Abstract
KIFC3 is a member of the Kinesin superfamily proteins (KIFs). The role of KIFC3 in non-small cell lung cancer (NSCLC) is unknown. This study aimed to elucidate the function of KIFC3 in NSCLC and the underlying mechanism. Immunohistochemistry indicated that KIFC3 was highly expressed in NSCLC tissues and correlated with the degree of differentiation, tumor size, lymph node metastasis and TNM stage. MTT, colony formation and Transwell assays demonstrated that KIFC3 overexpression promoted the proliferation, migration and invasion of NSCLC cells in vitro, while KIFC3 knockdown led to the opposite results. The protein expression levels of PI3Kp85α and p-Akt were increased after KIFC3 overexpression, meanwhile the downstream protein expression levels such as cyclin D1, CDK4, CDK6, RhoA, RhoC and MMP2 were increased. This promotion effect could be inhibited by a specific inhibitor of the PI3K/Akt pathway, LY294002. Co-immunoprecipitation assays confirmed the interaction between endogenous/exogenous KIFC3 and PI3Kp85α. Tumor formation experiments in nude mice confirmed that KIFC3 overexpression promoted the proliferation, migration and invasion of NSCLC cells in vivo and performed its biological function through the PI3K/Akt signaling pathway.In conclusion, KIFC3 promotes the malignant behavior of NSCLC cells through the PI3K/Akt signaling pathway.
Similar content being viewed by others
Introduction
Lung cancer is one of the main causes of cancer-related deaths worldwide, and the associated morbidity and mortality rates are increasing; thus, lung cancer has become a global public health problem1,2. Non-small cell lung cancer (NSCLC) accounts for approximately 85–90% of lung cancer cases3,4. Most patients are in an advanced stage at the time of treatment and generally have a poor prognosis, resulting in a substantial economic burden for patients' families and society5. In recent years, some new therapies have been developed that target factors related to lung cancer pathogenesis, such as EGFR mutation and ALK fusion6,7,8. However, the early diagnosis and treatment of NSCLC is still not satisfactory, and the 5 years survival rate is only 15–20%. The prognosis of patients with NSCLC is closely related to the proliferation, migration and invasion of tumor cells, especially the transcriptional regulation of some key genes related to these processes9,10. Therefore, it is particularly important to understand the molecular mechanism underlying the occurrence and development of NSCLC and further discover and recognize potential biomarkers and new therapeutic targets of lung cancer.
The PI3K/Akt signaling pathway is a key pathway in tumor occurrence and progression and is ubiquitous in a variety of human tumors11,12,13,14. The key to the PI3K/Akt signaling pathway is the phosphorylation of Akt and the activation of downstream transduction molecules, and PI3K promotes Akt phosphorylation by recruiting Akt to the cell membrane15. Phosphorylated Akt (p-Akt) is the activated form of Akt, and a large number of studies have reported that p-Akt is highly expressed in a variety of human cancers and is related to the prognosis of some cancers16,17,18,19. Once activated, Akt regulates the function of many downstream proteins and is involved in cell survival, proliferation, migration, metabolism and angiogenesis20,21.
Kinesin superfamily proteins (KIFs) were first discovered by Vale et al. in 198522, and at present, more than 45 kinds of human KIFs have been described23. KIFs facilitate the transport of mRNAs, protein complexes and organelles in an ATP- and microtubule-dependent manner24,25, which is necessary for cell mitosis and meiosis26,27. According to the location of the motor domain, KIFs are divided into three categories: NH2-terminal motor domain type, intermediate motor domain type and COOH-terminal motor domain type (called N-kinesin, M-kinesin and C-kinesin, respectively)28. KIFC3 is a member of the C-kinesin members of the KIFs family. The gene is located on chromosome 16q21 and encodes an 833-amino acid protein. KIFC3 is a minus-end microtubule-dependent KIF that is mainly localized in the centrosome. It has been shown that the balance of the KIFC3 and EG5 tetramer proteins controls the initiation of mitotic spindle assembly. At the beginning of mitosis, KIFC3 becomes the main driving force of centrosome cohesion to prevent the premature formation of the spindle and counteracts the separation driving force of EG5. EG5, also known as KIF11, is a member of the KIFs family and a plus-end microtubule-dependent KIF.A study showed that KIF11 overexpression had a correlation with advanced pathological grade, advanced T stage, presence of lymph node metastasis, and poor prognosis in lung cancer29. During mitosis, the timing of centrosome separation is critical for spindle formation and the accuracy of chromosome separation. Continuous centrosome aggregation can lead to chromosome misdivision30. Therefore, does the abnormal expression of KIFC3, which controls the timing of centrosome separation, contribute to the occurrence and development of tumors? After reviewing the relevant literature, we found that KIFC3 was positively correlated with biomarkers of liver cancer cell migration and invasion, and KIFC3 overexpression was associated with shorter overall survival (OS) in patients with liver cancer31. Another study showed that a chimeric DAXX-KIFC3 fusion protein formed by death domain-associated protein (DAXX) and KIFC3 promoted the occurrence of osteosarcoma32. However, the correlation between KIFC3 and lung cancer has not been reported. Therefore, there is potential value for the study of KIFC3 in NSCLC.
In this study, we investigated the expression of KIFC3 in NSCLC tissues, its correlation with clinicopathological parameters, and its influence on the biological behavior of NSCLC as well as the underlying mechanisms; we aimed to demonstrate that KIFC3 may be a new target for improved NSCLC therapy.
Results
KIFC3 is highly expressed in NSCLC tissues and associated with clinicopathological parameters
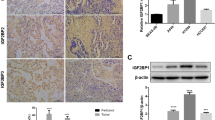
We collected NSCLC tissue samples from 109 patients for immunohistochemistry. The results showed that KIFC3 was mainly localized in the cytoplasm and nuclei and more highly expressed in NSCLC tissues than in para-carcinoma tissues (Fig. 1A). The expression of KIFC3 was correlated with clinicopathological parameters, including the degree of differentiation (P = 0.037), tumor size (P = 0.024), lymph node metastasis (P = 0.002), and TNM stage (P = 0.031) (Table 1). Kaplan‒Meier survival analysis showed that high KIFC3 protein expression was associated with short overall survival and poor prognosis in NSCLC patients (P = 0.031) (Fig. 1B). The above results suggested that KIFC3 is a carcinogenic factor in NSCLC.
Expression and location of KIFC3. (A) KIFC3 expression in NSCLC tissues analyzed by immunohistochemistry. (i) alveoli; (ii) normal bronchial epithelium; (iii) well-differentiated squamous cell carcinoma; (iv) well-differentiated adenocarcinoma; (v) poorly differentiated squamous cell carcinoma; (vi) poorly differentiated adenocarcinoma. Magnification, ×200. (B) Kaplan‒Meier survival analysis. High KIFC3 expression was associated with poor prognosis in NSCLC. (C) Western blot showing KIFC3 expression in 6 NSCLC cell lines and a normal human bronchial epithelial (HBE) cell line. Relative quantity analysis of protein expression with ImageJ software. (D) Immunofluorescence assay showed the localization of KIFC3 in 5 NSCLC cell lines. KIFC3 was mainly localized in the cytoplasm and nucleus. Magnification, ×400. NSCLC, non-small cell lung cancer.
KIFC3 expression and localization in NSCLC cell lines
Western blotting was performed to measure the expression of KIFC3 in 6 NSCLC cell lines (A549, LK2, SK-MS-1, H460, H1299, and H1975 cell lines) and a normal human bronchial epithelial (HBE) cell line. The results showed that KIFC3 was expressed at low levels in the HBE cell line, at high levels in the H460 cell line, at moderate levels in the A549 and H1299 cell lines, and at low levels in other NSCLC cell lines (Fig. 1C).
Immunofluorescence assays were conducted to determine the localization of KIFC3 in the A549, H1299, SK-MS-1, H460 and H1975 cell lines and showed that KIFC3 was mainly localized in the cytoplasm and nuclei, which further confirmed the results of immunohistochemistry (Fig. 1D).
KIFC3 promotes the proliferation of NSCLC cells
To further explore the effects of KIFC3 on the biological behavior of NSCLC cells in vitro, we used KIFC3-specific siRNA or a KIFC3 cDNA-expressing plasmid to knockdown or overexpress KIFC3 in A549 and H1299 cell lines, respectively. The transfection efficiency was determined by Western blot, and the results showed that KIFC3-specific siRNA effectively blocked KIFC3 expression, while the KIFC3 cDNA plasmid successfully overexpressed KIFC3 (Fig. 2A). Next, an MTT assay was used to assess the effect of KIFC3 on the proliferation of tumor cells. We found that KIFC3 knockdown suppressed the growth of the A549 and H1299 cell lines, whereas KIFC3 overexpression promoted the growth of these two cell lines (Fig. 2B). Moreover, KIFC3 knockdown inhibited colony formation ability in A549 and H1299 cell lines, while KIFC3 overexpression enhanced this ability (Fig. 2C). Altogether, these results suggested that KIFC3 promoted the proliferation ability of NSCLC cells in vitro.
KIFC3 promotes NSCLC cell proliferation. (A) The knockdown and overexpression efficiency of KIFC3 were determined by Western blotting. Relative quantity analysis of protein expression with ImageJ software. (B) MTT assay demonstrated that KIFC3 knockdown suppressed growth and KIFC3 overexpression promoted growth in A549 and H1299 cell lines. (C) Colony formation assay demonstrated that KIFC3 knockdown inhibited colony formation and KIFC3 overexpression promoted colony formation in A549 and H1299 cell lines. * P < 0.05, ** P < 0.01.
KIFC3 promotes the migration and invasion of NSCLC cells
The effects of KIFC3 on the migration of NSCLC cells were examined by Transwell migration assay. Transwell migration assays showed that when KIFC3 was knocked down, the number of tumor cells entering the lower chambers was significantly reduced compared with the control group, while KIFC3 overexpression led to the opposite results (Fig. 3A). The effects of KIFC3 on the invasion of NSCLC cells were examined by Transwell assay with Matrigel. The results showed that when KIFC3 was knocked down, the number of tumor cells crossing the Matrigel to the lower chambers was significantly reduced compared with the control group, whereas KIFC3 overexpression caused the opposite results (Fig. 3B). These results indicated that KIFC3 promoted the migration and invasion of NSCLC cells in vitro.
KIFC3 promotes NSCLC cell migration and invasion. (A) Transwell migration assay revealed that KIFC3 knockdown inhibited migration and KIFC3 overexpression promoted migration in A549 and H1299 cell lines. (B) Transwell invasion assays revealed that KIFC3 knockdown inhibited invasion and KIFC3 overexpression promoted invasion in A549 and H1299 cell lines. * P < 0.05, ** P < 0.01.
KIFC3 regulates the malignant behavior of NSCLC cells through the PI3K/Akt signaling pathway
After screening various signal transduction pathways, we speculated that KIFC3 might regulate the malignant behavior of NSCLC cells through the PI3K/Akt signaling pathway. Western blotting results showed that in the A549 and H1299 cell lines, the protein expression levels of PI3Kp85α and phosphorylated Akt (p-Akt) were stably and significantly decreased after KIFC3 knockdown. In contrast, the protein expression levels of PI3Kp85α and p-Akt were stably and significantly increased after KIFC3 overexpression. However, the protein expression of total Akt had no significant change after KIFC3 knockdown or overexpression (Fig. 4A). To further verify the above results, we examined changes in the expression of related proteins downstream of the PI3K/Akt signaling pathway after KIFC3 knockdown or overexpression. The results revealed that the protein expression of the proliferation-related proteins cyclin D1, CDK4 and CDK6, the migration-related proteins RhoA and RhoC, and the invasion-related protein MMP2 were decreased after KIFC3 knockdown, while KIFC3 overexpression caused the opposite results (Fig. 4B).
KIFC3 regulates the PI3K/Akt signaling pathway. (A) Western blotting demonstrated that KIFC3 knockdown reduced PI3Kp85α and p-Akt expression, and KIFC3 overexpression increased PI3Kp85α and p-Akt expression in A549 and H1299 cell lines. (B) Western blotting analysis demonstrated that KIFC3 knockdown reduced the expression of proteins associated with proliferation, migration and invasion downstream of the PI3K/Akt signaling pathway in A549 and H1299 cell lines, and KIFC3 overexpression caused the opposite results. Relative quantity analysis of protein expression with ImageJ software. * P < 0.05, ** P <0.01.
Furthermore, we used LY294002, a specific inhibitor of the PI3K/Akt signaling pathway, to verify the above results. We found that the increased expression of PI3Kp85α and p-Akt caused by KIFC3 overexpression was inhibited after adding LY294002. Additionally, the expression of related functional proteins downstream of this pathway was also inhibited, while the protein expression of total Akt was not significantly different after the inhibitor was added (Fig. 5). Next, we performed cell function experiments to verify the above changes. LY294002 was added to the experimental group, and an equal volume of DMSO was added to the control group. The results revealed that when KIFC3 was overexpressed, the colony formation of the experimental group was significantly reduced compared with that of the control group (Fig. 6A), and the number of tumor cells entering the lower chambers of the experimental group was significantly lower than that of the control group in the Transwell migration and invasion assay (Fig. 6B, C).
Effect of KIFC3 on protein expression in NSCLC cells treated with or without a PI3K/Akt inhibitor. Western blotting analysis demonstrated that the PI3K/Akt pathway inhibitor LY294002 reversed the KIFC3 overexpression-mediated increase in the expression of proteins in this pathway and related downstream proteins in A549 and H1299 cell lines. Relative quantity analysis of protein expression with ImageJ software. * P < 0.05, ** P < 0.01.
Effect of KIFC3 on the behavior of NSCLC cells treated with or without a PI3K/Akt inhibitor. (A) LY294002 was added to the experimental group, and an equal volume of DMSO was added to the control group. Colony formation assays revealed that when KIFC3 was overexpressed, the colony formation ability of the experimental group was reduced compared with that of the control group. (B) Transwell migration assay revealed that when KIFC3 was overexpressed, the migration of the LY294002 group was lower than that of the DMSO group. (C) Transwell invasion assay revealed that when KIFC3 was overexpressed, the invasion ability of the LY294002 group was lower than that of the DMSO group. (D) Co-immunoprecipitation assay verified an interaction between endogenous KIFC3 and PI3Kp85α. Cell lysates were immunoprecipitated with an anti-PI3Kp85α antibody or control IgG antibody. Western blotting demonstrated an interaction between endogenous KIFC3 and PI3Kp85α. (E) Co-immunoprecipitation assay verified an interaction between exogenous KIFC3 and PI3Kp85α. A549 and H1299 cells were transfected with the pCMV6-Myc-DDK-KIFC3 plasmid, and cell lysates were immunoprecipitated with an anti-DDK antibody or control IgG antibody. Western blotting demonstrated an interaction between exogenous KIFC3 and PI3Kp85α. (F) RT‒PCR revealed that the mRNA level of PI3K did not change after KIFC3 overexpression in A549 and H1299 cell lines. *P < 0.05, ** P <0.01.
All the above results proved that KIFC3 regulated the malignant behavior of NSCLC cells through the PI3K/Akt signaling pathway. Then, we verified whether there was an interaction between KIFC3 and PI3K by a co-immunoprecipitation assay. First, an anti-PI3Kp85α antibody was added to the cell lysate of the experimental group for immunoprecipitation, and the corresponding IgG antibody was added to the control group. The subsequent Western blotting results demonstrated an interaction between endogenous KIFC3 and PI3Kp85α (Fig. 6D). Then, A549 and H1299 cells were transfected with the pCMV6-Myc-DDK-KIFC3 plasmid, and the cell lysate was immunoprecipitated with anti-DDK antibody or control IgG antibody. The subsequent Western blotting results demonstrated an interaction between exogenous KIFC3 and PI3Kp85α (Fig. 6E).
To further explore whether KIFC3 affected the PI3K mRNA levels, we conducted RT‒PCR experiments, and the results revealed that the mRNA level of PI3K was not different after KIFC3 overexpression in A549 and H1299 cell lines (Fig. 6F). This result indicated that KIFC3 did not regulate PI3K at the mRNA level; therefore, we speculated that KIFC3 regulated PI3K at the protein level.
KIFC3 promotes the proliferation, migration and invasion of NSCLC cells in vivo
To further elucidate the biological function of KIFC3 and the underlying mechanism in vivo, we used A549 and H1299 cell lines to establish animal models of tumor formation in nude mice. The results of subcutaneous tumor formation in nude mice showed that, compared with the control group, subcutanous tumor volume, tumor weight and tumor growth rate were increased in the KIFC3 overexpression group (Fig. 7A, B, C). Total protein was extracted from tumor tissues for Western blotting analysis, and the results revealed that the protein expression levels of PI3Kp85α and p-Akt were increased after KIFC3 overexpression, while the protein expression of total Akt did not significantly change (Fig. 7D). The results of lung metastasis analysis in nude mice revealed that, compared with the control group, the number of pulmonary metastatic nodules was increased in the KIFC3 overexpression group (Fig. 7E). HE staining of lung metastatic tissues showed that the cancer nests in the KIFC3 overexpression group were larger than those in the control group (Fig. 7F). These results all confirmed that KIFC3 overexpression promoted the proliferation, migration and invasion of NSCLC cells in vivo and performed its biological function through the PI3K/Akt signaling pathway, which was consistent with the results of in vitro experiments.
KIFC3 promotes NSCLC cell proliferation, migration and invasion in vivo. (A, B, C) Subcutaneous tumor formation in nude mice showed that subcutaneous tumor volume, tumor weight and tumor growth rate increased when KIFC3 was overexpressed. (D) Total protein was extracted from subcutaneous tumor tissues. Western blotting analysis revealed that KIFC3 overexpression increased PI3Kp85α and p-Akt expression in vivo. Analysis of relative protein expression with ImageJ software. (E) Gross pictures of lung metastases in nude mice revealed that the number of pulmonary metastatic nodules increased when KIFC3 was overexpressed. (F) HE staining of lung metastatic tissues showed that the cancer nests in the KIFC3 overexpression group were larger than those in the control group. Magnification, ×200. HE, Hematoxylin and eosin. * P < 0.05, ** P < 0.01.
Discussion
Kinesin superfamily proteins (KIFs) is a huge molecular dynamics superfamily with 45 members.The superfamily uses the energy of ATP hydrolysis to move along the microtubule and supports a variety of cell functions, including mitosis, meiosis and material transport. Family members share extensive homology in the globular head domain containing microtubule and ATP-binding sites, which is the motor domain33. Microtubules are polymers composed of α and β tubulin, which have a rapidly growing end (plus end) and a slowly growing end (minus end)34. KIFs use microtubules as tracks to participate in various intracellular transport processes. According to the location of the motor domain, KIFs are divided into the N-kinesin, M-kinesin and C-kinesin subtypes. N-kinesin and M-kinesin are plus-end oriented, while C-kinesin is minus-end oriented. A study compared the expression of 32 kinds of KIFs in hepatocellular carcinoma (HCC) and paracancer tissues. The results showed that the expression of KIF2A and KIFC3 was positively correlated with biomarkers of HCC cell migration and invasion, and overexpression of KIF2A, KIF11, KIFC1 and KIFC3 was associated with shorter overall survival (OS) in HCC patients. This study revealed that most abnormal overexpression of KIFs was significantly associated with the progression and poor prognosis of HCC31.
KIFC3 is a C-kinesin type member of the KIFs family, and it is a minus-end microtubule-dependent motor protein. The motor domain of KIFC3 is located at the COOH-terminal and moves toward the minus end of microtubules. KIFC3 is involved in the apically targeted transport of insoluble membrane organelles35. It is also involved in adhesion between epithelial cells, which is required for zonula adherens maintenance36. As previously mentioned, the balance of KIFC3 and EG5 tetramer proteins controls the initiation of mitotic spindle assembly. Any abnormal mitosis can lead to cell death, gene deletion and even cancer37,38. Therefore, we speculated that overexpression of KIFC3 may promote the occurrence and progression of cancer. Previous literature showed that KIFC3 overexpression was significantly correlated with the progression and prognosis of hepatocellular carcinoma (HCC), suggesting that KIFC3 may be a potential biomarker for HCC31. Another study demonstrated that overexpression of KIFC3 can mediate docetaxel resistance in breast cancer cell lines39. Docetaxel bound to the β-tubulin subunit of microtubules and inhibited the exchange rate of free and bound tubulin40. This inhibition disrupted the formation of the mitotic spindle, inhibited cell division and led to cell death. Overexpression of KIFC3 increased the amount of free tubulin in breast cancer cells treated with docetaxel, suggesting that KIFC3 may antagonize the effect of docetaxel by promoting the partial separation of tubulin from microtubules. This may lead to new therapies, for example, therapies that exploit the compatible application of KIFC3 inhibitors and docetaxel and exert synergistic effects with docetaxel by interfering with the resistance mechanism. A recent study revealed that the increased expression levels of KIFC3 could enhance the proliferation, migration and invasion of colorectal cancer (CRC) cells41. Once again, KIFC3 was shown to promote the progression of malignant tumors.Currently, an increasing number of studies have shown that KIFC3 is closely related to the progression and prognosis of various malignant tumors.However, the correlation between KIFC3 and lung cancer has not been reported yet.Therefore, this study investigated the expression, effect on biological function and mechanism of KIFC3 in NSCLC.
In this study, we showed that the expression of KIFC3 in NSCLC tissues was higher than that in para-carcinoma tissues, and it was correlated with the degree of differentiation, tumor size, lymph node metastasis, TNM stage and prognosis. In vitro, cell function experiments demonstrated that KIFC3 overexpression promoted the proliferation, migration and invasion of NSCLC cells, while KIFC3 knockdown showed the opposite results. Moreover, we demonstrated that KIFC3 overexpression activated the PI3K/Akt signaling pathway in vitro, and the expression levels of proliferation-, migration- and invasion-related proteins downstream of this pathway were increased, while this promotion effect could be inhibited by a specific inhibitor of the PI3K/Akt pathway, LY294002. We confirmed the interaction between endogenous/exogenous KIFC3 and PI3Kp85α by a co-immunoprecipitation assay. Tumor formation experiments in nude mice confirmed that KIFC3 overexpression promoted the proliferation, migration and invasion of NSCLC cells in vivo and performed its biological function through the PI3K/Akt signaling pathway, which was consistent with the results of in vitro experiments.
Although our study confirmed an interaction between KIFC3 and PI3Kp85α, the exact molecular mechanism through which a domain of KIFC3 interacts with PI3Kp85α is still unclear. To determine whether it interacts with PI3Kp85α through the motor domain, we will construct a mutant in which the motor domain is deleted, and this will be the focus of our future research.
It has been confirmed that activation of the PI3K-Akt signaling pathway is involved in promoting cell proliferation, survival and metabolism in various malignant tumors. Although a variety of PI3K-Akt pathway inhibitors have been developed at present, few drugs are actually used in clinical practice due to poor tolerance and serious side effects. It is particularly important for the research and development of new drugs for lung cancer treatment to fully explore and utilize the interaction between proteins in the pathway. Our study demonstrated that KIFC3 promoted the proliferation, migration and invasion of NSCLC through the PI3K/Akt signaling pathway in vivo and in vitro, which may provide a research basis for the development of more effective, well-tolerated and low toxicity targeted therapies for NSCLC treatment in the future.
Materials and methods
Patients and specimens
This study collected pathological samples from 109 patients with NSCLC who visited the Department of Pathology of the First Affiliated Hospital of China Medical University from 2015 to 2019. None of the patients involved in this study received preoperative radiotherapy or chemotherapy. This study was approved by the medical research ethics committee of China Medical University, and informed consent was obtained from all patients.We confirm that all methods were performed in accordance with relevant guidelines and regulations.
Immunohistochemistry
The collected paraffin-embedded tissue specimens were sectioned successively at a thickness of 4 μm. The sections were deparaffinized in xylene, rehydrated in a series of gradient alcohol solutions, and repaired with pH = 6.0 citrate buffer at high temperature and high pressure for 2.5 min. The sections were then sequentially incubated with the reagents in the IHC kit (Maixin Bio, Fuzhou) according to the manufacturer's instructions, and they were incubated with anti-KIFC3 rabbit polyclonal antibody (1:200; ab154419; Abcam) overnight at 4 °C. After the color reaction, the sections were sealed with neutral resin. According to the staining intensity score, the expression level of KIFC3 was scored as 0 (no staining), 1 (weak staining), 2 (moderate staining), and 3 (high staining). The percentage of stained cells was scored as 1 (1–25%), 2 (26–50%), 3 (51–75%), and 4 (76–100%). The two scores were multiplied as the final score, with a score range of 0–12. Final scores ≤ 4 were considered to indicate low KIFC3 expression, and scores >4 were considered to indicate high KIFC3 expression.
Cell lines and cell culture
The A549, H1299, SK-MES-1, H460, H1975, and LK2 lung cancer cell lines were purchased from the Cell Bank of the Chinese Academy of Sciences (Shanghai).
Human bronchial epithelial (HBE) cells were obtained from the American Type Culture Collection (ATCC). The A549, H1299, H460, and H1975 cell lines were cultured in RPMI 1640 medium (Gibco). The SK-MES-1 cell line was cultured in minimal essential medium (Gibco). The HBE and LK2 cell lines were cultured in high-glucose Dulbecco's modified Eagle's medium (Gibco). All the media were supplemented with 10% FBS (Clark). The cells were incubated in a CO2 incubator at 37 °C.
Immunofluorescence
The cells were cultured in 24-well plates overnight, fixed with 4% paraformaldehyde for 20 min, incubated with 0.25% Triton X-100 for 15 min, blocked with 5% BSA at 37 °C for 2 h, and incubated with an anti-KIFC3 primary antibody (1:50; ab154419; Abcam) overnight at 4 °C. The next day, the cells were incubated with fluorescent secondary antibody (1:100; Beyotime) in the dark for 2 h, and then, the cell nuclei were stained with DAPI for 10 min. The cells were observed by laser confocal microscopy.
Plasmid construction and cell transfection
Lipofectamine 3000 (Invitrogen) was used as the transfection reagent. For KIFC3 knockdown, KIFC3-specific small interfering RNA (siRNA) and negative control siRNA were purchased from RiboBio. For KIFC3 overexpression, PCMV6-Myc-DDK-KIFC3 and pCMV6-Myc-DDK were purchased from GenePharma. Stable KIFC3 expression cell lines were established by adding G418 into the medium. LY294002 (MedChemExpress), a specific inhibitor of the PI3K/Akt signaling pathway, was used to inhibit the activity of this pathway, and the control group was treated with an equal volume of DMSO. The siRNA sequences used in this study are shown in Table S1.
Cell proliferation assay and colony formation assay
The MTT assay was used to evaluate cell proliferation. In a 96-well plate, 3000 cells in 100 µl of the corresponding medium were placed in each well. Then, 10 µl MTT reagent was added to each well in the dark at the same time every day. After 4 h of incubation, 100 µl DMSO was added to each well. Then, the absorbance of each well was measured by a microplate reader at a wavelength of 490 nm for 5 consecutive days. All the experiments were repeated three independent times.
Cell proliferation was also assessed by colony formation assay. A total of 500 cells in 4 ml of the corresponding medium were plated in each well of a 6-well plate. Then, the cells were incubated in a CO2 incubator at 37 °C for 10–14 days, fixed with precooled methanol, and stained with crystal violet. All the experiments were repeated three independent times.
Cell migration and invasion analysis
The Transwell migration assay was carried out in a 24-well plate. A549 and H1299 cells were placed in the upper chamber at densities of 80,000/200 μl and 50,000/200 μl, respectively. The medium in the upper chamber contained 2% FBS.
Then, 600 μl of medium containing 20% FBS was added to the lower chamber, and the plates were incubated in an incubator at 37 °C for 18 h, followed by fixation with precooled methanol and staining with crystal violet. The cells that entered the lower chamber were counted. All the experiments were repeated three independent times.
For the Transwell invasion assay, 100 μl Matrigel (1:9 dilution; BD Biosciences) was added to the upper chamber one day before the experiment, and the remaining steps were the same as those in the Transwell migration assay.
Western blotting
Total protein was extracted from cells and tissues with lysis buffer (Beyotime) and phenylmethylsulfonyl fluoride (PMSF) (Beyotime). Total protein was separated by 10% SDS‒PAGE and transferred to polyvinylidene difluoride (PVDF) membranes (Millipore). The membranes were blocked in 5% skim milk at 37 °C for 2 h and then incubated with different primary antibodies (Table S2) overnight at 4 °C. The next day, the membranes were incubated with an HRP-conjugated anti-mouse/rabbit IgG (1:2000; Proteintech) at 37 °C for 2 h. Finally, the membranes were incubated with enhanced chemiluminescence reagent (Beyotime) and visualized by a Bio-Imaging system (Bio-Rad). GAPDH was used as the loading control. All the experiments were repeated three independent times.The molecular weight of our target proteins is relatively dispersed, and there is a significant difference in transfer time. If they are transferred together, some may be over-transferred,while others may not reach the specified time, so we didn't incubate them on one membrane.
Co-immunoprecipitation assay
Total protein was extracted from cells with lysis buffer (Beyotime) and PMSF (Beyotime). Total protein was added to Protein A + G agarose beads (Beyotime) and blocked at 4 °C for 2 h. The agarose beads were removed by centrifugation, and the supernatant was divided into an experimental group and a control group. The experimental group was incubated with an anti-DDK antibody (HT201-01; TransGen Biotech) or an anti-PI3Kp85α antibody (1:1000; 4257; Cell Signaling Technology) overnight at 4 °C, and the control group was incubated with an anti-IgG antibody (1:2000; ZSGB-BIO). The next day, protein A + G agarose beads were added to the mixture at 4 °C for 6 h, the supernatant was removed after centrifugation, and the precipitate was washed with lysis buffer. The collected samples were heated in boiling water for 10 min and used for Western blotting. All the experiments were repeated three independent times.
RNA extraction and real-time PCR
Total RNA was extracted from cells with TRIzol reagent (TransGen Biotech) and reverse transcribed into cDNA using the PrimeScript RT Master Mix kit (Takara) according to the manufacturer’s instructions. The cDNA samples were amplified by quantitative PCR using the TB Green Premix EX Taq II kit (Takara) according to the manufacturer’s instructions. The reaction conditions were as follows: 95 °C for 30 s, followed by 40 cycles at 95 °C for 5 s and 60 °C for 30 s. The relative expression level of the target gene was determined by the 2−ΔΔCT method with GAPDH as the internal control. The primer sequences used are shown in Table S3. All experiments were repeated three independent times.
Tumor formation in nude mice
The animal protocol in this study was approved by the Institutional Animal Care and Use Committee (IACUC) of China Medical University (CMU2022133).We confirm that all methods were performed in accordance with relevant guidelines and regulations.
Four-week-old female BALB/c nude mice were purchased from Beijing Vital River Laboratory Animal Technology Company and maintained under specific pathogen-free (SPF) conditions for one week before the experiment. For subcutaneous tumor formation, the mice were injected subcutaneously in the right axilla with 1 × 107 lung cancer cells/200 µl; KIFC3-transfected A549 or H1299 cells were used as the experimental group and empty vector-transfected A549 or H1299 cells served as the control group. The size of subcutaneous tumors was measured at 6, 9, 12, 15, 18, 21 and 24 days after injection. The vernier caliper was used to measure the longest diameter(length,L) of the tumor and the longest transverse diameter(width, W) perpendicular to the longest diameter. The formula V = ½ × L × W2 was used to calculate the volume (V) of the tumor. And the growth curve was plotted based on the tumor volume.The mice were euthanized by cervical dislocation 24 days after injection, the tumors were resected, the tumor size and weight were measured, and the total protein of the tumor was extracted and analyzed by Western blotting. For pulmonary metastatic tumors, the mice were injected with 1 × 106 lung cancer cells/100 µl (KIFC3-transfected A549 and H1299 cells or the corresponding empty vector-transfected A549 and H1299 cells) through the tail vein. The mice were euthanized by cervical dislocation 8 weeks after injection, and autopsy was performed to examine the dissemination of pulmonary tumors. Then, the tumor tissue was fixed with formaldehyde, embedded in paraffin, sectioned successively, stained with hematoxylin and eosin, and exa mined under a microscope.
Statistical analysis
SPSS 21.0 and GraphPad Prism 8.0 software were used for statistical analysis. The chi-square test was used to analyze the correlation between the expression of KIFC3 and clinicopathological parameters. Student’s t-test was used to analyze the differences between two groups. One-way analysis of variance was used to compare differences between multiple groups. A P value < 0.05 was considered statistically significant.
Institutional review board statement
This study was approved by the medical research ethics committee of China Medical University, and informed consent was obtained from all patients. The animal protocol in this study was approved by the Institutional Animal Care and Use Committee (IACUC) of China Medical University (CMU2022133).
Conclusions
-
(1)
The high expression of KIFC3 was correlated with the degree of differentiation, tumor size, lymph node metastasis, TNM stage and prognosis of NSCLC.
-
(2)
In vitro experiments showed that KIFC3 promoted the proliferation, migration and invasion of NSCLC cells.
-
(3)
KIFC3 regulated the malignant behavior of NSCLC cells through PI3K/Akt signaling pathway in vitro.
-
(4)
KIFC3 promoted the proliferation, migration and invasion of NSCLC through PI3K/Akt signaling pathway in vivo.
Data availability
All data generated or analyzed during this study are included in this article. Further details are available from the corresponding author upon request.
References
Yang, D. et al. Epidemiology of lung cancer and lung cancer screening programs in China and the United States. Cancer Lett 468, 82–87 (2020).
Thai, A. A. et al. Lung cancer. Lancet 398(10299), 535–554 (2021).
Sung, H. et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 71(3), 209–249 (2021).
Siegel, R. L. et al. Cancer statistics, 2022. CA Cancer J. Clin. 72(1), 7–33 (2022).
Cang, S. et al. Assessment of plasma amino acids, purines, tricarboxylic acid cycle metabolites, and lipids levels in NSCLC patients based on LC-MS/MS quantification. J. Pharm. Biomed. Anal. 221, 114990 (2022).
Wang, H. et al. Comprehensive analyses of genomic features and mutational signatures in adenosquamous carcinoma of the lung. Front. Oncol. 12, 945843 (2022).
Shaw, A. et al. Crizotinib versus chemotherapy in advanced ALK-positive lung cancer. N. Engl. J. Med. 368(25), 2385–2394 (2013).
Wang, T. et al. TGFβ1/integrin β3 positive feedback loop contributes to acquired EGFR TKI resistance in EGFR-mutant lung cancer. J. Drug Target. 31, 1–13 (2022).
Zhong, D. et al. SMYD3 regulates the abnormal proliferation of non-small-cell lung cancer cells via the H3K4me3/ANO1 axis. J. Biosci. 47, 53 (2022).
Yang, J. et al. SPI1 mediates transcriptional activation of TPX2 and RNF2 to regulate the radiosensitivity of lung squamous cell carcinoma. Arch. Biochem. Biophys. 730, 109425 (2022).
Kumar, S. & Agnihotri, N. Piperlongumine, a piper alkaloid targets Ras/PI3K/Akt/mTOR signaling axis to inhibit tumor cell growth and proliferation in DMH/DSS induced experimental colon cancer. Biomed. Pharmacother. 109, 1462–1477 (2019).
Gohr, K. et al. Inhibition of PI3K/Akt/mTOR overcomes cisplatin resistance in the triple negative breast cancer cell line HCC38. BMC Cancer 17(1), 711 (2017).
Muñoz, J. et al. Tobacco exposure enhances human papillomavirus 16 oncogene expression via EGFR/PI3K/Akt/c-Jun signaling pathway in cervical cancer cells. Front. Microbiol. 9, 3022 (2018).
Li, J. et al. Long non-coding RNA PICART1 inhibits cell proliferation by regulating the PI3K/AKT and MAPK/ERK signaling pathways in gastric cancer. Eur. Rev. Med. Pharmacol. Sci. 23(2), 588–597 (2019).
Nussinov, R. et al. Phosphorylation and driver mutations in PI3Kα and PTEN autoinhibition. Mol. Cancer Res. 19(4), 543–548 (2021).
Gan, Y. et al. TDRG1 regulates chemosensitivity of seminoma TCam-2 cells to cisplatin via PI3K/Akt/mTOR signaling pathway and mitochondria-mediated apoptotic pathway. Cancer Biol. Ther. 17(7), 741–750 (2016).
Lonetti, A. et al. Improving nelarabine efficacy in T cell acute lymphoblastic leukemia by targeting aberrant PI3K/AKT/mTOR signaling pathway. J. Hematol. Oncol. 9(1), 114 (2016).
Yao, Z. et al. Prognostic role of the activated p-AKT molecule in various hematologic malignancies and solid tumors: A meta-analysis. Front. Oncol. 10, 588200 (2020).
Liu, C. et al. Correction: Lapatinib inhibits CIP2A/PP2A/p-Akt signaling and induces apoptosis in triple negative breast cancer cells. Oncotarget 8(6), 10760 (2017).
Chu, Y. et al. Co-culture with chorionic villous mesenchymal stem cells promotes endothelial cell proliferation and angiogenesis via ABCA9-AKT pathway. FASEB J. 36(10), e22568 (2022).
Guerrero-Zotano, A., Mayer, I. & Arteaga, C. PI3K/AKT/mTOR: Role in breast cancer progression, drug resistance, and treatment. Cancer Metastasis Rev. 35(4), 515–524 (2016).
Vale, R., Reese, T. & Sheetz, M. Identification of a novel force-generating protein, kinesin, involved in microtubule-based motility. Cell 42(1), 39–50 (1985).
Thankachan, J. M. & Setty, S. R. G. KIF13A-A key regulator of recycling endosome dynamics. Front. Cell Dev. Biol. 10, 877532 (2022).
Gao, H. et al. KIF2A regulates ovarian development via modulating cell cycle progression and vitollogenin levels. Insect Mol. Biol. 30(2), 165–175 (2021).
Li, X. et al. Kinesin family members KIF2C/4A/10/11/14/18B/20A/23 predict poor prognosis and promote cell proliferation in hepatocellular carcinoma. Am. J. Transl. Res. 12(5), 1614–1639 (2020).
Vicente, J. & Wordeman, L. Mitosis, microtubule dynamics and the evolution of kinesins. Exp. Cell Res. 334(1), 61–69 (2015).
Liang, W. T. et al. Prognostic significance of KIF23 expression in gastric cancer. World J. Gastrointest. Oncol. 12(10), 1104–1118 (2020).
Hirokawa, N. & Takemura, R. Kinesin superfamily proteins. In Encyclopedia of Biological Chemistry 2nd edn 679–687 (Academic Press, Waltham, 2013).
Ling, J. J. et al. KIF11, a plus end-directed kinesin, as a key gene in benzo(a) pyrene-induced non-small cell lung cancer. Environ. Toxicol. Pharmacol. 89, 103775 (2022).
Hata, S. et al. The balance between KIFC3 and EG5 tetrameric kinesins controls the onset of mitotic spindle assembly. Nat. Cell Biol. 21(9), 1138–1151 (2019).
Chen, J. et al. Kinesin superfamily protein expression and its association with progression and prognosis in hepatocellular carcinoma. J. Cancer Res. Ther. 13(4), 651–659 (2017).
Mason-Osann, E. et al. Identification of a novel gene fusion in ALT positive osteosarcoma. Oncotarget 9(67), 32868–32880 (2018).
Hirokawa, N. et al. Submolecular domains of bovine brain kinesin identified by electron microscopy and monoclonal antibody decoration. Cell 56(5), 867–878 (1989).
Jakobs, M. A. H., Zemel, A. & Franze, K. Unrestrained growth of correctly oriented microtubules instructs axonal microtubule orientation. Elife 11, e77608 (2022).
Noda, Y. et al. KIFC3, a microtubule minus end-directed motor for the apical transport of annexin XIIIb-associated Triton-insoluble membranes. J. Cell Biol. 155(1), 77–88 (2001).
Sako-Kubota, K. et al. Minus end-directed motor KIFC3 suppresses E-cadherin degradation by recruiting USP47 to adherens junctions. Mol. Biol. Cell 25(24), 3851–3860 (2014).
Zhang, F. et al. The genomic stability regulator PTIP is required for proper chromosome segregation in mitosis. Cell Div. 17(1), 5 (2022).
Bočkaj, I. et al. The H3.3K27M oncohistone affects replication stress outcome and provokes genomic instability in pediatric glioma. PLoS Genet. 17(11), e1009868 (2021).
Tan, M. H. et al. Specific kinesin expression profiles associated with taxane resistance in basal-like breast cancer. Breast Cancer Res Treat. 131(3), 849–858 (2012).
Alfano, A. et al. SRC kinase-mediated tyrosine phosphorylation of TUBB3 regulates its stability and mitotic spindle dynamics in prostate cancer cells. Pharmaceutics 14(5), 932 (2022).
Liao, H. et al. KIFC3 promotes proliferation, migration, and invasion in colorectal cancer via PI3K/AKT/mTOR signaling pathway. Front. Genet. 13, 848926 (2022).
Acknowledgements
We would appreciate the language editing services provided by American Journal Experts.
Author information
Authors and Affiliations
Contributions
Yue Ma, Renyi Zhou and Xueshan Qiu conceived and designed the experiments; Yue Ma and Renyi Zhou prepared the manuscript; Yue Ma, Yao Zhang, Xizi Jiang, Jingqian Guan and Huanxi Wang performed the experiments; Jiameng Zhang and Yue Tong analyzed the data.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical statement
This study is in accordance with ARRIVE guidelines.
Informed consent
Informed consent was obtained from all subjects involved in the study.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Ma, Y., Zhang, Y., Jiang, X. et al. KIFC3 promotes the proliferation, migration and invasion of non-small cell lung cancer through the PI3K/AKT signaling pathway. Sci Rep 14, 20471 (2024). https://doi.org/10.1038/s41598-024-71602-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-71602-0
- Springer Nature Limited