Abstract
The transcription factor NKX2-1/TTF-1 is involved in lung pathophysiology, including breathing, innate defense and tumorigenesis. To understand the mechanism by which NKX2-1 regulates genes involved in such pathophysiology, we have previously performed ChIP-seq and identified genome-wide NKX2-1-binding sites, which revealed that NKX2-1 binds to not only proximal promoter regions but also multiple intra- and inter-genic regions of the genes regulated by NKX2-1. However, the roles of such regions, especially non-proximal ones, bound by NKX2-1 have not yet been determined. Here, using CRISPRi (CRISPR/dCas9-KRAB), we scrutinize the functional roles of 19 regions/sites bound by NKX2-1, which are located in genes involved in breathing and innate defense (SFTPB, LAMP3, SFTPA1, SFTPA2) and lung tumorigenesis (MYBPH, LMO3, CD274/PD-L1). Notably, the CRISPRi approach reveals that a portion of NKX2-1-binding sites are functionally indispensable while the rest are dispensable for the expression of the genes, indicating that functional roles of NKX2-1-binding sites are unequally yoked.
Similar content being viewed by others
Introduction
Ever since it was first shown that Nkx2-1 (also known as TTF-1) deleted mice had impaired lung development, which results in death at birth due to a breathing defect1, the molecular mechanisms by which NKX2-1 regulates genes involved in lung development and physiology have been extensively pursued2. One of the genes critical for breathing at birth is SFTPB (also known as surfactant protein B gene), since Sftpb-deleted mice also succumb to death at birth because of a breathing defect due to collapsed lungs3. Using gel-shift (also known as EMSA [Electrophoretic mobility shift assay]) and plasmid-based reporter assays, NKX2-1 has been shown to bind to the proximal promoter region of human SFTPB and stimulate its promoter activity4,5. A microarray analysis to determine the genome-wide NKX2-1 downstream target genes in human lung epithelial cells also identified SFTPB as well as LAMP3 (also known as Lysosomal Associated Membrane Protein 3), both of which co-localize at lamellar bodies (a cellular organelle) that process and secrete surfactant to reduce lung surface tension, which is required for proper lung function6. These studies indicate that NKX2-1 is indispensable for lung development involved in breathing in part through the regulation of SFTPB and LAMP3 expression.
The microarray analysis also identified that SFTPA (also known as surfactant protein A gene), including SFTPA1 and SFTPA2 (a paralog of SFTPA1), is induced by NKX2-1 in human lung epithelial cells6, in part through the proximal promoters of SFTPA1 and SFTPA2 bound by NKX2-1, which has been reported using gel-shift and plasmid-based reporter assays7,8,9. Although SFTPA1/SFTPA2 belong to the same surfactant protein gene family as SFTPB, Sftpa1-deleted mice (of note, no Sftpa2 in mouse) are viable at birth and have no apparent lung abnormality10. However, Sftpa1-deleted mice are susceptible to bacterial, viral, and fungal infection11,12,13,14,15, indicating that NKX2-1 is involved in innate defense by the clearance of respiratory bacteria, virus, and fungus through SFTPA.
Development of a monoclonal NKX2-1 antibody (8G7G3/1) revealed that NKX2-1 is expressed in normal human lung epithelium as well as in human lung carcinomas, especially lung adenocarcinoma and small-cell lung carcinoma, but not colon or breast carcinoma16. Since then, the antibody has been used extensively in the clinic to distinguish primary lung adenocarcinomas from lung metastases17,18. Notably, NKX2-1 is not only highly expressed in primary human lung adenocarcinoma, but also its chromosomal locus (14q13.3) is highly amplified in ~10% of human lung adenocarcinomas19,20,21,22. Discovery of the 14q13.3 amplification led to subsequent studies to understand the functional roles of NKX2-1 and its downstream target genes that influence lung tumorigenesis. Importantly, such studies have revealed that NKX2-1 functions as a tumor promoter or a tumor suppressor in a context-dependent fashion. For example, NKX2-1 promotes EGFR-mutant lung tumorigenesis while it suppresses KRAS-mutant lung tumorigenesis23,24,25,26,27,28,29. Among the downstream target genes induced by NKX2-1, MYBPH, LMO3, and CD274 (also known as PD-L1) are of particular interest since they directly influence lung tumorigenesis24,28,30. MYBPH (also known as myosin binding protein H) inhibits cell motility and metastasis of lung adenocarcinoma24 while LMO3 promotes cell survival28. CD274/PD-L1, a suppressor of adaptive immunity, expressed on lung tumor cells binds to PD-1 on T cells and inhibits the anti-tumorigenic activity of T cells31. These reports indicate that NKX2-1 influences lung tumorigenesis in part through the regulation of MYBPH, LMO3, and CD274/PD-L1 expression.
The regulatory mechanisms by which NKX2-1 activates expression of these genes had previously been investigated by focusing on the proximal promoter (upstream) regions using gel-shift and plasmid-based reporter assays, which do not provide a chromatin-based and genome-wide mechanism of gene regulation. Subsequently, we used the ChIP (chromatin immunoprecipitation)-seq assay to determine unbiased genome-wide NKX2-1 binding sites in human lung epithelial cells27. ChIP-seq analysis by us and others indicated that NKX2-1 bound to not only proximal upstream regions but also distal upstream and/or downstream regions as well as intronic regions and intergenic regions in the genome27,28,29,32. Although ChIP-seq can reveal where NKX2-1 binds in the genome, it does not indicate whether NKX2-1-binding sites are functionally important or not. The ability to determine the functional relevance of transcription factor-binding sites has recently been achieved using CRISPR/Cas9 technology. Mansour and colleagues using a CRISPR/Cas9-mediated deletion approach, deleted a transcription factor MYB-binding site (identified by ChIP-seq) between TAL1 and STIL, and showed that the MYB-binding site is required for the expression of TAL133. We have previously used the same approach and determined that a transcription factor SPDEF-binding site (identified by ChIP-seq) between MUC5AC and MUC5B is required for the expression of MUC5B30. We have also recently used a modified CRISPR/Cas9 technology (CRISPRi) that carries a deactivated Cas9 fused to the repressor KRAB (CRISPR/dCas9-KRAB) to repress the transcription of genes without creating DNA double-stranded breaks. Gene-regulatory regions in the genome that are targeted by CRISPRi are selected by 20-mer sgRNA target sequences34,35. We thus targeted the SPDEF-binding region and found that the CRISPRi approach is more streamlined than the CRISPR/Cas9-mediated deletion approach since cell cloning is not required in CRISPRi36. In the present study, using the CRISPRi approach, we set out to determine whether NKX2-1-binding sites that were previously identified by ChIP-seq are indispensable for the expression of NKX2-1 downstream target genes, including SFTPB, LAMP3, SFTPA1, SFTPA2, MYBPH, LMO3, and CD274/PD-L1, that are involved in breathing, innate defense, and tumorigenesis (Fig. 1).
NKX2-1-binding sites identified by ChIP-seq were targeted by CRISPRi that carries dCas9-KRAB and sgRNAs, which match to each binding site sequence, and the expression of NKX2-1 downstream target genes (SFTPB, LAMP3, SFTPA1, SFTPA2, MYBPH, LMO3, and CD274/PD-L1) was assessed. The putative lung function of each gene is shown. TSS transcription start site.
Results
First intron of SFTPB bound by NKX2-1 is most critical for the expression of SFTPB induced by NKX2-1
Our previous ChIP-seq data from an A549 human lung epithelial cell line stably expressing NKX2-127,30 indicate that NKX2-1 binds to five regions (upstream, first intron, ninth intron, proximal downstream, distal downstream) of the locus of SFTPB, which is an indispensable gene for breathing (Fig. 2a). Notably, ATAC-seq (Assay for Transposase-Accessible Chromatin using sequencing) data from A549 cells obtained from ENCODE37 indicated that an ATAC peak was not observed at an NKX2-1-binding site in the first intronic region, suggesting that NKX2-1 is able to access genomic regions that are considered to be inaccessible by ATAC-seq (Figs. 2a, 3a, and Supplementary Data 1). In order to understand which NKX2-1-binding sites in the five regions are important for the expression of SFTPB induced by NKX2-1, we created A549 cells stably expressing dCas9-KRAB with or without NKX2-1 (Supplementary Figs. 1 and 2) and transfected synthetic sgRNAs targeting individual DNA elements within or near NKX2-1-binding motifs (CTTG/CAAG) in the five regions of SFTPB (Fig. 2a and Supplementary Fig. 3). Of note, A549 cells do not express endogenous NKX2-1 (Supplementary Figs. 1 and 4). We then assessed the expression of SFTPB, LAMP3, SFTPA1, SFTPA2, MYBPH, LMO3, and CD274/PD-L1. This is a CRISPRi approach that we previously reported36. Notably, sgRNAs targeting the upstream region and first and ninth introns of the SFTPB locus significantly suppressed the expression of SFTPB more than two-fold (P < 0.05, red), indicating that these three regions (upstream, first, and ninth introns) are critical gene-regulatory regions for the expression of SFTPB induced by NKX2-1. The sgRNA targeting the first intronic region most extensively suppressed the expression of SFTPB compared to those targeting the upstream region and ninth intron (98.0% vs. 54.8 and 66.0%; Fig. 2b). The specificity of CRISPRi targeting this region was further confirmed by genome-wide RNA-seq (Fig. 3b and Supplementary Data 2). The sgRNAs targeting the proximal and distal downstream regions of SFTPB did not suppress the expression of SFTPB. Unexpectedly, the sgRNA targeting the first intronic region also suppressed the expression of LAMP3 more than two-fold (P < 0.05) (Fig. 2b) though, according to the CRISPOR off-target search38 (Supplementary Data 3), the sgRNA sequence does not match a genomic DNA sequence at the LAMP3 locus. Since the siRNA-mediated inhibition of endogenous SFTPB did not affect the expression of LAMP3 (Supplementary Fig. 5), the first intronic region may regulate the expression of LAMP3 in trans (of note, SFTPB and LAMP3 are located at chromosomes 2 and 3, respectively). Hi-C data from A549 cells obtained by ENCODE data37 indicate that the SFTPB locus on chromosome 2 interacts with other loci in different chromosomes, including chromosome 3, supporting a possibility of such trans inter-chromosomal interactions (Supplementary Fig. 6).
a Shown are ATAC-seq and ChIP-seq data indicating that NKX2-1 binds to upstream (#1), first (#2), ninth intronic (#3), proximal (#4), and distal downstream (#5) regions of human SFTPB locus. ChIP-seq was previously performed using an A549 lung epithelial cell line stably expressing NKX2-127,30. ATAC-seq was obtained from the ENCODE database37. NKX2-1-binding sites are shown as peaks obtained from bam files. Significant peaks compared to input peaks are shown as side bars obtained from bed files. b Shown is the gene expression of NKX2-1 downstream target genes affected by CRISPRi from biologically independent triplicates, in which synthetic sgRNAs targeting NKX2-1-binding sites (#) located at the regions described in (a) are transiently transfected into A549 cells expressing dCas9-KRAB with or without NKX2-1 (PGK.Nkx2-1 or PGK vector control), or H441 cells with or without dCas9-KRAB. The sgRNA targeting NKX2-1-binding sites located at the upstream (#1) and first intronic (#2) regions significantly suppressed the expression of SFTPB in both A549 and H441 cells. The sgRNA targeting NKX2-1-binding sites located at the first intronic (#2) region also significantly suppressed the expression of LAMP3 induced by NKX2-1 in A549 cells. Of note, endogenous NKX2-1 is expressed in H441 cells but not in A549 cells (see Supplementary Fig. 1). Results are expressed as the mean ± SD of the triplicates for each group. Only P < 0.05 and more than two-fold suppression are considered significant CRISPRi-mediated suppression (highlighted in red). Unt., untransfected. Cont., non-targeted control sgRNA.
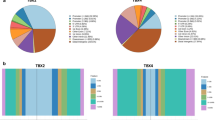
a Shown are pie charts that indicate the number of NKX2-1 ChIP-seq peaks and ATAC-seq peaks in A549 cells. A portion (but not all) of the NKX2-1-binding sites are located in ATAC-accessible genomic regions. b Shown is an MA plot that indicates differential gene expression regulated by CRISPRi targeting the first intron (#2) of SFTPB, which was analyzed by RNA-seq using RNA from A549 cells that express dCas9-KRAB and NKX2-1 with sgRNA #2 or the sgRNA control as described in Fig. 2b. Biologically independent triplicates were used for each group (sgRNA #2 or sgRNA control). Red points represent genes with log2 fold change ≦1 or ≧−1; padj <0.05. The expression of SFTPB was most significantly repressed (log2 fold change −5.96; p value 2.06E−257; padj 3.06E−253) while the expression of LAMP3 was weakly repressed (log2 fold change −1.85; p value 2.64E−03; padj 1.00E + 00).
We also created an H441 human lung epithelial cell line that stably expresses dCas9-KRAB to complement the above study using A549 cells. Of note, H441 cells express endogenous NKX2-1 while A549 cells do not (Supplementary Fig. 1). We assessed which NKX2-1 downstream target genes identified by a gain-of-function analysis using A549 cells27 were regulated by NKX2-1 in H441 cells by a loss-of-function approach using siRNA. The expression of SFTPB, SFTPA1, SFTPA2, MYBPH, and LMO3 was reduced by two independent siRNAs targeting endogenous NKX2-1, while the expression of LAMP3 and CD274/PD-L1 was not, suggesting that the expression of SFTPB, SFTPA1, SFTPA2, MYBPH, and LMO3 (but not that of LAMP3 and CD274/PD-L1) is regulated by NKX2-1 in H441 cells (Supplementary Fig. 7). As described above, we then transfected H441 cells that express dCas9-KRAB with the same sgRNAs and assessed the expression levels of the respective genes (Fig. 2b). Notably, sgRNAs targeting the upstream and first intron of the SFTPB locus significantly suppressed the expression of SFTPB (Fig. 2b), which is consistent with the results using A549 cells (Fig. 2b); however, the sgRNA targeting the ninth intron of SFTPB, which was suppressive in A549 cells, was not suppressive in H441 cells, suggesting a cell type-specific gene regulation. Conversely, while sgRNA targeting the distal downstream region of SFTPB did not suppress the expression of SFTPB in A549 cells, expression was suppressed in H441 cells. These results suggest that the NKX2-1-binding sites located at the upstream and first intronic regions of SFTPB are essential to NKX2-1-mediated expression of SFTPB regardless of cell type; however, other regions function in a cell type-dependent fashion.
Both the third and fifth introns of LAMP3 bound by NKX2-1 are critical for the expression of LAMP3 induced by NKX2-1
The ChIP-seq data from an A549 lung epithelial cell line stably expressing NKX2-127,30 also indicate that NKX2-1 binds to two regions (third and fifth introns) of the locus of LAMP3, a product of which associates with SFTPB (Fig. 4a). Notably, these two NKX2-1-binding sites are not in the ATAC-accessible regions (Fig. 4a), again suggesting that NKX2-1 is able to bind to genomic regions that are considered to be inaccessible by ATAC-seq (Fig. 3a and Supplementary Data 1). In order to understand which NKX2-1-binding sites in the two regions are important for the expression of LAMP3 induced by NKX2-1, we took the same CRISPRi approach as described above except that sgRNAs targeting the two regions of LAMP3 were used (Supplementary Fig. 3). Notably, both sgRNAs targeting their respective regions suppressed the expression of LAMP3 more than twofold (P < 0.05) (Fig. 4b). Unexpectedly, the sgRNA targeting the third intronic region also suppressed the expression of SFTPB and MYBPH more than twofold (P < 0.05) (Fig. 4b) though they are located at different chromosomes from LAMP3. Of note, LAMP3, SFTPB, and MYBPH are located at chromosomes 3, 2, and 1, respectively. The CRISPOR off-target search38 does not indicate that the sgRNA sequence matches the genomic DNA sequences of the SFTPB and MYBPH loci (Supplementary Data 4), suggesting that the third intronic region may regulate the expression of LAMP3 in cis and those of SFTPB and MYBPH in trans. Since the sgRNA targeting the fifth intronic region suppressed the expression of LAMP3 at a similar level to the one targeting the third intronic region but did not suppress the expression of the other genes, the suppressed expression of SFTPB and MYBPH may not occur through the reduced expression of LAMP3 mRNA and/or protein. As expected, these sgRNAs did not affect the expression of LAMP3 or other NKX2-1 downstream target genes in H441 cells (Fig. 4b) since the siRNA-mediated loss-of-function experiment indicates that NKX2-1 does not largely regulate the expression of LAMP3 in H441 cells (Supplementary Fig. 7), indicating a cell type-specific gene regulation by NKX2-1.
a Shown are ATAC-seq and ChIP-seq data indicating that NKX2-1 binds to the third (#1) and fifth (#2) intronic regions of the human LAMP3 locus. Significant peaks are described as in Fig. 2a. b Shown is gene expression of NKX2-1 downstream target genes affected by CRISPRi as described in Fig. 2b. The sgRNA targeting NKX2-1-binding sites located at the third (#1) and fifth (#2) regions significantly suppressed the expression of LAMP3 induced by NKX2-1 in A549 cells but not in H441 cells. The sgRNA targeting NKX2-1-binding sites located at the third (#1) intronic region also significantly suppressed the expression of SFTPB and MYBPH induced by NKX2-1. Results are expressed as the mean ± SD of the triplicates for each group. Only P < 0.05 and more than twofold suppression are considered significant CRISPRi-mediated suppression (highlighted in red). Unt. untransfected, Cont. non-targeted control sgRNA.
The intergenic region between SFTPA1 and SFTPA2 carrying two NKX2-1-binding sites functions as a gene-regulatory region in cis
SFTPA1 and its paralog SFTPA2, products of which are critical for innate defense against respiratory bacteria, viruses, and fungi, are located at chromosome 10q22.3. The ChIP-seq data27,30 indicate that NKX2-1 binds to a proximal upstream region of SFTPA1 and a distal upstream region of SFTPA1 and SFTPA2, which are located at ATAC-inaccessible regions (Fig. 5a and Supplementary Data 1). As described above, we performed the CRISPRi approach (Supplementary Fig. 3 and Supplementary Data 5) and assessed the gene-regulatory functions of these two regions in A549 cells. Notably, both sgRNAs targeting proximal and distal regions significantly suppressed the expression of SFTPA1; however, only the sgRNA targeting the distal region suppressed the expression of SFTPA2 more than twofold (P < 0.05) (Fig. 5b). These sgRNAs did not affect the expression of other genes, including SFTPB, LAMP3, MYBPH, LMO3, and CD274/PD-L1, suggesting the two regions do not function in trans at least for these genes. Consistent with the results in A549 cells, the same pattern of gene regulation was observed in H441 cells (Fig. 5b).
a Shown are ATAC-seq and ChIP-seq data indicating that NKX2-1 binds to an intergenic (#1 and #2) region of human SFTPA1 and SFTPA2. Significant peaks are described as in Fig. 2a. MBL3P and SFTPA3P are pseudogenes. b Shown is gene expression of NKX2-1 downstream target genes affected by CRISPRi as described in Fig. 2b. The sgRNA targeting an NKX2-1-binding site located at the proximal upstream (#1) region of SFTPA1 significantly suppressed the NKX2-1-induced expression of SFTPA1 but not SFTPA2, while the one targeting an NKX2-1-binding site located at the distal upstream (#2) region of SFTPA1 and SFTPA2 significantly suppressed the NKX2-1-induced expression of both SFTPA1 and SFTPA2. Results are expressed as the mean ± SD of the triplicates for each group. Only P < 0.05 and more than twofold suppression are considered significant CRISPRi-mediated suppression (highlighted in red). Unt. untransfected, Cont. non-targeted control sgRNA.
Since NKX2-1 binds to a number of intragenic regions, we employed a CRISPRi approach that influences gene-regulatory activity without deleting a part of the genes. However, since the distal upstream region of SFTPA1 and SFTPA2, which is bound by NKX2-1, is in an intergenic region (opposed to intragenic) that is ~20 kb away from both SFTPA1 and SFTPA2, we also employed the CRISPR/Cas9-mediated approach to delete the distal upstream region and assessed the expression of SFTPA1, SFTPA2, and other NKX2-1 downstream target genes. Consistent with the results obtained by the CRISPRi approach, deletion of the distal upstream region by CRISPR/Cas9 most significantly repressed the expression of both SFTPA1 and SFTPA2 in A549 cells (Supplementary Fig. 8).
Four regions at the MYBPH locus bound by NKX2-1 are all critical for the expression of MYBPH induced by NKX2-1
The ChIP-seq data27,30 indicate that NKX2-1 binds to five regions (far distal, distal and proximal upstream, and third and sixth introns) of the locus of MYBPH, a product of which is involved in cell motility and metastasis of lung adenocarcinoma (Fig. 6a). Notably, two NKX2-1-binding sites located at the distal upstream regions (but not the three other regions) are in ATAC-accessible regions (Fig. 6a and Supplementary Data 1). As described above, the CRISPRi approach was conducted to assess the importance of these five regions (Supplementary Fig. 3). Notably, all sgRNAs targeting the five regions significantly suppressed the expression of MYBPH more than twofold (P < 0.05) in A549 cells (Fig. 6b), indicating that all five regions bound by NKX2-1 are essential for the expression of MYBPH that is induced by NKX2-1. The suppression of MYBPH was also confirmed at the protein level (Supplementary Fig. 4). Of note, the sgRNA targeting a region in the sixth intron significantly suppressed the expression of SFTPA1 more than twofold (P < 0.05) (Fig. 6b) though the sgRNA sequence does not match the genomic DNA sequence at the SFTPA1 locus according to the CRISPOR off-target search38 (Supplementary Data 6), suggesting that the sixth intron may regulate the expression of SFTPA1 in trans. However, the sixth intron did not regulate the expression of either MYBPH or SFTPA1 in H441 cells (Fig. 6b), suggesting a cell type-specific gene regulation by NKX2-1.
a Shown are ATAC-seq and ChIP-seq data indicating that NKX2-1 binds to far distal (#1), distal (#2) and proximal (#3) upstream regions and the third (#4), and sixth (#5) intronic regions of the human MYBPH locus. Significant peaks are described as in Fig. 2a. b Shown is the gene expression of NKX2-1 downstream target genes affected by CRISPRi as described in Fig. 2b. The sgRNA targeting NKX2-1-binding sites located at three upstream (#1, #2, and #3) and one intronic (#4) regions significantly suppressed the expression of MYBPH. The sgRNA targeting NKX2-1-binding sites located at the sixth intronic (#5) region also significantly suppressed the expression of SFTPA1 induced by NKX2-1 in A549 cells. Results are expressed as the mean ± SD of the triplicates for each group. Only P < 0.05 and more than twofold suppression are considered significant CRISPRi-mediated suppression (highlighted in red). Unt. untransfected, Cont. non-targeted control sgRNA.
Three regions at the second intron of the LMO3 locus bound by NKX2-1 are all critical for the expression of LMO3 induced by NKX2-1
The ChIP-seq data27,30 indicate that NKX2-1 binds to three distinct regions (ATAC inaccessible) at the second intron of LMO3, a product of which is involved in the survival of lung adenocarcinoma cells (Fig. 7a). As described above, CRISPRi was performed to assess whether these three distinct intronic regions influence the expression of LMO3 (Supplementary Fig. 3 and Supplementary Data 7). Notably, all sgRNAs targeting the three regions significantly suppressed the expression of LMO3 more than 2-fold (P < 0.05) in both A549 and H441 cells (Fig. 7b), indicating that all three regions at the second intron bound by NKX2-1 function as gene-regulatory regions for the expression of LMO3 that is induced by NKX2-1, regardless of cell type.
a Shown are ATAC-seq and ChIP-seq data indicating that NKX2-1 binds to three distinctive (#1, #2, and #3) regions of the second intron of human LMO3. Significant peaks are described as in Fig. 2a. b Shown is gene expression of NKX2-1 downstream target genes affected by CRISPRi as described in Fig. 2b. The sgRNAs targeting an NKX2-1-binding site located at the three distinctive (#1, #2, and #3) regions of the second intron of human LMO3 significantly suppressed the expression of LMO3, respectively. Results are expressed as the mean ± SD of the triplicates for each group. Only P < 0.05 and more than twofold suppression are considered significant CRISPRi-mediated suppression (highlighted in red). Unt. untransfected, Cont. non-targeted control sgRNA.
An upstream region but not the first intronic region bound by NKX2-1 is critical for the expression of CD274/PD-L1 induced by NKX2-1
The ChIP-seq data27,30 indicate that NKX2-1 binds to two regions (upstream and the first intron; ATAC inaccessible) of the locus of CD274/PD-L1, a product of which inactivates the anti-tumorigenic function of T cells (Fig. 8a). As described above, the CRISPRi approach was used to assess the gene-regulatory function of these two regions (Supplementary Figs. 3, 4 and Supplementary Data 8). Notably, an sgRNA targeting the upstream region significantly suppressed the expression of CD274/PD-L1 more than twofold at the mRNA level (P < 0.05) and also at the protein level in A549 cells. In contrast, the sgRNA targeting the first intron did not suppress the mRNA expression of CD274/PD-L1 (Fig. 8b), indicating that the upstream region bound by NKX2-1 functions as a gene-regulatory region for the expression of CD274/PD-L1 and is indispensable while the intronic region in A549 cells is not. Although the siRNA-mediated loss-of-function study indicates that NKX2-1 does not regulate the expression of CD274/PD-L1 in H441 cells, the sgRNA targeting the first intron of the CD274/PD-L1 locus (an NKX2-1-binding site) significantly suppressed the expression of CD274/PD-L1 more than twofold (P < 0.05) in H441 cells (Fig. 8b), suggesting that the first intronic region of CD274/PD-L1 acts a critical gene-regulatory region in a cell-type-dependent fashion.
a Shown are ATAC-seq and ChIP-seq data indicating that NKX2-1 binds to the upstream (#1) and first intronic (#2) regions of human CD274/PD-L1. Significant ChIP-seq peaks are described as in Fig. 2a. b Shown is gene expression of NKX2-1 downstream target genes affected by CRISPRi as described in Fig. 2b. The sgRNA targeting an NKX2-1-binding site located at the upstream (#1) region significantly suppressed the expression of CD274/PD-L1 induced by NKX2-1 in A549 cells while the sgRNA targeting an NKX2-1-binding site located at the first intronic (#2) region significantly suppressed the expression of CD274/PD-L1 in H441 cells. Results are expressed as the mean ± SD of the triplicates for each group. Only P < 0.05 and more than twofold suppression are considered significant CRISPRi-mediated suppression (highlighted in red). Unt. untransfected, Cont. non-targeted control sgRNA.
Discussion
In the past decade, ChIP-seq has identified far more transcription factor-binding sites than gel-shift and reporter assays, and further has revealed that transcription factors bind not only proximal promoter regions but also non-proximal regions, including intronic regions and distal intergenic regions. However, none of these techniques can determine the functional roles of such regions. It is essential to determine whether such regions indeed influence the mRNA expression of genes located nearby and/or distally. The recent discovery of the CRISPR/Cas9 technology has resolved this issue by providing the means to delete or repress specific regions in order to determine their functional roles. Here, using the CRISPR/Cas9 technology, we determined the function of critical gene-regulatory regions bound by NKX2-1, a lineage-specific transcription factor that is essential for lung function and that has been most extensively studied in the lung field.
SFTPB is the first lung epithelial gene that was reported as an NKX2-1 downstream target gene. Using gel-shift and reporter assays, NKX2-1 was shown to bind and activate a proximal promoter region located at the first intron of SFTPB4,5, the data of which are consistent with our ChIP-seq and CRISPRi results (Fig. 2). Of note, this first intronic region contains a SNP (rs3024791) that is associated with chronic obstructive pulmonary disease (COPD)39, suggesting that this region is a critical region regulated by NKX2-1 that influences disease severity by affecting the expression of SFTPB. In addition to this region located at the first intron, using the ChIP-seq with CRISPRi approach, we identified another functionally important NKX2-1-binding site in the upstream region (Fig. 2). Likewise, SNPs located in this region may affect NKX2-1-binding and in turn influence the expression of SFTPB, thereby modifying the lung disease severity. Using the ChIP-seq assay, we previously identified three additional NKX2-1-binding sites in the ninth intronic, proximal, and distal downstream regions; however, the CRISPRi analysis in the present study indicated that two regions were cell-type-specific regulatory regions and one region was functionally redundant for the expression of SFTPB (Fig. 2), suggesting that all of the NKX2-1-binding sites identified by ChIP-seq are not always functionally critical in every cell type.
LAMP3 is one of the lung epithelial genes highly induced by NKX2-16,27. However, the molecular mechanism by which NKX2-1 regulates the expression of LAMP3 has not been previously determined. Our ChIP-seq data indicate that NKX2-1 does not bind to a conventional binding region such as upstream or the first intronic region; instead, it binds to the third and fifth intronic regions that are more than ~10 kb away from the transcription start site (TSS). Furthermore, the CRISPRi data indicate that both regions are critically important for the expression of LAMP3 induced by NKX2-1 (Fig. 4), indicating that ChIP-seq with CRISPRi is superior in identifying the gene-regulatory functions of these far distal regions compared to the conventional approach using gel-shift and reporter assays since they are normally limited to targeting regions within 5 kb from the TSS. Importantly, such a gene-regulatory mechanism was found only in one lung epithelial cell line (A549 cells) but not in the other lung epithelial cell line (H441 cells), indicating a cell-type-specific lung gene regulation.
The transcriptional regulation of SFTPA/Sftpa by NKX2-1 was initially studied using gel-shift and reporter assays to analyze mouse and baboon proximal promoter regions40,41. Later, human SFTPA1 and SFTPA2 gene regulation by NKX2-1 was reported by our group and others, focusing on their proximal promoter regions, using gel-shift and reporter assays7,8,9. Our ChIP-seq data indicate that NKX2-1 bound to the same proximal promoter region located at the upstream region of SFTPA1 (Fig. 5a) and the CRISPRi data indicate that the proximal promoter region is indispensable for the expression of SFTPA1 (Fig. 5b), which is consistent with our previous report using gel-shift and reporter assays8 and also suggests that this proximal region is conserved as an important gene-regulatory region among species. Unexpectedly, our ChIP-seq data also indicate that NKX2-1 binds to a distal upstream region of SFTPA1 and SFTPA2 (intergenic region between SFTPA1 and SFTPA2), which is ~20 kb away from both genes (Fig. 5a). Importantly, this distal region was critical for the expression of both SFTPA1 and SFTPA2, while the proximal region close to SFTPA1 was critical only for the expression of SFTPA1 but not SFTPA2 (Fig. 5b), which is consistent with our previous finding36 that some gene-regulatory regions function to control the expression of a single gene while the others function to control that of multiple genes. Notably, these gene-regulatory mechanisms of SFTPA1 and SFTPA2 by NKX2-1 were seen in both A549 and H441 lung epithelial cell lines, suggesting that some lung gene-regulatory regions are highly conserved regardless of cell type.
Since the functional relevance of NKX2-1 in lung tumorigenesis other than as a biomarker was reported19,20,21,22, downstream target genes regulated by NKX2-1 that influence lung tumorigenesis have been actively sought (e.g., MYBPH, LMO3, and CD274/PD-L1). MYBPH is induced by NKX2-1 and acts as a tumor suppressor24. The molecular mechanism by which NKX2-1 regulates the expression of MYBPH was previously studied using reporter assay and ChIP-qPCR focusing on its promoter region located at the proximal upstream region, which indicates that NKX2-1 activates the promoter region24. This result is consistent with our ChIP-seq and CRISRPi data targeting the same region (Fig. 6). Importantly, our ChIP-seq and CRISRPi data indicate that not only the proximal upstream region but also three other regions located at distal upstream and intronic regions that are bound by NKX2-1 are functionally critical for the expression of MYBPH (Fig. 6). The sixth intronic region that is bound by NKX2-1 was critical in A549 cells but not in H441 cells. These results suggest again that an unbiased approach using ChIP-seq and CRISRPi in multiple cell types, which can search beyond the proximal regions near the TSS, helps to better understand the entire molecular mechanism by which transcription factors, including NKX2-1, regulate the expression of genes.
In contrast to MYBPH, which acts as a tumor suppressor, LMO3 (an NKX2-1 downstream target gene) promotes the survival of lung tumor cells28, indicating a context-dependent function of NKX2-1 in lung tumorigenesis. The molecular mechanism by which NKX2-1 regulates the expression of LMO3 was previously studied using gel-shift and ChIP-seq assays, which indicated that NKX2-1 bound to a region located at the first intron of LMO328, which is not consistent with our ChIP-seq data, indicating that NKX2-1 binds to three regions at the second intron of LMO3 (Fig. 7a). This discrepancy might be due to the cell line that we used for ChIP-seq, which carries different oncogenes (H3122 cells carrying an ALK fusion [28] vs. A549 cells carrying KRASG12S [27]). Our CRISPRi data indicate that all three regions located at the second intron are essential for the expression of LMO3 in both A549 and H441 cells (Fig. 7b; of note, H441 cells also carry a KRAS mutation, KRASG12V, but not an ALK fusion), suggesting that the same CRISPRi approach targeting the region located at the first intron in H3122 cells would address the functional relevance of the region.
CD274/PD-L1 that is induced by NKX2-1 has been a hallmark of cancer immunotherapy along with its receptor PD-1 (also known as PDCD1), which can be targeted by therapeutic antibodies that activate anti-tumorigenic T cells by blocking the interaction between CD274/PD-L1 and PDCD1/PD-131. The gene-regulatory mechanism of CD274/PD-L1 has been described previously in reviews42,43. Our ChIP-seq data indicate that NKX2-1 binds to two regions of the CD274/PD-L1 locus, ~ 2.7 kb upstream region and +4.8 kb first intron (Fig. 8a). To our knowledge, the upstream region is bound by only NKX2-1 while the intronic region is bound by not only NKX2-1 but also the AP-1 transcription factor44. Our CRISPRi data indicate that the upstream region is critical for the expression of CD274/PD-L1 induced by NKX2-1 in A549 cells while the intronic region is critical for the expression of CD274/PD-L1 in H441 cells (Fig. 8b). Since the siRNA-mediated loss-of-function analysis indicates that NKX2-1 is redundant for the expression of CD274/PD-L1, the intronic region bound by AP-1 might be critical for the expression of CD274/PD-L1 in H441 cells.
In the present study, taking advantage of ENCODE database that harbors ATAC-seq datasets from A549 cells, we assessed whether NKX2-1 binds to chromatin-accessible genomic regions that are defined as such by ATAC-seq. To our surprise, more than half of the NKX2-1-binding sites were located at genomic regions that are considered to be chromatin inaccessible by ATAC-seq (Fig. 3a). To our knowledge, NKX2-1 is not considered to be a pioneer transcription factor that is able to bind to chromatin-inaccessible genomic regions45. Future studies are required to understand the mechanism by which NKX2-1 accesses such chromatin-inaccessible genomic regions or to develop a means to assess chromatin accessibility other than ATAC-seq.
Here, we investigated the roles of NKX2-1-binding sites in the expression of NKX2-1-regulated genes in human lung epithelial cells in vitro. Among such NKX2-1-regulated genes, the in vivo roles of SFTPB and SFTPA1/SFTPA2 in lung pathophysiology have been particularly well studied using Sftpb- or Sftpa1-knockout mice3,10,11,12,13,14,15. Notably, a region bound by NKX2-1 in the first intron of human SFTPB is highly conserved with the one in the upstream region of mouse Sftpb (84% similarity). Likewise, a region bound by NKX2-1 in the proximal upstream region of SFTPA1 is highly conserved with the one in the upstream region of mouse Sftpa1 (74% similarity). Using CRISPR/Cas9-mediated deletion, Simeonov et al. have assessed the in vivo role of a region in the first intron of mouse Il2ra in blood cells, a region of which is highly conserved with a gene-regulatory enhancer region of human IL2RA46. In future studies, targeting such conserved gene-regulatory regions in mice using CRISPR/Cas9 and/or dCas9 will further determine the importance of NKX2-1-binding sites as gene-regulatory regions in lung pathophysiology in vivo.
Methods
Processing and visualization of ChIP-seq and ATAC-seq data
ChIP-seq of NKX2-1 in an A549 human lung epithelial (carcinoma) cell line, which we previously performed twice independently27,30 (GSE86959), was visualized using the UCSC Genome Browser47. Supplementary Figs. 9–14 describe both independent replicates with different gene isoforms. Peak files of three replicates of ENCODE ATAC-seq on A549 cell line (ENCODE accession#: ENCSR220ASC) were downloaded from GSE114202. ATAC peak genome coordinates were converted from GRCh38 to hg19 using the UCSC liftOver tool and visualized using the UCSC Genome Browser. Peak overlap was performed using HOMER mergePeaks function with “-d given” parameter48. Overlapped peaks among the three ATAC-seq replicates were considered as ATAC-seq peaks, and overlapped peaks between the two NKX2-1 ChIP-seq replicates were considered as NKX2-1 ChIP-seq peaks.
Processing and visualization of Hi-C data
Mapping quality thresholded chromatin interactions (.hic files) of four replicates of ENCODE Hi-C on an A549 cell line (ENCODE accession#: ENCSR662QKG) were downloaded from ENCODE web portal. Inter-chromosomal interactions with the SFTPB locus (including 10 kb upstream of the transcription start site and 10 kb downstream of the transcription end site) were extracted at the resolution of 50 kb using the “dump” function of Juicer tools (version 1.22)49. Inter-chromosomal interactions with at least two observed counts were visualized in a circular layout using BioCircos50.
Synthetic sgRNA
Synthetic sgRNAs targeting each locus (SFTPB, LAMP3, SFTPA1, SFTPA2, MYBPH, LMO3, and CD274/PD-L1; Supplementary Fig. 3 and Supplementary Data 3-8) were designed using CRISPOR37 and generated using the Invitrogen custom TrueGuide gRNA (sgRNA) ordering tool (ThermoFisher, Waltham, MA). Non-targeted gRNA (sgRNA) was used as a negative control (cat# A35526, ThermoFisher).
Cells
A549 lung carcinoma cells and H441 lung epithelial (papillary adenocarcinoma) cells (obtained from ATCC in Manassas, VA) that stably express dCas9-KRAB were created and maintained as ATCC instructed36. A549 cells stably expressing dCas9-KRAB were infected by lentivirus carrying either rat Nkx2-1 or backbone control (PGK-IRES-EGFP)51. A549 and H441 cells expressing dCas9-KRAB were transfected with synthetic sgRNAs (50 nM at the final concentration) or siRNAs targeting human NKX2-1 (Silencer Select siRNA, ID: s224731 for #1, ID: s14154 for #2) or human SFTPB (Silencer Select siRNA, ID: s12757 for #1, ID: s227103 for #2) (60 nM at the final concentration; cat# 4390843 for negative control [siNeg]) using Lipofectamine RNAiMAX Reagent (cat# 13778075, ThermoFisher).
Gene expression analyses
Gene expression analyses were performed using TaqMan gene expression analysis36 using probes (Hs00167036 for SFTPB, Hs01111316 for LAMP3, Hs00831305 for SFTPA1, Hs00359837 for SFTPA2, Hs00192226 for MYBPH, Hs00998696 for LMO3, Hs01125301 for CD274/PD-L1, Rn01512482 for rat Nkx2-1, Hs00968940 for human NKX2-1 and Hs02758991 for GAPDH as control from ThermoFisher). A probe for dCas9-KRAB was custom designed by ThermoFisher as described in ref. 36.
RNA-seq analysis
RNA-seq36 was performed, in which RNA was obtained from A549 cells constitutively expressing dCas9-KRAB that were transiently transfected with synthetic sgRNA targeting the first intron of SFTPB (#2) or non-targeted control gRNA. After quality assessment and pre-processing of RNA-seq reads36 were performed, gene expression was measured using htseq-count. Differential expression analysis for each comparison was performed using Bioconductor DESeq2 package. Differential expression with at least log2 fold change ≧1 or ≦−1 and padj<0.05 was considered significant.
CRISPR/Cas9-mediated deletion of an NKX2-1-binding site
An NKX2-1-binding site located at the intergenic region of SFTPA1 and SFTPA2 was deleted in A549 cells using CRISPR/Cas930, in which sgRNA target sequences described in Supplementary Fig. 8b were used. The deletion was confirmed by PCR amplification (Supplementary Fig. 8c) and Sanger sequencing by the DNA Sequencing and Genotyping Core at CCHMC using primers described in Supplementary Fig. 8b.
Immunoblotting (IB)
Immunoblot assays52 were performed using rabbit anti-NKX2-1 (TTF-1) antibody (1:5,000, cat# WRAB-1231; Seven Hills Bioreagents, Cincinnati, OH), rabbit anti-MYBPH (1:5,000, cat# HPA061383; MilliporeSigma, St. Louis, MO), rabbit anti-CD274 (PD-L1) antibody (1:5,000; clone E1L3N; cat# 13684, Cell Signaling Technology, Danvers, MA), rabbit anti-Cas9 (dCas9) antibody (1:5,000; EPR18991; cat# 189380, Abcam, Cambridge, UK), and rabbit anti-ACTA1 antibody (1:5,000; cat# A2066, MilliporeSigma).
Statistics and reproducibility
Realtime qPCR results were normalized against non-targeted sgRNA samples in independent biological triplicates. Results are expressed as the mean ± SD of the triplicates for each group. Undetermined Ct values due to low or no expression were set as Ct=40. Statistical relevance was determined using a two-sided Student’s t-test on transfected vs. control sgRNA transfected cells' relative expression with a minimum P value < 0.05. GraphPad Prism 8 was used for graphing and statistical analysis. Raw numbers for all replicates that generated the results are shown in Supplementary Data 9.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
The authors declare that all data supporting the findings of this study are available within the article and supplementary data, including Supplementary Figs and Supplementary Data. The RNA-seq raw data are available in the GEO repository (Gene Expression Omnibus, https://www.ncbi.nlm.nih.gov/geo/, accession number: GSE162186).
References
Kimura, S. et al. The T/ebp null mouse: thyroid-specific enhancer-binding protein is essential for the organogenesis of the thyroid, lung, ventral forebrain, and pituitary. Genes Dev. 10, 60–69 (1996). 1.
Maeda, Y., Davé, V. & Whitsett, J. A. Transcriptional control of lung morphogenesis. Physiol. Rev. 87, 219–244 (2007).
Clark, J. C. et al. Targeted disruption of the surfactant protein B gene disrupts surfactant homeostasis, causing respiratory failure in newborn mice. Proc. Natl Acad. Sci. USA 92, 7794–7798 (1995). 15.
Bohinski, R. J., Di Lauro, R. & Whitsett, J. A. The lung-specific surfactant protein B gene promoter is a target for thyroid transcription factor 1 and hepatocyte nuclear factor 3, indicating common factors for organ-specific gene expression along the foregut axis. Mol. Cell Biol. 14, 5671–5681 (1994).
Yan, C., Sever, Z. & Whitsett, J. A. Upstream enhancer activity in the human surfactant protein B gene is mediated by thyroid transcription factor 1. J. Biol. Chem. 270, 24852–24857 (1995). 20.
Kolla, V. et al. Thyroid transcription factor in differentiating type II cells: regulation, isoforms, and target genes. Am. J. Respir. Cell Mol. Biol. 36, 213–225 (2007).
Islam, K. N. & Mendelson, C. R. Glucocorticoid/glucocorticoid receptor inhibition of surfactant protein-A (SP-A) gene expression in lung type II cells is mediated by repressive changes in histone modification at the SP-A promoter. Mol. Endocrinol. 22, 585–596 (2008).
Fukazawa, T., Maeda, Y., Sladek, F. M. & Owen-Schaub, L. B. Development of a cancer-targeted tissue-specific promoter system. Cancer Res 64, 363–369 (2004). 1.
Liu, D., Yi, M., Smith, M. & Mendelson, C. R. TTF-1 response element is critical for temporal and spatial regulation and necessary for hormonal regulation of human surfactant protein-A2 promoter activity. Am. J. Physiol. Lung Cell Mol. Physiol. 295, L264–L271 (2008).
Korfhagen, T. R. et al. Altered surfactant function and structure in SP-A gene targeted mice. Proc. Natl Acad. Sci. USA 93, 9594–9599 (1996). 3.
LeVine, A. M. et al. Surfactant protein A-deficient mice are susceptible to group B streptococcal infection. J. Immunol. 158, 4336–4340 (1997). 1.
LeVine, A. M. et al. Surfactant protein-A-deficient mice are susceptible to Pseudomonas aeruginosa infection. Am. J. Respir. Cell Mol. Biol. 19, 700–708 (1998).
LeVine, A. M. et al. Surfactant protein-A enhances respiratory syncytial virus clearance in vivo. J. Clin. Invest. 103, 1015–1021 (1999).
Harrod, K. S., Trapnell, B. C., Otake, K., Korfhagen, T. R. & Whitsett, J. A. SP-A enhances viral clearance and inhibits inflammation after pulmonary adenoviral infection. Am. J. Physiol. 277, L580–L588 (1999).
Linke, M. J. et al. Immunosuppressed surfactant protein A-deficient mice have increased susceptibility to Pneumocystis carinii infection. J. Infect. Dis. 183, 943–952 (2001). 15.
Holzinger, A. et al. Monoclonal antibody to thyroid transcription factor-1: production, characterization, and usefulness in tumor diagnosis. Hybridoma 15, 49–53 (1996).
Kaufmann, O. & Dietel, M. Thyroid transcription factor-1 is the superior immunohistochemical marker for pulmonary adenocarcinomas and large cell carcinomas compared to surfactant proteins A and B. Histopathology 36, 8–16 (2000).
Compérat, E. et al. Variable sensitivity and specificity of TTF-1 antibodies in lung metastatic adenocarcinoma of colorectal origin. Mod. Pathol. 18, 1371–1376 (2005).
Tanaka, H. et al. Lineage-specific dependency of lung adenocarcinomas on the lung development regulator TTF-1. Cancer Res 67, 6007–6011 (2007). 1.
Kendall, J. et al. Oncogenic cooperation and coamplification of developmental transcription factor genes in lung cancer. Proc. Natl Acad. Sci. USA 104, 16663–16668 (2007). 16.
Weir, B. A. et al. Characterizing the cancer genome in lung adenocarcinoma. Nature 450, 893–898 (2007). 6.
Kwei, K. A. et al. Genomic profiling identifies TITF1 as a lineage-specific oncogene amplified in lung cancer. Oncogene 27, 3635–3640 (2008). 5.
Saito, R. A. et al. Thyroid transcription factor-1 inhibits transforming growth factor-beta-mediated epithelial-to-mesenchymal transition in lung adenocarcinoma cells. Cancer Res. 69, 2783–2791 (2009). 1.
Hosono, Y. et al. MYBPH, a transcriptional target of TTF-1, inhibits ROCK1, and reduces cell motility and metastasis. EMBO J. 31, 481–493 (2012). 18.
Winslow, M. M. et al. Suppression of lung adenocarcinoma progression by Nkx2-1. Nature 473, 101–104 (2011). 5.
Yamaguchi, T. et al. NKX2-1/TITF1/TTF-1-Induced ROR1 is required to sustain EGFR survival signaling in lung adenocarcinoma. Cancer Cell 21, 348–361 (2012). 20.
Maeda, Y. et al. Kras(G12D) and Nkx2-1 haploinsufficiency induce mucinous adenocarcinoma of the lung. J. Clin. Invest. 122, 4388–4400 (2012).
Watanabe, H. et al. Integrated cistromic and expression analysis of amplified NKX2-1 in lung adenocarcinoma identifies LMO3 as a functional transcriptional target. Genes Dev. 27, 197–210 (2013). 15.
Snyder, E. L. et al. Nkx2-1 represses a latent gastric differentiation program in lung adenocarcinoma. Mol. Cell. 50, 185–199 (2013). 25.
Guo, M. et al. Gene signature driving invasive mucinous adenocarcinoma of the lung. EMBO Mol. Med 9, 462–481 (2017).
Chen, L. & Han, X. Anti-PD-1/PD-L1 therapy of human cancer: past, present, and future. J. Clin. Invest. 125, 3384–3391 (2015).
Isogaya, K. et al. A Smad3 and TTF-1/NKX2-1 complex regulates Smad4-independent gene expression. Cell Res. 24, 994–1008 (2014).
Mansour, M. R. et al. Oncogene regulation. An oncogenic super-enhancer formed through somatic mutation of a noncoding intergenic element. Science 346, 1373–1377 (2014). 12.
Gilbert, L. A. et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 154, 442–451 (2013).
Thakore, P. I. et al. Highly specific epigenome editing by CRISPR-Cas9 repressors for silencing of distal regulatory elements. Nat. Methods 12, 1143–1149 (2015).
Stuart, W. D. et al. CRISPRi-mediated functional analysis of lung disease-associated loci at non-coding regions. NAR Genom. Bioinform. 2, lqaa036 (2020).
ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 (2012). 6.
Haeussler, M. et al. Evaluation of off-target and on-target scoring algorithms and integration into the guide RNA selection tool CRISPOR. Genome Biol. 17, 148 (2016). 5.
Foreman, M. G. et al. Polymorphic variation in surfactant protein B is associated with COPD exacerbations. Eur. Respir. J. 32, 938–944 (2008).
Bruno, M. D., Bohinski, R. J., Huelsman, K. M., Whitsett, J. A. & Korfhagen, T. R. Lung cell-specific expression of the murine surfactant protein A (SP-A) gene is mediated by interactions between the SP-A promoter and thyroid transcription factor-1. J. Biol. Chem. 270, 6531–6536 (1995). 24 Erratum in: J. Biol. Chem. 270, 16482 (1995).
Li, J., Gao, E. & Mendelson, C. R. Cyclic AMP-responsive expression of the surfactant protein-A gene is mediated by increased DNA binding and transcriptional activity of thyroid transcription factor-1. J. Biol. Chem. 273, 4592–4600 (1998). 20.
Dong, P., Xiong, Y., Yue, J., Hanley, S. J. B. & Watari, H. Tumor-intrinsic PD-L1 signaling in cancer initiation, development and treatment: beyond immune evasion. Front. Oncol. 8, 386 (2018). 19.
Zerdes, I., Matikas, A., Bergh, J., Rassidakis, G. Z. & Foukakis, T. Genetic, transcriptional and post-translational regulation of the programmed death protein ligand 1 in cancer: biology and clinical correlations. Oncogene 37, 4639–4661 (2018).
Green, M. R. et al. Constitutive AP-1 activity and EBV infection induce PD-L1 in Hodgkin lymphomas and posttransplant lymphoproliferative disorders: implications for targeted therapy. Clin. Cancer Res. 18, 1611–1618 (2012). 15.
Iwafuchi-Doi, M. & Zaret, K. S. Pioneer transcription factors in cell reprogramming. Genes Dev. 28, 2679–2692 (2014). 15.
Simeonov, D. R. et al. Discovery of stimulation-responsive immune enhancers with CRISPR activation. Nature 549, 111–115 (2017). 7.
Kent, W. J. et al. The human genome browser at UCSC. Genome Res 12, 996–1006 (2002).
Heinz, S. et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010).
Durand, N. C. et al. Juicer provides a one-click system for analyzing loop-resolution Hi-C experiments. Cell Syst. 3, 95–98 (2016).
Cui, Y. et al. BioCircos.js: an interactive Circos JavaScript library for biological data visualization on web applications. Bioinformatics 32, 1740–1742 (2016). 1.
Maeda, Y. et al. Airway epithelial transcription factor NK2 homeobox 1 inhibits mucous cell metaplasia and Th2 inflammation. Am. J. Respir. Crit. Care Med 184, 421–429 (2011). 15.
Maeda, Y. et al. PARP-2 interacts with TTF-1 and regulates expression of surfactant protein-B. J. Biol. Chem. 281, 9600–9606 (2006). 7.
Acknowledgements
We thank J. Whitsett, T. Fukazawa, and M. Durbin for materials and discussion. This work was supported by the NIH grants (U01HL134745, R01CA240317, LRRC Young Investigator Pilot Project Grant), Cincinnati Children’s Hospital Medical Center (Trustee Award Grant, CF-RDP Pilot & Feasibility Grant, GAP funding), and the Rotary Foundation Global Grant.
Author information
Authors and Affiliations
Contributions
Y.M. conceived the original idea and designed the study. W.D.S., I.M.F-B., K.T., and Y.M. conducted the experiments. M.G. conducted the bioinformatical analysis. All authors were involved in interpreting the findings, revising the drafts, and approving the final version.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Stuart, W.D., Fink-Baldauf, I.M., Tomoshige, K. et al. CRISPRi-mediated functional analysis of NKX2-1-binding sites in the lung. Commun Biol 4, 568 (2021). https://doi.org/10.1038/s42003-021-02083-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s42003-021-02083-4
- Springer Nature Limited