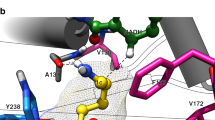
Abstract
The asymmetric reductive amination of ketones enables the one-step synthesis of chiral amines from readily available starting materials. Here we report the discovery of a family of native NAD(P)H-dependent amine dehydrogenases (nat-AmDHs) competent for the asymmetric reductive amination of aliphatic and alicyclic ketones, adding significantly to the biocatalytic toolbox available for chiral amine synthesis. Studies of ketone and amine substrate specificity and kinetics reveal a strong preference for aliphatic ketones and aldehydes, with activities of up to 614.5 mU mg−1 for cyclohexanone with ammonia, and 851.3 mU mg−1 for isobutyraldehyde with methylamine as the amine donor. Crystal structures of three nat-AmDHs (AmDH4, MsmeAmDH and CfusAmDH) reveal the active site determinants of substrate and cofactor specificity and enable the rational engineering of AmDH4 for the generated activity towards pentan-2-one. Analysis of the three-dimensional catalytic site distribution among bacterial biodiversity revealed a superfamily of divergent proteins with representative specificities ranging from amino acid substrates to hydrophobic ketones.





Similar content being viewed by others
Data availability
Crystallographic data that support the findings of this study have been deposited in the Protein Data Bank (PDB) under accession codes 6G1H, 6G1M, 6IAU and 6IAQ. All the other data supporting the findings of this study are available within the paper and its Supplementary Information and Supplementary Data or from the corresponding authors upon reasonable request.
References
Bornscheuer, U. T. Biocatalysis: successfully crossing boundaries. Angew. Chem. Int. Ed. 55, 4372–4373 (2016).
Ghislieri, D. & Turner, N. J. Biocatalytic approaches to the synthesis of enantiomerically pure chiral amines. Top. Catal. 57, 284–300 (2014).
Sharma, M., Mangas-Sanchez, J. & Grogan, G. NAD(P)H-dependent dehydrogenases for the asymmetric reductive amination of ketones: structure, mechanism, evolution and application. Adv. Synth. Catal. 359, 2011–2025 (2017).
Grogan, G. Synthesis of chiral amines using redox biocatalysis. Curr. Opin. Chem. Biol. 43, 15–22 (2017).
Vidal, L. S., Kelly, C. L., Mordaka, P. M. & Heap, J. T. Review of NAD(P)H-dependent oxidoreductases: properties, engineering and application. Biochim. Biophys. Acta Protiens Proteomics 1866, 327–347 (2017).
Höhne, M. & Bornscheuer, U. T. Biocatalytic routes to optically active amines. ChemCatChem 1, 42–51 (2009).
Patil, M. D., Grogan, G., Bommarius, A. S. & Yun, H. Oxidoreductase-catalyzed synthesis of chiral amines. ACS Catal. 8, 10985–11015 (2018).
Abrahamson, M. J. et al. Development of an amine dehydrogenase for synthesis of chiral amines. Angew. Chem. Int. Ed. 51, 3969–3972 (2012).
Abrahamson, M. J., Wong, J. W. & Bommarius, A. S. The evolution of an amine dehydrogenase biocatalyst for the asymmetric production of chiral amines. Adv. Synth. Catal. 355, 1780–1786 (2013).
Bommarius, A. S., Abrahamson, M. J. & Bommarius, B. Engineered amine dehydrogenases and methods of use thereof. US patent 8,835,136, B2 (2014).
Au, S. K., Bommarius, B. R. & Bommarius, A. S. Biphasic reaction system allows for conversion of hydrophobic substrates by amine dehydrogenases. ACS Catal. 4, 4021–4026 (2014).
Pushpanath, A. et al. Understanding and overcoming the limitations of Bacillus badius and Caldalkalibacillus thermarum amine dehydrogenases for biocatalytic reductive amination. ACS Catal. 7, 3204–3209 (2017).
Ye, L. J. et al. Engineering of amine dehydrogenase for asymmetric reductive amination of ketone by evolving Rhodococcus phenylalanine dehydrogenase. ACS Catal. 5, 1119–1122 (2015).
Lowe, J., Ingram, A. A. & Groger, H. Enantioselective synthesis of amines via reductive amination with a dehydrogenase mutant from Exigobacterium sibiricum: substrate scope, co-solvent tolerance and biocatalyst immobilization. Bioorg. Med. Chem. 26, 1387–1392 (2017).
Chen, F. et al. Reshaping the active pocket of amine dehydrogenases for asymmetric synthesis of bulky aliphatic amines. ACS Catal. 8, 2622–2628 (2018).
Wetzl, D. et al. Asymmetric reductive amination of ketones catalyzed by imine reductases. ChemCatChem 8, 2023–2026 (2016).
Agard, N. J. et al. Engineered imine reductases and methods for the reductive amination of ketone and amine compounds. US patent 2015132807 A1 (2015).
Itoh, N., Yachi, C. & Kudome, T. Determining a novel NAD+-dependent amine dehydrogenase with a broad substrate range from Streptomyces virginiae IFO 12827: purification and characterization. J. Mol. Catal. B 10, 281–290 (2000).
Wang, S. & Fang, B. Method for preparing chiral amine through asymmetric reduction under catalysis of marine strain OTI. Chinese patent 103224963B (2013).
Aleku, G. A. et al. A reductive aminase from Aspergillus oryzae. Nat. Chem. 9, 961–969 (2017).
France, S. P. et al. Identification of novel bacterial members of the imine reductase enzyme family that perform reductive amination. ChemCatChem 10, 1–6 (2018).
Fonknechten, N. et al. A conserved gene cluster rules anaerobic oxidative degradation of l-ornithine. J. Bacteriol. 191, 3162–3167 (2009).
Mayol, O. et al. Asymmetric reductive amination by a wild-type amine dehydrogenase from the thermophilic bacteria Petrotoga mobilis. Catal. Sci. Technol. 6, 7421–7428 (2016).
Zaparucha, A., de Berardinis, V. & Vaxelaire-Vergne, C. in Modern Biocatalysis: Advances Towards Synthetic Biological Systems Ch. 1 (RSC, Cambridge, 2018).
Knaus, T., Bohmer, W. & Mutti, F. G. Amine dehydrogenases: efficient biocatalysts for the reductive amination of carbonyl compounds. Green Chem. 19, 453–463 (2017).
Godoy-Alcántar, C., Yatsimirsky, A. K. & Lehn, J. M. Structure–stability correlations for imine formation in aqueous solution. J. Phys. Org. Chem. 18, 979–985 (2005).
Scheller, P. N. et al. Imine reductase-catalyzed intermolecular reductive amination of aldehydes and ketones. ChemCatChem 7, 3239–3242 (2015).
Reddy, S. G., Sacchettini, J. C. & Blanchard, J. S. Expression, purification, and characterization of Escherichia coli dihydrodipicolinate reductase. Biochemistry 34, 3492–3501 (1995).
Cirilli, M. et al. The three-dimensional structure of the ternary complex of Corynebacterium glutamicum diaminopimelate dehydrogenase-NADPH-l-2-amino-6-methylene-pimelate. Protein Sci. 9, 2034–2037 (2000).
Liu, W. et al. Structural and mutational studies on the unusual substrate specificity of meso-diaminopimelate dehydrogenase from Symbiobacterium thermophilum. Chembiochem 15, 217–222 (2014).
Vanhooke, J. L. et al. Phenylalanine dehydrogenase from Rhodococcus sp. M4: high-resolution X-ray analyses of inhibitory ternary complexes reveal key features in the oxidative deamination mechanism. Biochemistry 38, 2326–2339 (1999).
Sharma, M. et al. A mechanism for reductive amination catalyzed by fungal reductive aminases (RedAms). ACS Catal. 8, 11534–11541 (2018).
Lichman, B. R. et al. Structural evidence for the dopamine-first mechanism of norcoclaurine synthase. Biochemistry 56, 5274–5277 (2017).
Bastard, K. et al. Revealing the hidden functional diversity of an enzyme family. Nat. Chem. Biol. 10, 42–49 (2014).
Sigrist, C. J. et al. New and continuing developments at PROSITE. Nucleic Acids Res. 41, D344–D347 (2013).
Cosgrove, S. C. et al. Imine reductases, reductive aminases, and amine oxidases for the synthesis of chiral amines: discovery, characterization, and synthetic applications. Methods Enzymol. 608, 131–149 (2018).
Furnham, N. et al. Large-scale analysis exploring evolution of catalytic machineries and mechanisms in enzyme superfamilies. J. Mol. Biol. 428, 253–267 (2016).
Kabsch, W. XDS. Acta Cryst. D Biol. Cryst. 66, 125–132 (2010).
Winter, G. xia2: an expert system for macromolecular crystallography data reduction. J. Appl. Crystallogr. 43, 186–190 (2009).
Vagin, A. & Teplyakov, A. Molrep: an automated program for molecular replacement. J. Appl. Crystallogr. 30, 1022–1025 (1997).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. Sec. D: Biol. Crystallogr. 60, 2126–2132 (2004).
Murshudov, G. N., Vagin, A. & Dodson, E. J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D53, 240–255 (1997).
Nestl, B. M. et al. Structural and functional insights into asymmetric enzymatic dehydration of alkenols. Nat. Chem. Biol. 13, 275–281 (2017).
de Melo-Minardi, R. C., Bastard, K. & Artiguenave, F. Identification of subfamily-specific sites based on active sites modeling and clustering. Bioinformatics 26, 3075–3082 (2010).
Le Guilloux, V., Schmidtke, P. & Tuffery, P. Fpocket: an open source platform for ligand pocket detection. BMC Bioinformatics 10, 168 (2009).
Tomita, H., Katsuyama, Y., Minami, H. & Ohnishi, Y. Identification and characterization of a bacterial cytochrome P450 monooxygenase catalyzing the 3-nitration of tyrosine in rufomycin biosynthesis. J. Biol. Chem. 292, 15859–15869 (2017).
Acknowledgements
The authors thank M. Salanoubat for supporting the project, N. Fonknechten for fruitful discussions and assistance with genomic context analysis, C. Pelle and P. Sirvain for large-scale purification and analytical gel filtration of the described enzymes, O. Maciejak (University Val d’Essonne) for NMR assistance, the Region Ile de France for financial support of the 600 MHz spectrometer, S. Hart and J Agirre for assistance with X-ray data collection and data analysis respectively, and the Diamond Light Source for access to beamlines I04 and I04-1 under proposal no. mx-9948. This work was supported by Commissariat à l’énergie atomique et aux énergies alternatives (CEA), the CNRS and the University of Evry Val d’Essonne. The authors thank GlaxoSmithKline for the award of a part studentship to A.F., the Brazilian Government for a fellowship to L.B. under the Coordination for the Improvement of Higher Education Personnel (CAPES) scheme and the COST Action CM1303 ‘System Biocatalysis’ for STSM of O.M in G.G.’s laboratory.
Author information
Authors and Affiliations
Contributions
C.V.V. conceived the project and directed it with G.G. C.V.V., G.G., O.M., A.Z. and V.d.B. supervised the project. V.d.B. and J.-L.P. performed the candidate enzyme selection. A.D., V.P. carried out the gene cloning, the protein expression and purification on a small scale and the enzymatic screening with input from A.M. and J.-L.P.. O.M., V.P. and C.V.V. conducted the specific activity measurements. G.G., J.P.T., A.F. and L.B. conducted all the structural resolution. K.B. conceived and conducted the structural bioinformatics analysis with input from O.M., C.V.V. and G.G. O.M. carried out the biochemical experiments under the supervision of A.P. O.M. and C.V.V. performed the analytical and semi-preparative scale reactions. C.V.V., G.G., K.B. and O.M. wrote the manuscript with input from A.P., A.Z. and V.d.B.
Corresponding authors
Ethics declarations
Competing interest
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figures 1–32, Supplementary Tables 1–9, Supplementary Methods, Supplementary Note 1, Supplementary Discussion, Supplementary References
Supplementary Data 1
model of AmDH4 (closed form) in complex with NAD+, and product 2,4-DAP
Supplementary Data 2
model of AmDH4 (closed form) in complex with NAD+, ammonia and substrate (2R)−2A4OP
Supplementary Data 3
model of AmDH4 variant N135V/N163V/R161M/H264L in complex with NAD+, ammonia and pentan-2-one
Supplementary Data 4
model of CfusAmDH in complex with NADP+, ammonia and cyclohexanone
Supplementary Data 5
model of MsmeAmDH in complex with NADP+, ammonia and cyclohexanone
Supplementary Data 6
list of the 5313 proteins (Uniprot ID) used in the Sequence Similarity Network
Supplementary Data 7
multiple alignment on the whole sequences of proteins from G3 and G4 groups
Rights and permissions
About this article
Cite this article
Mayol, O., Bastard, K., Beloti, L. et al. A family of native amine dehydrogenases for the asymmetric reductive amination of ketones. Nat Catal 2, 324–333 (2019). https://doi.org/10.1038/s41929-019-0249-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41929-019-0249-z
- Springer Nature Limited
This article is cited by
-
Modification of the substrate specificity of leucine dehydrogenase by site-directed mutagenesis based on biocomputing strategies
Systems Microbiology and Biomanufacturing (2023)
-
Redox cascade reaction for kinetic resolution of racemic α-methylbenzylamine and biosynthesis of α-phenylethanol
Applied Microbiology and Biotechnology (2023)
-
Multifunctional biocatalyst for conjugate reduction and reductive amination
Nature (2022)
-
The Reductive Amination of Carbonyl Compounds Using Native Amine Dehydrogenase from Laribacter hongkongensis
Biotechnology and Bioprocess Engineering (2021)
-
The combination of asymmetric hydrogenation of olefins and direct reductive amination
Nature Communications (2020)