE tumors compared to human RMS, our data support the notion that Neu1 haploinsufficiency in the NPE model promotes the development of a more poorly differentiated phenotype, as demonstrated by their histologically pleomorphic appearance. Enrichment analysis of NPE/PE and ERMS correlative genes identified pathways involved in metabolism, cell signaling, and differentiation (Supplementary Fig. 4a, b, Supplementary Data 2). This analysis also distinguished two small clusters of genes in NPE and PE tumors that correlated with the expression signatures of PAX3-FOXO1 ARMS (r = 0.67 and r = 0.68, respectively) and PAX7-FOXO1 ARMS (r = 0.65 and r = 0.66, respectively) (Fig. 2h, orange boxes). These clusters included PAX7 and CTHRC1 (collagen triple helix repeat containing 1), which encode proteins involved in cell differentiation and tissue remodeling. Together, this comparison highlights the genetic resemblance between the murine NPE and PE tumors with human RMS, in particular with the ERMS subtype.
Low NEU1 expression correlates with plasma membrane redistribution of LAMP1 and increased lysosomal exocytosis
To assess the molecular events that might contribute to the aberrant histopathology and fibrotic state of NPE tumors, we focused on exacerbated lysosomal exocytosis. Testable readouts of this dysregulated process are an accumulation of a long-lived, sialylated LAMP1 due to low NEU1 activity, and LAMP1 redistribution at the cell surface22,25. We exploited both these parameters to measure the expression levels of LAMP1 as well as the fraction of this protein at the cell surface in human TMAs from several RMS patients. Matched immunostained tissue cores from RMS patients showed that very low expression of NEU1 was accompanied by an overall increase in LAMP1 (Fig. 3a, Supplementary Fig. 5a, b). Using a membrane immunoreactivity computer learning algorithm applied to TMA cores, we quantified LAMP1 positive staining at the cell membrane (LAMP1PMpos) (Fig. 3b–d). We found that in most RMS samples, independent of their subtype, low NEU1 expression correlated with increased LAMP1 at the cell surface (Fig. 3d), a potential indicator of the cells’ enhanced exocytic activity. Additionally, analysis of LAMP1 immunoreactivity in each TMA via a density heatmap identified areas of dense immunoreactivity for LAMP1 that mostly coincided with tumor-stromal borders and areas of pleomorphism (Fig. 3b, c). This patchy LAMP1 positive staining also highlighted the extent of cell heterogeneity within each RMS sample (Fig. 3c).
a Representative core of human RMS TMAs immunostained for LAMP1. Scale bar: 100 µm. b Density map of algorithm applied to a LAMP1-stained core showing LAMP1PM minimal and maximum density and location. Scale bar: 100 µm. c Heatmap representing the intensity of LAMP1PM staining of the core shown in a. Scale bar: 100 µm. d Correlation between low NEU1 and high LAMP1PM immunoreactivity in paired cores of human RMS TMAs. Two TMAs were used; Paired t-test; n = 41 biologically independent samples. e Representative micrographs of increased Lamp1 immunostaining in NPE compared with PE RMS. Punctuated lysosomal staining, typical for Lamp1. Scale bar: 25 µm, inset scale bar: 10 µm. f Flow cytometry histogram showing higher Lamp1PMpos expression in cells in NPE RMS versus PE RMS. g Increased Lamp1PM intensity quantified by flow cytometry in NPE versus PE RMS. Mean ± s.d.; Paired t-test; n = 9 biologically independent tumors. h Increased β-hexosaminidase activity in the interstitial fluid from NPE versus PE RMS. Mean ± s.d.; Student’s (unpaired) t-test; n = 7 biologically independent tumors.
Increased LAMP1 levels in human RMS were paralleled by a similarly altered expression pattern of this protein in NPE compared with PE RMS (Fig. 3e). To assess how many mouse tumor cells had increased Lamp1PMpos, as a proxy for enhanced lysosomal exocytosis, we performed flow cytometric analysis of non-permeabilized NPE and PE dissociated tumor cells (Fig. 3f, g). This identified a population of NPE tumor cells with high Lamp1PMpos, which was distinct from that in PE tumors (Fig. 3f, g). To prove that NPE RMS had increased lysosomal exocytosis, interstitial fluids from both NPE and PE tumors were assayed for the lysosomal enzyme β-hexosaminidase22,37. Significantly higher activity of β-hexosaminidase was measured in the interstitial fluid of several NPE RMS compared with PE samples (Fig. 3h), confirming that NPE tumors were more exocytic.
Single-cell multiplex flow cytometry underscores the complex cell heterogeneity in Neu1 +/− /Ptch1 +/− /ETV7 TG+/− tumors
Based on the combined features of increased Lamp1PMpos, lysosomal exocytosis, and pleomorphism in limb and trunk NPE compared with PE RMS, we designed a multiplex high parameter flow cytometry strategy that evaluated Lamp1PMpos cells separately from Lamp1PMneg cells within three main cell populations: cancer, stroma, and hematopoietic cells (Fig. 4a). Protein expression data were generated by using Lamp1PM in combination with markers selective for each cell population35,38,39,40,41,42,43,44,45,46,47,48,49,50, followed by tSNE analysis. The distribution and density of cells in limb and trunk RMS from NPE and PE mice highlighted the extent of cell heterogeneity (Fig. 4b). To uncover distinct cell populations within each tumor sample, we narrowed the tSNE analysis to tumor and stromal cells by excluding hematopoietic cells, using CD45 (Supplementary Fig. 6). Several discrete cell clusters were identified, which differed not only between NPE and PE RMS, independently of their CD45 status, but also between limb and trunk tumors (Fig. 4c). This may be linked to the cell of origin that supports the development of trunk versus limb tumors, but a more comprehensive investigation would be necessary to confirm these differences.
a Flow chart of multiplex flow cytometry analysis used to identify different cell populations in NPE and PE RMS. b tSNE density heatmaps of limb and trunk RMS from PE and NPE mice depicting areas of low and high cell number, based on the expression of the different markers (in a). c tSNE graphs of limb and trunk RMS from PE and NPE mice indicating the distribution of CD45neg and CD45pos cells.
Lamp1 at the plasma membrane marks highly exocytic and undifferentiated cell populations in Neu1 +/− /Ptch1 +/− /ETV7 TG+/− tumors
tSNE analyses of the selected CD45neg cell populations in limb and trunk tumors of both NPE and PE mice (Fig. 4b, c) again revealed distinct cell density and distribution (Fig. 5a, b). These cell populations were further analyzed by using Lamp1PM expression in combination with a set of canonical markers distinct for cancer and stromal cells (i.e., desmin, Ki67, Sca1, Sma, Pcam1, CD44, Lyve1, and EpCam) (Fig. 5c–h). This allowed for the annotation of 26 and 22 different cell clusters in limb and trunk tumors, respectively (numbered in Fig. 5c–h). Within the annotated populations, the total percentage of Lamp1PMpos exocytic cells was 45.95% in limb (cell clusters 1–12) and 11.14% in trunk (cell clusters 1–10) RMS (Fig. 5e–h). In both limb and trunk RMS a clearly larger percentage of Lamp1PMpos cells was detected in NPE (cluster 1–12) than in PE (cell clusters 1–10) tumors (Fig. 5e, f). Most importantly, the majority of Lamp1PMpos cells showed pleomorphic features (cell clusters 1–6) and expressed simultaneously the terminal (myo)differentiation marker desmin and markers of progenitor or undifferentiated cells, such as Ki67 (proliferation), Sca1 (progenitor), Sma (smooth muscle actin/mesenchymal), Pcam1 and CD44 (endothelial), and Lyve1 (lymphatic endothelial) (Fig. 5c–f). These exocytic, pleomorphic subpopulations accounted for 21% and 15% of cancer cells in the limb and trunk (cell clusters 1–6), respectively, and were more abundant in NPE than PE RMS (Fig. 5e–h). The combination of pleomorphic and exocytic cells enriched in NPE tumors indicates that Neu1 haploinsufficiency promotes the poorly differentiated state of the tumors, albeit an in-depth explanation of this phenomenon may require further investigation.
a, b tSNE graphs showing density and distribution of CD45neg cells from limb (a) and trunk (b) RMS of PE (blue) and NPE (red) mice. c, d tSNE of single-cell analysis of limb (c) and trunk (d) representation of the distinct cell clusters identified in RMS (numbered and colored) with the markers used with CD45neg cells from PE and NPE mice. e, f Annotated limb (e) and trunk (f) RMS CD45neg exocytic Lamp1PMpos and Lamp1PMneg cell populations from PE and NPE mice. Bubble map indicates the percentages of cells from PE and NPE contributing to a cell cluster. Heatmap shows the signal intensity of expression of the different markers used. g, h Pie graph of CD45neg cells from limb (g) and trunk (h) PE and NPE RMS showing the percentage of each cell cluster (numbered and colored matched).
Similarly, within the stromal CD45neg and Desneg cell populations, the majority of Lamp1PMpos cells included cancer-associated fibroblasts (SMA) (cell cluster 7–9) and epithelial cells (cell clusters 10–11 in limb and 10 in trunk), but also expressed markers typical of myofibroblasts/mesenchymal cells, e.g., Ki67, Sca1, Sma, Pcam1, CD44, Lyve1, and EpCam (epithelial) (Fig. 5e–h). Again, these proliferative, exocytic stromal cells were mostly enriched in NPE limb (cell cluster 7) and trunk RMS (Fig. 5e–h). Curiously, a small percentage of endothelial cells with progenitor features (Pcam1pos, Sca1pos) was almost exclusively present in NPE tumors (cell cluster 22 in limb and 15 in trunk) (Fig. 5e–h). Expression levels of markers defining tumor and stromal cells were also assessed by tSNE heatmaps that demonstrated the higher signal intensity of some of the markers (Des, Sma, Sca, Pcam1, Lamp1, and Ki67), specifically in NPE RMS (Supplementary Fig. 7a, b). These tumors also contained a higher percentage of CD45pos macrophages, T cells, and B cells, than PE tumors (Supplementary Fig. 7c–f), likely contributing to an inflammatory pro-tumorigenic microenvironment. However, a more detailed analysis of the immune infiltration would be required to tease out its function in these tumor models. These results, based on single-cell protein expression, suggest that in NPE tumors, not only cancer cells but also stromal cells are maintained in an intermediate state of transdifferentiation that could be the basis of their distinct pleomorphic phenotype.
Pleomorphic Neu1 +/– /Ptch1 +/– /ETV7 TG +/– RMS develop an adipocytic metabolic signature
Although the cell composition of NPE and PE limb and trunk tumors was similar, there was a distinctly higher percentage of cell populations expressing specific combinations of markers in NPE vs PE tumors (Fig. 5e, f). Taking the latter into account, we queried microarray data from individual NPE and PE limb and trunk RMS in search of distinct gene expression signatures that would add to their respective protein expression profiles and support pleiomorphism. The microarray analysis allowed for the detailed identification of differentially expressed genes between NPE and PE tumors. The volcano plot and heatmap (Fig. 6a, Supplementary Fig. 8a) comprising a cohort of limb and trunk RMS showed that, compared with PE, NPE tumors had 491 upregulated and 287 downregulated genes (Fig. 6a, Supplementary Fig. 8a, Supplementary Data 3). Using Enrichr51,52, we found that most of the upregulated genes in NPE RMS belong to pathways of lipid metabolism, myogenesis, and epithelial-to-mesenchymal transition (EMT) (Supplementary Fig. 8b, Supplementary Data 4), whereas genes downregulated in NPE belong to the following pathways: matrix metalloproteinases, cytokine-cytokine receptor interaction, HIF-1 signaling pathway and lung fibrosis (Supplementary Fig. 8c, Supplementary Data 5). GSEA (geneset enrichment analysis) of the total genetic landscape of limb and trunk RMS from NPE and PE mice identified the following geneset libraries: the KEGG PPAR (peroxisome proliferator-activated receptor), the Gene Ontology (GO) fatty acid metabolic process, the GO brown fat cell differentiation, and the KEGG vascular smooth muscle contraction (Fig. 6b). The leading edge of all four libraries/pathways positively correlated with NPE upregulated genes (Fig. 6b). White, beige and brown adipogenesis, and fatty acid metabolism are regulated by the essential transcription factor PPARγ53. In three out of the four aforementioned GSEA pathways, Pparg was one of the most upregulated genes in NPE RMS (Fig. 6b, Supplementary Data 6), together with some of its direct target genes, e.g., Fabp4, (fatty-acid-binding protein 4 or adipocyte protein 2), Cd36 (cluster of differentiation 36), AdipoQ (adiponectin), and its receptor AdipoR1, but not AdipoR2 (Fig. 6a, c, Supplementary Data 6). The derivation of brown adipocytes from skeletal muscle precursors or beige adipocytes from white adipocytes is orchestrated by PPARγ recruitment of PRDM16 (PR (PRD1-BF1_RIZ1 homologous)-domain-containing 16), which together form a core transcriptional complex53. PPARγ also recruits EBF2 (the early B-cell factor 2), a selective determinant of brown and beige adipocyte precursors54, and coactivates the expression of UCP1 (uncoupling protein 1 of brown adipocytes), PPARA, and PRDM1653. We found that in NPE RMS Pparg, Prdm16, and Ebf2 were upregulated together with their target, Ucp1 (Fig. 6c, Supplementary Data 6). It is well documented that beige adipocytes that transdifferentiate from white adipocytes are derived from mesodermal stem cells expressing Pdgfra, Pdgfrb, Acta2, and Myh11, whereas brown adipocytes share with skeletal muscle cells the combined markers of dermomyotome precursors, Myf5 and Pax755,56. In our NPE RMS, only Acta2 and Myh11 were significantly upregulated but not Pdgfra, Pdgfrb, Myf5, or Pax7 (Fig. 6d, Supplementary Data 6), suggesting a preferential presence of white and beige adipocytes.
a Volcano plot comparing differentially expressed genes in NPE versus PE RMS. Several genes enriched in the pathways identified by GSEA are annotated in the graph. Genes annotated in green were confirmed to be highly increased by qRT-PCR analysis. b Main KEGG and GO pathways identified by GSEA of differentially expressed genes in NPE vs PE RMS. FDR < 0.25. c–e Tumor qRT-PCR analysis showing features of adipogenesis (c); Pparg (PE: n = 10; NPE: n = 7), Prdm16 and Fabp4 (PE: n = 7; NPE: n = 12), Ucp1 (n = 6), Adipoq (PE: n = 9; NPE: n = 7), AdipoR1 (n = 10), AdipoR2 (PE: n = 10; NPE: n = 9), CD36 (PE: n = 6; NPE: n = 11), myogenesis (d); Pax3 (PE: n = 6; NPE: n = 7), Pax7 (PE: n = 8; NPE: n = 7), Myf5 (PE: n = 8; NPE: n = 6), Myod1 and Myocd (PE: n = 8; NPE: n = 9), Myog (PE: n = 10; NPE: n = 9), Myh11 (PE: n = 8; NPE: n = 11), Des (n = 9) and tumor aggressiveness (e); Myc (PE: n = 9; NPE: n = 12), Gli1 and Gli2 (PE: n = 7; NPE: n = 12), Snai1 (PE: n = 6; NPE: n = 8), Snai2 and Vacm1 (PE: n = 5; NPE: n = 7), Upk1b (PE: n = 6; NPE: n = 7), Prss35 (n = 7) in NPE versus PE RMS. qRT-PCR results are from normalized mRNA expression relative to PE RMS samples. Mean ± s.d.; Welch (unpaired) t-test; biologically independent tumors.
The earliest progenitor marker of myogenesis, Pax357 was downregulated in NPE vs PE RMS, while Pax7 and Myf5 showed no significant difference in expression (Fig. 6d). The other upstream regulators of myogenesis, MyoD and Myog, were significantly increased in NPE RMS (Fig. 6d), while the terminal differentiation gene Des was downregulated (Fig. 6d). Additionally, Myocd and Myh11, both smooth muscle specification genes, were highly upregulated in the NPE compared to PE RMS (Fig. 6d, Supplementary Data 6); Myh11 was also found in the leading edge of the KEGG vascular smooth muscle contraction pathway (Fig. 6b, Supplementary Data 6). Notably, human MYH11 protein, which is normally not expressed in RMS, is a marker of sarcoma pleomorphism58,59, and an effector of lysosomal exocytosis downstream of Neu1 downregulation19. So far, these results suggest that NPE RMS are already committed to a myogenic fate without reaching full differentiation. Moreover, the upregulation Myh11 reinforces the notion that these tumors are both pleomorphic and exocytic.
Although NPE and PE RMS depend on the combination of Ptch1+/− and ETV7TG+/− for tumor specification and growth, the addition of Neu1 haploinsufficiency drastically increased Myc expression, the marker of proliferation, and the downstream effectors of the SHH pathway, Gli1 and Gli2, known to be associated with metastatic growth (Fig. 6e). In agreement with the status of partial differentiation and smooth muscle/mesenchymal characteristics of NPE RMS, genes involved in epithelial-to-mesenchymal transition (EMT) and metastasis i.e., Snai1, Snai2, Vcam1, Upk1b, and Prss35, were also significantly upregulated (Fig. 6e). It is noteworthy that the top-upregulated gene in NPE vs PE RMS is H2Eb1, encoding the mouse histocompatibility class II antigen E beta (MCH II), which is involved in antigen presentation, CD4+ T cell recognition and activation of the immune response60 (Fig. 6a). The relevance of this observation will require extensive future study exploring the immune response component of these tumors, which is beyond the scope of this work.
To exclude the possibility that Neu1 haploinsufficiency by itself can cause the significant changes in gene expression observed between PE and NPE tumors, we performed RT-qPCR analysis of the same set of genes in normal muscle tissue from both NPE and PE mice and found no differences (Supplementary Fig. 9a–c). Overall, our results indicate that NPE RMS maintains a status of intermediate differentiation and pleomorphism fueled by a strong adipogenic component, likely linked to Neu1 haploinsufficiency.
Pleomorphic Neu1 +/– /Ptch1 +/– /ETV7 TG +/– RMS share an adipogenic signature with human ARMS and ERMS
Based on these data, we revisited the H&E and Masson’s trichrome-stained PE and NPE RMS and identified a slight increase in the percentage of adipose cells in the NPE sections, although this difference was not statistically significant (Supplementary Fig. 10a). These were especially enriched in the pleomorphic foci and the invasive fronts, showing extensive collagen deposition and ECM remodeling (Fig. 1h asterisk, Fig. 2b). To confirm the gene expression data, we probed NPE and PE tumor sections with adiponectin the product of the AdipoQ gene that was highly upregulated in the NPE tumors and compared its expression pattern with that in human RMS TMAs (Fig. 7a, b). Adiponectin, the essential secreted adipokine in adipocyte signaling, was highly expressed in NPE tumors compared to PE, and was detected mostly at the cell surface of adipocytes and tumor cells, including large multinucleated pleomorphic cells (Fig. 7a). In TMAs from RMS patients, adiponectin immunoreactivity was positive in all cores, albeit of low intensity, and localized to the cytoplasm of neoplastic cells. However, in a few cores, adiponectin intensity was moderate to high (Fig. 7b). Notably, white adipose tissue was detected in 25% (10/40) of RMS cores. Querying the human RNAseq PCGP dataset, we found a group of five RMS patients with high expression of ADIPOQ, that was clearly above the standard deviation of the median expression of all analyzed samples (n = 44) (Fig. 7c). These patients were all diagnosed with ARMS, the highest expression was from recurrent disease, and one had an undefined RMS subtype (Fig. 7c). All five patients with ADIPOQ upregulation also showed higher than median levels of PPARG, CD36, and MYH11, but expressed lower levels of UCP1 (Fig. 7c–g). Most importantly, we found that ARMS patients expressed significantly higher levels of ADIPOQ, PPARG and MYH11 when compared with ERMS patients (Fig. 7h–l). Using GSEA we also compared ADIPOQ expression in RMS patients with geneset profiles from the MSigDB collection (Supplementary Fig. 10b). This analysis confirmed that the RMS patients with the highest ADIPOQ expression also showed upregulation of genes belonging to the pathways of adipogenesis, vascular smooth muscle contraction, and ECM receptor interaction, but not myogenesis (Supplementary Fig. 10b, Supplementary Data 7). These results support the presence of an adipose component associated with undifferentiated characteristics in some of the aggressive forms of human RMS.
a Representative micrographs of increased adiponectin immunostaining in NPE versus PE RMS. Increased cell surface positive staining is depicted in the zoom inset of NPE RMS. Scale bar: 25 µm. b Representative micrograph of strong adiponectin immunostaining in a core of an undifferentiated human RMS from 2 TMAs; n = 40 biologically independent samples. Intense staining is depicted in the zoom inset. Scale bar: 25 µm. c–g Representation of genetic expression of ADIPOQ (c), PPARG (d), CD36 (e), MYH11 (f), and UCP1 (g) across human patient RMS RNAseq data (PCGP). Gene expression is represented in relation to the mean value. All samples of ERMS (blue), ARMS (orange) and undefined RMS (black) analyzed are shown first on the x-axis (RMS), followed by the representation of the cluster of the top five samples with the highest ADIPOQ expression (Top 5). n = 44 biologically independent samples. h–l Comparison of ADIPOQ (h), PPARG (i), CD36 (j), MYH11 (k), and UCP1 (l) expression between ERMS and ARMS patients. Mean ± s.d; Mann–Whitney t-test used to compare ERMS and ARMS; n = 27 and n = 14 biologically independent samples, respectively.
Discussion
Pleomorphic RMS is a rare and aggressive form of sarcoma, occurring mostly in adults that is often difficult to diagnose and treat4. These tumors present with a mixture of large, atypical pleomorphic rhabdomyoblasts, often multinucleated, and partially differentiated spindle-like cells35.
By lowering the expression levels of the lysosomal sialidase Neu1 in the Ptch1+/−/ETV7TG+/− genetic background, we have successfully developed a spontaneously occurring mouse model of pleomorphic RMS without the use of conditional alleles. It is important to emphasize that the effect of Neu1+/− is not in the generation of RMS, but rather in its pleomorphic transformation once the tumor is initiated; while ETV7 expression is promoting a high incidence of RMS formation in Ptch1+/− mice32. Although NPE tumors arise preferentially in the limbs and trunk and only express 10% of the genes that trended with human ARMS, their genetic features are much closer to ERMS. Unfortunately, the unavailability of compatible genetic data from human pleomorphic RMS that we could compare to our NPE datasets, limits our current evaluation of NPE tumors to their histological characteristics, and further investigation is required to confirm NPE mice as a model of human pleomorphic RMS.
Exploiting the low NEU1 > high lysosomal exocytosis axis, we developed a single-cell approach that relied on the increased levels of the Neu1 substrate Lamp1 at the PM of cancer and stromal cells. This approach allowed us to pinpoint tumor heterogeneity, consisting of specific cell populations that are highly exocytic and likely responsible for initiating/perpetuating RMS transformation and ECM remodeling. The ensuing fibrotic microenvironment surrounding pleomorphic foci in NPE RMS is typical of the most aggressive and chemo-resistant tumors in humans61.
The presence of mature adipocytes together with adipose-like cells in both murine and human RMS could be explained by a shared developmental ancestry between skeletal muscle and brown fat62,63. However, brown adipocytes originate from early dermomyotomes (Pax7+/En1+/Myf5+), while distinct somite populations expressing MyoD downstream of Myf5 are already committed to myogenesis. Therefore, fate determination between brown adipocytes and committed somite populations should happen prior to MyoD expression56. Given the high levels of MyoD detected in NPE RMS, we can infer that these tumors have reduced brown adipocytes. In contrast, they might be enriched in beige adipocytes, since these derive from progenitors expressing Sma, Myh11, Pdgfra, or Pdgfrb, and transdifferentiate from mature white adipocytes stimulated, among others, by PPARγ activation and release of adipokines, including adiponectin56,64,65. In this scenario, tumor cells, cancer-associated fibroblasts, and cancer-associated adipocytes may together sustain cancer progression and chemo-resistance by fueling metabolism and maintaining a pro-fibrotic microenvironment downstream of low NEU1 activity. These combined features, largely depending on posttranslational modifications of glycans, appear to be common between our NPE model and human RMS, given that increased adipocytes have been detected in poorly differentiated, human metastatic or recurrent RMS, as well as in chemotherapy-treated RMS66,67,68.
We propose that the NPE mice can be exploited as a pre-clinical model of pleomorphic RMS to understand the pathology of these aggressive tumors and explore new therapeutic opportunities in pathways controlled by NEU1. Furthermore, the overlooked adipose component in these tumors underscores the importance of combining the use of adipose markers during the diagnosis of this type of RMS.
Methods
Mice
All procedures were performed following NIH guidelines and animal protocols approved by the St Jude Children’s Research Hospital Institutional Animal Care and Use Committee (IACUC); animal protocol #235–100636. Neu1+/− FVB/NJ69 mice were crossed with Ptch1+/−/ETV7TG/+/− (129sv/C57BL/6) mice32,33. The mice, both males, and females (1 month–24 months), used in this study were obtained (over a timespan of 2 years) from the same strategic breeding schedule: Neu1+/−/Ptch1+/−/ETV7TG+/− X Neu1+/−/Ptch1+/−/ETV7TG+/+, but PE and NPE mice with tumors not necessarily came from the same litter. Mice presenting with masses were observed for lethargy and sacrificed when moribund, or if they displayed symptoms such as paralysis or when the tumor volume reached 20% of body mass in accordance with IACUC-approved guidelines.
Real-time quantitative polymerase chain reaction
Total RNA was isolated from mouse tumors using the PureLink RNA Kit (Life Technologies), and DNA contaminants were removed on a deoxyribonuclease I column (Life Technologies), according to the manufacturer’s protocol. RNA quantity and purity were measured using a NanoDrop Lite spectrophotometer (Thermo Fisher Scientific). Complementary DNA was synthesized using 0.5–5 μg of total RNA with RT2 First Strand Kit (QIAGEN). RT-qPCR was performed using RT2 SYBR Green qPCR Mastermix, 1 μl (8, 16, or 50 ng) of complementary DNA, 10 μM primer, and ribonuclease-free water in a 25 μl reaction volume on an iQ5 or CFX96 real-time PCR machine (Bio-Rad). Samples were normalized to 18 S ribosomal RNA. The specific primers used are summarized in Supplementary Table 1.
Western blotting and co-immunoprecipitations
Frozen tumors were powdered by grinding in liquid nitrogen and homogenized in 1× Cell Signaling lysis buffer [20 mM tris-HCl, (pH 7.5), 150 mM NaCl, 1 mM Na2EDTA, 1 mM EGTA, 1% Triton X-100, 2.5 mM sodium pyrophosphate, 1 mM β-glycerophosphate, 1 mM Na3VO4, and leupeptin (1 μg/ml) (Cell Signaling Technologies) supplemented with 1 mM phenylmethylsulfonyl fluoride (PMSF), for 2 min at 30 Hz in the Omni Prep Multi-Sample homogenizer (QIAGEN). Lysates were spun through a QIAshredder column (Qiagen), and freeze-thawed three times. After centrifugation at 20,000 × g for 30 min at 4 °C, the protein concentration in the supernatant was determined using a BCA protein assay kit (Bio-Rad). Per sample, 750 μg of protein was subjected to ETV7 IP. Antibody (2 μg) was added to 500 μl of lysate, and samples were rotated overnight at 4 °C. Protein G–coated Dynabeads (Life Technologies) was added (10 μl per sample), and samples were rotated at 4 °C for 90 min. Bead-antibody-protein complexes were captured using a DynaMag-2 magnet (Life Technologies) and washed four times with CHAPS lysis buffer [40 mM Hepes (pH 7.4), 1 mM EDTA, 120 mM NaCl, 10 mM sodium pyrophosphate, 10 mM β-glycerophosphate, 0.3% CHAPS, 50 mM NaF, 1.5 mM NaVO, 1 mM PMSF, and one tablet of EDTA-free protease inhibitors (Roche) per 10 ml solution]. Antibody-protein complexes were retrieved from the beads by heating at 70 °C in 1.25× LDS loading buffer (Life Technologies) in CHAPS lysis buffer and loaded on precast 4–12% bis-tris protein gels (Life Technologies). Proteins were transferred onto nitrocellulose membranes using the iBLOT system (Life Technologies) following the manufacturer’s protocol. Membranes were blocked with 5% milk and 0.1% Tween 20 in tris-buffered saline (TBS) and incubated with the appropriate antibodies (Supplementary Table 2) in 5% bovine serum albumin in TBS with 0.1% Tween 20 overnight at 4 °C. All primary antibody incubations were followed by incubation with secondary horseradish peroxidase (HRP)–conjugated antibody (Pierce) in 5% milk and 0.1% Tween 20 in TBS and visualized with SuperSignal West Pico or Femto Chemiluminescent Substrate (ThermoScientific), using a Biorad ChemiDoc MP imaging system.
Hematoxylin & Eosin (H&E) staining and Immunohistochemistry (IHC)
Murine tumor tissues were fixed in 10% buffered formalin and embedded in paraffin. Sections of 6 µm were cut and deparaffinized. H&E staining was performed following standard procedures. For immunohistochemistry, after deparaffinization, antigen retrieval was performed using citrate buffer [10 mM Tris-sodium citrate, pH 6.0, 0.05% Tween 20] (for Neu1 and LAMP1), or Tris-EDTA [10 mM Tris Base, 1 mM EDTA pH9, 0.05% Tween 20] (for MyoD, Myogenin and Adiponectin). Endogenous peroxidase was removed by incubation with 3% hydrogen peroxidase in methanol for 5 min. For Neu1, Lamp1, and adiponectin, we used the ImmPress HRP polymer system following the manufacturer’s instructions (VECTOR Laboratories cat# MP-7801) with overnight incubations with primary antibodies (Supplementary Table 2). Mouse anti-MyoD and anti-Myogenin antibodies were biotinylated with the ARK (Animal Research Kit), Peroxidase (DAKO K3954) following the manufacturer’s instructions. Signal detection was performed with the Vectastain Elite ABC HRP (Vector Laboratories Cat# PK6100) followed by Stable DAB chromogen (Invitrogen Cat#75018). Slides were counterstained with hematoxylin, dehydrated, and mounted with a xylene-based mounting medium. Formalin-fixed paraffin-embedded human RMS TMAs were obtained from the St Jude Pathology/Laboratory Medicine Department. Specimens were de-identified, and the study was approved by the Institutional Review Board (IRB approval Pro00008511) as non-human subject research.
Human RMS TMAs were obtained from the St Jude Pathology/Laboratory Medicine Department (approved by IRB Pro00008511).
Masson’s Trichrome staining
Masson’s Trichrome staining was done as previously described70. In brief, FFPE (formalin-fixed paraffin-embedded) murine RMS samples and patient TMAs on slides were fixed in Bouin’s solution for 1 h at 60 °C. Sections were then stained sequentially at room temperature with Weigert’s iron hematoxylin, Biebrich scarlet-acid fuchsin, phosphotungstic/phosphomolybdic acid, and aniline blue. Sections were washed, dehydrated, and mounted with a xylene-based mounting medium. Trichrome Masson’s-stained slides were scanned to ×20 scalable images with an Aperio Scanscope XT (Leica Biosystems, Inc.). The Aperio color deconvolution and colocalization algorithms and the ImageScope software v12.4.3 (Leica Biosystems, Inc.) were used to quantify ECM/collagen (blue) deposition in the microenvironment and within tumor cells.
Nuclear morphometry analysis
H&E-stained murine RMS samples were scanned to ×20 scalable images with an Aperio ScanScope XT (Leica Biosystems, Inc.) and annotated using ImageScope software v12.4.3 (Leica Biosystems, Inc.). The Genie tissue classifier was trained to identify tumor nuclei and quantify their numbers and sizes.
Quantification of LAMP1 staining
IHC stained images from RMS TMAs were scanned with a Pannoramic 250 Flash III (3DHistech, Inc.) to ×32 scalable images. Images from tissue cores from the whole slide were then manually annotated using HALO v3.2.1851.354 software (Indica Labs). A Membrane v1.7 algorithm was trained to identify LAMP1 immunoreactivity at the membrane level. After positive immunoreactivity was identified with the algorithm, a density heatmap spatial analysis was performed on the analyzed samples to determine the minimal and maximal intercellular density and spacing of immune-positive neoplastic cells within a 25 µm radius in each core using the Spatial Analysis Module HALO 3.2 (Indica labs).
β-Hexosaminidase activity in tumor’s interstitial fluid
Frozen tumors were powdered by grinding in liquid nitrogen and homogenized in PBS for 2 min at 30 Hz in the Omni Prep Multi-Sample homogenizer (QIAGEN). Interstitial fluid was purified by sequential centrifugation steps at 300 × g for 10 min, 2000 × g for 10 min, and 10,000 × g for 30 min to remove cells and cell debris. The supernatant/interstitial fluid was spun in an ultracentrifuge at 100,000 × g for 2 h (SW32Ti rotor) to remove small vesicles and exosomes. All steps were performed at 4 °C. Interstitial fluid was diluted 5 times in PBS and spun through a Sephadex column at pH 5.5 for 2 min at 850 × g. Beta-hexosaminidase activity19 was measured with 4-Methylumbelliferyl N-acetyl-β-D-glucosaminide fluorometric substrate (3 mmol/L in citrate phosphate buffer pH4.4 (6 mmol/L citric acid, 10 mmol/L disodium hydrogen phosphate) (Sigma M2133). Ten µL of interstitial fluid, or when needed a 1:10 dilution, was incubated at 37 °C with 10 uL of substrate for 1 h. The reaction was stopped with 200 mL carbonate stop buffer (0.5 M Na2CO3 with the pH set to 10.7 by adding 0.5 M NaHCO3). The amount of fluorescence generated from each reaction indicates cleavage of the fluorescent tag from its peptide substrate and is proportional to the amount of enzyme present. The mean fluorescence of samples given assay buffer was subtracted from the mean fluorescence of samples given assay buffer plus the specific substrate. Mean fluorescence values were interpolated to a standard curve, adjusted for dilution and length of incubation, and normalized to protein by BCA. The final units of activity were reported as nanomoles per hour per milligram.
Gene expression analyses
For microarray analysis of NPE and PE trunk and limb RMS, total RNA (100 ng) was converted into biotin-labeled cRNA (Ambion WT Expression Kit, Affymetrix Inc) and hybridized to Clariom S Mouse GeneChip (Affymetrix Inc) and signals summarized by RMA (Affymetrix Expression Console v1.1). Probe signals from arrays were normalized and transformed into log2 transcript expression values using the Robust Multiarray Average algorithm (Partek Genomics Suite v6.6). Patient’s RMS gene expression data from the Pediatric Cancer Genome Project (PCGP) were also used. For the comparison of murine versus human RMS, principal component analysis (PCA) and quality-control metrics removed outliers from both PCGP RNAseq and murine microarray data. We used data from 8 ARMS with PAX3/FOXO1 fusion, 4 ARMS with PAX7/FOXO1 fusion and 22 ERMS (accession# EGAS00001000256)71. Metadata were also supplemented for fusion calls from public RMS data (PeCan https://pecan.stjude.cloud/home). Correlations were calculated with Pearson’s correlation of deciles of the mean log2 FPKM (for RNAseq) and RMA values for the microarray’s expression by class. There were 91 genes that passed the false discovery rate (FDR) at 5%, having a difference of 5 or more deciles. GSEA (https://www.gsea-msigdb.org/gsea/index.jsp)72,73 was also performed using the curated pathways from MSigDB72,74. Differentially expressed transcripts were identified by ANOVA, and the FDR was estimated. Functional enrichment analysis of gene lists was performed using the DAVID bioinformatics databases (https://david.ncifcrf.gov/). Additional gene sets for transcriptional analysis were identified using Enrichr51,52,75. For adipogenic gene expression analysis, gene level feature counts were downloaded from the St. Jude PCGP for 44 patient samples of RMS (14 ARMS, 27 ERMS, and 3 RMS undefined). These counts were normalized, and log2 transformed using voom-limma, and then log2CPM was graphed in Prism. The normalized, transformed counts were also used for GSEA analysis using a combination of experimentally derived gene sets, as well as established gene sets from MSigDB72,74. Normalized RMA microarray values were compared using voom-limma in R and displayed in a volcano plot.
Multiplex flow cytometry
NPE and PE tumors were dissociated with the Mouse Dissociation kit and gentle MACS Octo Dissociator with heaters following the manufacturer’s instructions (MACS, Miltenyi Biotec). Large aggregates persisting in the cell preparation were then removed by filtering them through 70 μm cell strainers. The resulting single-cell suspensions were stained with fluorochrome-conjugated monoclonal antibodies as noted in Supplementary Table 2, and analyzed using a BD FACSymphony A9 analyzer (BD, San Jose) equipped with 355, 400, 440, 488, 561, and 640 nm lasers for excitation and an array of 30 detectors with the appropriate light filters for resolving the specified fluorochromes. Self-organizing maps for visualizing and interpreting cytometry data, FlowSOM, was used76 on 60,000 cells from limb and trunk NPE and PE RMS to generate density, heatmaps, and tSNE graphs. Cellular clusters were identified and annotated based on the differential expression of markers. Additional two-dimensional analyses were conducted using both FlowJo and FACS Diva software suites.
Statistics and reproducibility
Quantitative data are presented as mean ± s.d. of at least six biologically independent samples/tumors. A number of replicates are noted in the figure legends. Statistical analyses were performed using GraphPad Prism. Student’s t-test for unpaired and paired (two-tailed), Welch’s t-test for unpaired t-test, one-way ANOVA t-test, Mann–Whitney for unpaired (two-tailed) t-test, all tests have a 95% confidence interval, and log-rank test for trend was performed to ascertain statistical significance and are noted in the figure legends. Mean differences were considered significant when P < 0.05. Source data underlying the main figures are presented in Supplementary Data 1.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
The data that support the findings of this study are available from the corresponding authors on reasonable request. Uncropped and unedited blot images are provided in Supplementary Fig. 2. Microarray data in this publication are deposited in NCBI’s functional genomic data repository Gene Expression Omnibus (GEO) and are assecible through GEO series accession number GSE212378. Previously published datasets used in this study were deposited in the European Bioinformatics Institue (EMBL-EBI) and accessible through accession number EGAS00001000256. Source data underlying Fig. 1b, d, I, j, 2d–g, 3d, g, h, 6c–e, 7c–l and Supplementary Fig. 1a, 3a, b, d, e, 9a–c, 10a are provided within this paper (Supplementary Data 1). Patient’s RMS gene expression data can be accessed through StJude open resource page (PeCan https://pecan.stjude.cloud/home).
References
Dagogo-Jack, I. & Shaw, A. T. Tumour heterogeneity and resistance to cancer therapies. Nat. Rev. Clin. Oncol. 15, 81–94 (2018).
Marusyk, A., Janiszewska, M. & Polyak, K. Intratumor heterogeneity: the rosetta stone of therapy resistance. Cancer Cell 37, 471–484 (2020).
Oberlin, O. et al. Prognostic factors in metastatic rhabdomyosarcomas: results of a pooled analysis from United States and European cooperative groups. J. Clin. Oncol. 26, 2384–2389 (2008).
Kashi, V. P., Hatley, M. E. & Galindo, R. L. Probing for a deeper understanding of rhabdomyosarcoma: insights from complementary model systems. Nat. Rev. Cancer 15, 426–439 (2015).
Marshall, A. D. & Grosveld, G. C. Alveolar rhabdomyosarcoma—the molecular drivers of PAX3/7-FOXO1-induced tumorigenesis. Skelet. Muscle 2, 25 (2012).
Kohashi, K. et al. Alterations of RB1 gene in embryonal and alveolar rhabdomyosarcoma: special reference to utility of pRB immunoreactivity in differential diagnosis of rhabdomyosarcoma subtype. J. Cancer Res. Clin. Oncol. 134, 1097–1103 (2008).
Chen, Y. et al. Mutations of the PTPN11 and RAS genes in rhabdomyosarcoma and pediatric hematological malignancies. Genes Chromosomes Cancer 45, 583–591 (2006).
Shern, J. F. et al. Comprehensive genomic analysis of rhabdomyosarcoma reveals a landscape of alterations affecting a common genetic axis in fusion-positive and fusion-negative tumors. Cancer Discov. 4, 216–231 (2014).
Hahn, H. et al. Rhabdomyosarcomas and radiation hypersensitivity in a mouse model of Gorlin syndrome. Nat. Med. 4, 619–622 (1998).
Paulson, V. et al. High-resolution array CGH identifies common mechanisms that drive embryonal rhabdomyosarcoma pathogenesis. Genes Chromosomes Cancer 50, 397–408 (2011).
Martinelli, S. et al. RAS signaling dysregulation in human embryonal Rhabdomyosarcoma. Genes Chromosomes Cancer 48, 975–982 (2009).
Taylor, A. C. et al. P53 mutation and MDM2 amplification frequency in pediatric rhabdomyosarcoma tumors and cell lines. Med. Pediatr. Oncol. 35, 96–103 (2000).
Buckingham, M. Gene regulatory networks and cell lineages that underlie the formation of skeletal muscle. Proc. Natl Acad. Sci. USA 114, 5830–5837 (2017).
Keller, C. & Guttridge, D. C. Mechanisms of impaired differentiation in rhabdomyosarcoma. FEBS J. 280, 4323–4334 (2013).
Tiffin, N., Williams, R. D., Shipley, J. & Pritchard-Jones, K. PAX7 expression in embryonal rhabdomyosarcoma suggests an origin in muscle satellite cells. Br. J. Cancer 89, 327–332 (2003).
Hettmer, S. et al. Sarcomas induced in discrete subsets of prospectively isolated skeletal muscle cells. Proc. Natl Acad. Sci. USA 108, 20002–20007 (2011).
Rubin, B. P. et al. Evidence for an unanticipated relationship between undifferentiated pleomorphic sarcoma and embryonal rhabdomyosarcoma. Cancer Cell 19, 177–191 (2011).
Hatley, M. E. et al. A mouse model of rhabdomyosarcoma originating from the adipocyte lineage. Cancer Cell 22, 536–546 (2012).
Machado, E. et al. Regulated lysosomal exocytosis mediates cancer progression. Sci. Adv. 1, e1500603 (2015).
d’Azzo, A., Machado, E. & Annunziata, I. Pathogenesis, emerging therapeutic targets and treatment in Sialidosis. Expert Opin. Orphan Drugs 3, 491–504 (2015).
Platt, F. M., d’Azzo, A., Davidson, B. L., Neufeld, E. F. & Tifft, C. J. Lysosomal storage diseases. Nat. Rev. Dis. Prim. 4, 27 (2018).
Yogalingam, G. et al. Neuraminidase 1 is a negative regulator of lysosomal exocytosis. Dev. Cell 15, 74–86 (2008).
Kima, P. E., Burleigh, B. & Andrews, N. W. Surface-targeted lysosomal membrane glycoprotein-1 (Lamp-1) enhances lysosome exocytosis and cell invasion by Trypanosoma cruzi. Cell Microbiol. 2, 477–486 (2000).
Machado, E. R., Annunziata, I., van de Vlekkert, D., Grosveld, G. C. & d’Azzo, A. Lysosomes and cancer progression: a malignant liaison. Front. Cell Dev. Biol. 9, 642494 (2021).
Andrews, N. W. Detection of lysosomal exocytosis by surface exposure of Lamp1 luminal epitopes. Methods Mol. Biol. 1594, 205–211 (2017).
van de Vlekkert, D. et al. Excessive exosome release is the pathogenic pathway linking a lysosomal deficiency to generalized fibrosis. Sci. Adv. 5, eaav3270 (2019).
Zanoteli, E. et al. Muscle degeneration in neuraminidase 1-deficient mice results from infiltration of the muscle fibers by expanded connective tissue. Biochim. Biophys. Acta 1802, 659–672 (2010).
Hahn, H. et al. Genetic mapping of a Ptch1-associated rhabdomyosarcoma susceptibility locus on mouse chromosome 2. Genomics 84, 853–858 (2004).
Ng, J. M. & Curran, T. The Hedgehog’s tale: developing strategies for targeting cancer. Nat. Rev. Cancer 11, 493–501 (2011).
Goodrich, L. V., Milenkovic, L., Higgins, K. M. & Scott, M. P. Altered neural cell fates and medulloblastoma in mouse patched mutants. Science 277, 1109–1113 (1997).
Qi, X. & Li, X. Mechanistic insights into the generation and transduction of hedgehog signaling. Trends Biochem. Sci. 45, 397–410 (2020).
Harwood, F. C. et al. ETV7 is an essential component of a rapamycin-insensitive mTOR complex in cancer. Sci. Adv. 4, eaar3938 (2018).
Numata, M., Klein Geltink, R. I. & Grosveld, G. C. Establishment of a transgenic mouse to model ETV7 expressing human tumors. Transgenic Res. 28, 115–128 (2019).
Potter, M. D., Buijs, A., Kreider, B., van Rompaey, L. & Grosveld, G. C. Identification and characterization of a new human ETS-family transcription factor, TEL2, that is expressed in hematopoietic tissues and can associate with TEL1/ETV6. Blood 95, 3341–3348 (2000).
Furlong, M. A., Mentzel, T. & Fanburg-Smith, J. C. Pleomorphic rhabdomyosarcoma in adults: a clinicopathologic study of 38 cases with emphasis on morphologic variants and recent skeletal muscle-specific markers. Mod. Pathol. 14, 595–603 (2001).
Winkler, J., Abisoye-Ogunniyan, A., Metcalf, K. J. & Werb, Z. Concepts of extracellular matrix remodelling in tumour progression and metastasis. Nat. Commun. 11, 5120 (2020).
Rodriguez, A., Webster, P., Ortego, J. & Andrews, N. W. Lysosomes behave as Ca2+-regulated exocytic vesicles in fibroblasts and epithelial cells. J. Cell Biol. 137, 93–104 (1997).
Chen, C., Zhao, S., Karnad, A. & Freeman, J. W. The biology and role of CD44 in cancer progression: therapeutic implications. J. Hematol. Oncol. 11, 64 (2018).
Han, C., Liu, T. & Yin, R. Biomarkers for cancer-associated fibroblasts. Biomark. Res. 8, 64 (2020).
Hyun, K. A. et al. Epithelial-to-mesenchymal transition leads to loss of EpCAM and different physical properties in circulating tumor cells from metastatic breast cancer. Oncotarget 7, 24677–24687 (2016).
Lee, E., Pandey, N. B. & Popel, A. S. Crosstalk between cancer cells and blood endothelial and lymphatic endothelial cells in tumour and organ microenvironment. Expert Rev. Mol. Med. 17, e3 (2015).
Misharin, A. V., Morales-Nebreda, L., Mutlu, G. M., Budinger, G. R. & Perlman, H. Flow cytometric analysis of macrophages and dendritic cell subsets in the mouse lung. Am. J. Respir. Cell Mol. Biol. 49, 503–510 (2013).
Mukai, K. et al. Critical role of P1-Runx1 in mouse basophil development. Blood 120, 76–85 (2012).
Park, J. W., Park, J. M., Park, D. M., Kim, D. Y. & Kim, H. K. Stem cells Antigen-1 enriches for a cancer stem cell-like subpopulation in mouse gastric cancer. Stem Cells 34, 1177–1187 (2016).
Perot, G. et al. Smooth muscle differentiation identifies two classes of poorly differentiated pleomorphic sarcomas with distinct outcome. Mod. Pathol. 27, 840–850 (2014).
Sahin, A. A. et al. Tumor proliferative fraction in solid malignant neoplasms. A comparative study of Ki-67 immunostaining and flow cytometric determinations. Am. J. Clin. Pathol. 96, 512–519 (1991).
Sapino, A. et al. Expression of CD31 by cells of extensive ductal in situ and invasive carcinomas of the breast. J. Pathol. 194, 254–261 (2001).
Truong, L. D. et al. The diagnostic utility of desmin. A study of 584 cases and review of the literature. Am. J. Clin. Pathol. 93, 305–314 (1990).
Yui, M. A. & Rothenberg, E. V. Developmental gene networks: a triathlon on the course to T cell identity. Nat. Rev. Immunol. 14, 529–545 (2014).
Zhang, Y. Y. et al. CD31 regulates metastasis by inducing epithelial-mesenchymal transition in hepatocellular carcinoma via the ITGB1-FAK-Akt signaling pathway. Cancer Lett. 429, 29–40 (2018).
Chen, E. Y. et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinforma. 14, 128 (2013).
Kuleshov, M. V. et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 44, W90–W97 (2016).
Ma, X., Wang, D., Zhao, W. & Xu, L. Deciphering the roles of PPARgamma in adipocytes via dynamic change of transcription complex. Front. Endocrinol. (Lausanne) 9, 473 (2018).
Wang, W. et al. Ebf2 is a selective marker of brown and beige adipogenic precursor cells. Proc. Natl Acad. Sci. USA 111, 14466–14471 (2014).
Seale, P. et al. PRDM16 controls a brown fat/skeletal muscle switch. Nature 454, 961–967 (2008).
Ikeda, K., Maretich, P. & Kajimura, S. The common and distinct features of brown and beige adipocytes. Trends Endocrinol. Metab. 29, 191–200 (2018).
Murphy, M. & Kardon, G. Origin of vertebrate limb muscle: the role of progenitor and myoblast populations. Curr. Top. Dev. Biol. 96, 1–32 (2011).
Nicolas, M. M., Tamboli, P., Gomez, J. A. & Czerniak, B. A. Pleomorphic and dedifferentiated leiomyosarcoma: clinicopathologic and immunohistochemical study of 41 cases. Hum. Pathol. 41, 663–671 (2010).
Oda, Y. et al. Pleomorphic leiomyosarcoma: clinicopathologic and immunohistochemical study with special emphasis on its distinction from ordinary leiomyosarcoma and malignant fibrous histiocytoma. Am. J. Surg. Pathol. 25, 1030–1038 (2001).
Couture, A. et al. HLA-class II artificial antigen presenting cells in CD4(+) T cell-based immunotherapy. Front Immunol. 10, 1081 (2019).
Henke, E., Nandigama, R. & Ergun, S. Extracellular matrix in the tumor microenvironment and its impact on cancer therapy. Front. Mol. Biosci. 6, 160 (2019).
Atit, R. et al. Beta-catenin activation is necessary and sufficient to specify the dorsal dermal fate in the mouse. Dev. Biol. 296, 164–176 (2006).
Kajimura, S. et al. Initiation of myoblast to brown fat switch by a PRDM16-C/EBP-beta transcriptional complex. Nature 460, 1154–1158 (2009).
Cao, Y. Adipocyte and lipid metabolism in cancer drug resistance. J. Clin. Investig. 129, 3006–3017 (2019).
Dirat, B. et al. Cancer-associated adipocytes exhibit an activated phenotype and contribute to breast cancer invasion. Cancer Res. 71, 2455–2465 (2011).
Quincey, C., Banerjee, S. S., Eyden, B. P. & Vasudev, K. S. Lipid rich rhabdomyosarcoma. J. Clin. Pathol. 47, 280–282 (1994).
Zuppan, C. W., Mierau, G. W. & Weeks, D. A. Lipid-rich rhabdomyosarcoma-a potential source of diagnostic confusion. Ultrastruct. Pathol. 15, 353–359 (1991).
Balitzer, D., McCalmont, T. H. & Horvai, A. E. Adipocyte-like differentiation in a posttreatment embryonal rhabdomyosarcoma. Case Rep. Pathol. 2015, 406739 (2015).
de Geest, N. et al. Systemic and neurologic abnormalities distinguish the lysosomal disorders sialidosis and galactosialidosis in mice. Hum. Mol. Genet. 11, 1455–1464 (2002).
Van De Vlekkert, D., Machado, E. & d’Azzo, A. Analysis of generalized fibrosis in mouse tissue sections with Masson’s trichrome staining. Bio Protoc. 10, e3629 (2020).
Chen, X. et al. Targeting oxidative stress in embryonal rhabdomyosarcoma. Cancer Cell 24, 710–724 (2013).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA 102, 15545–15550 (2005).
Mootha, V. K. et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 34, 267–273 (2003).
Liberzon, A. et al. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 1, 417–425 (2015).
Xie, Z. et al. gene set knowledge discovery with Enrichr. Curr. Protoc. 1, e90 (2021).
Van Gassen, S. et al. FlowSOM: using self-organizing maps for visualization and interpretation of cytometry data. Cytom. A 87, 636–645 (2015).
Acknowledgements
We thank I. Annunziata for the critical reading and input. A.d’A. holds the Jewelers for Children Endowed Chair in Genetics and Gene Therapy. G.C.G. holds the Albert and Rosemary Joseph Endowed Chair in Genetic Research. This work was funded in part by grants from the by NIH grants CA021764 and 5R01GM104981, CCSG St. Jude Developmental Funds, the Assisi Foundation of Memphis, and the American Lebanese Syrian Associated Charities (ALSAC).
Author information
Authors and Affiliations
Contributions
E.M. designed, performed, and analyzed all the experiments, and wrote the manuscript. D.v.d.V. performed all the RT-qPCRs, isolated interstitial fluid, did statistical analyses, analyzed data, assembled the figures, and edited the manuscript. H.T. diagnosed and analyzed all formalin-fixed, paraffin-embedded samples of mouse tumors and TMAs of RMS patients, and performed all computational algorithm analyses. S.P. helped design, performed, and analyzed all FACS and FlowSOM data. S.M.D. helped analyze microarray data, performed GSEA, and analyzed PCGP human RMS data and helped with the final figures pertaining to these analyses. J.L. and R.A. performed the FACS and FlowSOM analyses. D.F. performed correlation analyses of murine RMS vs human RMS genes. G.N. performed and analyzed the microarray data. H.H. embedded, processed, and cut sections of all mouse tumors. F.H. performed mTORC3/ETV7 co-immunoprecipitations. S.K. diagnosed TMAs of RMS patients. G.C.G. designed experiments, analyzed data, edited, and wrote the manuscript. A.d’A. conceived the project designed experiments, analyzed data, supervised the study, edited, and wrote the manuscript. All authors approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Biology thanks Heidi Hahn and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editor: George Inglis. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Machado, E.R., van de Vlekkert, D., Sheppard, H.S. et al. Haploinsufficiency of the lysosomal sialidase NEU1 results in a model of pleomorphic rhabdomyosarcoma in mice. Commun Biol 5, 992 (2022). https://doi.org/10.1038/s42003-022-03968-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s42003-022-03968-8
- Springer Nature Limited