Abstract
Endometrial cancer (EC), one of the most prevalent carcinomas in females, is associated with increasing mortality. We identified the CHD4 R975H mutation as a high-frequency occurrence in EC patients through a comprehensive survey of EC databases. Computational predictions suggest that this mutation profoundly impacts the structural and functional integrity of CHD4. Functional assays revealed that the CHD4 R975H mutation enhances EC cell invasion, proliferation, and colony formation, promoting a cancer stem cell (CSC)-like phenotype. RNA-seq analysis of cells expressing CHD4 R975H mutant revealed a transcriptomic landscape marked by the activation of several cancer-promoting signaling pathways, including TNF-α signaling via NF-κB, KRAS, P53, mTOR, TGF-β, EGFR, Myc and growth factor signaling. Validation assays confirmed the activation of these pathways, further demonstrating that CHD4 R975H mutation induces stemness in EC cells and M2-like polarization of tumor-associated macrophages (TAMs). Our study elucidated the oncogenic role of CHD4 R975H mutation, highlighting its dual impact on facilitating cancer stemness and transforming TAMs into an immunosuppressive subtype. These findings contribute valuable insights into the molecular mechanisms driving EC progression and open avenues for targeted therapeutic interventions.
Similar content being viewed by others
Introduction
Endometrial cancer (EC), which is derived from endometrium, the inner layer of the uterus, has been regarded as a growing health concern with increasing incidence and cancer-related mortality rates. It is the sixth most prevalent cancer in females, with 417,367 cases diagnosed globally in 2020. Patients in the advanced stages of EC have grim prognoses and increased mortality rates1. Currently, effective therapies for EC are elusive, underscoring the urgency of obtaining a deeper understanding of the molecular mechanisms driving its occurrence and progression. Cancer stemness, delineated by the capacity for self-renewal and initiation of carcinomas by cancer stem cells (CSCs), encompasses a spectrum of cancerous traits propelled by CSC-associated signaling pathways. Its implications include cancer proliferation, invasion, and metastasis2,3. In tumor microenvironment (TME), tumor-associated macrophages (TAMs) predominantly exhibit an immunosuppressive M2-like phenotype, also contributing to the promotion of tumor cell growth, invasion and metastasis4. Previous investigations have underscored the reciprocal relationship between CSCs and TAMs, wherein CSCs have been shown to stimulate M2 polarization of TAMs. This interaction between TAMs and cancer cells not only fosters an immunosuppressive microenvironment but also plays a pivotal role in driving carcinogenesis5,6 Consequently, unravelling the intricacies of the regulatory mechanisms governing cancer stemness and TAMs M2-like polarization in EC patients has profound implications for the development of clinically effective treatments targeting CSCs.
Although the molecular intricacies of EC development remain largely elusive, a recent review revealed frequent mutations in a set of tumor-related genes, including PTEN (64–80% in G1–G3 endometrioid endometrial cancer (EEC)), TP53 (59–93% in serous endometrial cancer (SEC)), PIK3CA (22–59% in G1–G3 EEC), and KRAS (19–43% in G1–G3 EEC), among others7. Notably, PTEN has phosphatase activity and functions as a tumor suppressor by inhibiting PI3K-AKT pathway. It converts phosphatidylinositol-3,4,5-trisphosphate (PIP3) to phosphatidylinositol-4,5-bisphosphate (PIP2). Mutation of PTEN results in loss of function, leading to aberrant AKT activation and subsequent EC progression8. Similarly, mutations in p53, another critical tumor suppressor, yield a p53 mutant that exerts carcinogenic effects by disrupting the function of wild-type p53, which is essential for DNA repair and apoptosis9. This underscores the pivotal role of cancer-associated mutations, wherein specific genetic alterations engender both functional perturbations and acquisition of oncogenic functions.
Chromodomain helicase DNA-binding protein 4 (CHD4) is a pivotal subunit of the nucleosome remodelling and histone deacetylation (NuRD) complex that features helicase/ATPase domains crucial for ATP-dependent chromatin remodelling and transcriptional regulation10. Its multifaceted roles extend beyond chromatin dynamics to encompass DNA damage repair, where CHD4 orchestrates NuRD recruitment to DNA damage sites and cell cycle modulation through p53 deacetylation11. Moreover, the involvement of CHD4 in cellular processes such as differentiation, stem cell renewal, and tumor progression underscore its diverse functions12. In the intricate landscape of cancer development, CHD4 plays dual roles, exhibiting both promotive and repressive functions13,14,15. Notably, in ovarian cancer, CHD4 disruption in BRCA2-deficient cells confers protection against replication forks, leading to chemoresistance13. In colorectal cancer, CHD4 recruits DNA methyltransferases to silence tumor suppressor genes, maintaining their transcriptional repression14. Conversely, depletion of CHD4 upregulates stemness marker genes, inducing a CSC phenotype in EC cells15. Intriguingly, in endometrial cancer, CHD4 mutations, notably in the SEC subtype (10–19%)8, have eluded a clear understanding of their functional implications.
To address this gap, our study combines experimental approaches and transcriptomic analyses to elucidate the role of the CHD4 R975H mutant in endometrial cancer cells. Our findings revealed that the CHD4 R975H mutation assumes an oncogenic function, activating cancer-promoting pathways and concomitantly augmenting both cancer stemness and the M2-like polarization of TAMs. This study provides a novel perspective on the impact of CHD4 mutations in endometrial cancer, shedding light on the intricate role of CHD4 mutations in orchestrating critical events in tumor progression.
Materials and methods
Cell culture
Human endometrial cancer cell line Ishikawa and AN3-CA was purchased from the European Collection of Cells Cultures (Public Health England, UK). Human kidney cell line 293 T was purchased from Takara (Lenti-X™ 293 T Cell Line, Takara). Human leukemia monocytic cell line THP-1 was a kind gift from Dr. Rio Sugimura’s laboratory. Ishikawa and 293 T cell lines were cultured in Dulbecco's modified Eagle medium (DMEM, high glucose, Gibco). THP-1 cell line was cultured in Roswell Park Memorial Institute 1640 Medium (RPMI 1640 Medium, Gibco). Both medium were supplemented with 10% foetal bovine serum (FBS, qualified, Gibco) and an antibiotic–antimycotic mixture (Gibco). All cell lines were maintained at 37 °C and 5% CO2 in the Heracell VIOS 160i CO2 incubator (Thermo).
RNA isolation and qRT‒PCR
Total RNA was isolated from the cell lines using TRIzol reagent (Thermo). RNA was quantified via a Nanodrop2000 (Thermo). Total RNA was reverse transcribed into cDNA using the RevertAid RT Reverse Transcription Kit (Thermo). Samples were prepared using iTaq Universal SYBR Green Supermix (Bio-Rad) and subjected to qRT‒PCR on the ABI StepOne Plus real-time PCR system (Applied Biosystems). The primer sequences are listed in Supplementary Table 1. Relative mRNA levels of the genes were normalized to those of the endogenous control gene (GAPDH or RPL13A). Data were analysed using the 2−ΔΔCT method.
Vector construction and mutagenesis
CHD4 coding sequence (CDS) was manually separated into two pieces, and each was amplified from 293 T cDNA by PCR. Both pieces were cocloned and inserted into pSBtet-Bla (60510, Addgene) using NEBuilder HiFi DNA Assembly Master Mix (NEB) to generate an integral CHD4-pSBtet-Bla vector. For CHD4 R975H mutant, overlapping primers flanking the mutation site were designed based on the CHD4 vector. CHD4 R975H CDS and pSBtet-Bla vector sequence were manually separated into two pieces, and each was amplified from the CHD4 vector by PCR. The cloning method was the same as that for CHD4 wild type (CHD4 WT). CHD4 shRNA oligo was purchased from Integrated DNA Technologies (IDT, Hong Kong). CHD4 shRNA oligo was annealed and inserted into pLV-H1TetO-GFP-Bsd (Biosettia). The sequences of the primers and oligo used for cloning are listed in Supplementary Table 2.
Lentivirus packaging and infection
293 T cell line was cultured in a 60 mm plate (Corning) and cotransfected with 2 μg of shCHD4 lentiviral vector, 2 μg of psPAX2 (12260; Addgene) and 1 μg of pMD2. G (12259; Addgene) using LipoD293 (SignaGen). The supernatant was collected 48 h and 72 h later and concentrated using PEG-it Virus Precipitation Solution (System Biosciences). The concentrated virus was used to infect cells with 8 μg/ml polybrene. 24 h after infection, the cells were subjected to further assays. Doxycycline (1 μg/ml) was added to the medium to induce the expression of the lentiviral vector.
Stable transfection cell line construction
The Sleeping Beauty (SB) transposable system was adopted to construct a stable transfection cell line16. Ishikawa cell line was cultured in 60 mm plates and cotransfected with 2.85 μg of the CHD4 wild-type or CHD4 R975H vector and 0.15 μg of the SB100X vector (34879, Addgene). AN3-CA CHD4 R975H cell line was conducted using the same method. 24 h after transfection, 15 μg/ml blasticidin was added to the medium, after which the cells were cultured for 14 days. After selection, Ishikawa CHD4 WT and CHD4 R975H cell lines were subjected to further assays and AN3-CA CHD4 R975H cell line was only adopted in spheroid assay. Doxycycline (1 μg/ml) was added to the medium to induce the expression of the CHD4 WT and CHD4 R975H vectors.
Transwell assay
Transwell inserts with 8 μm pore sizes were precoated with Geltrex (Gibco) and added to 24-well plates (Corning). For each well, 700 μl of DMEM containing 10% FBS was added to the lower chamber. A total of 1 × 105 cells were suspended in 300 μl of serum-free DMEM and added to the upper chamber. After 24 h, the inserts were harvested, and the cells in the upper chamber that had not migrated were scraped away with a cotton swab. The inserts were fixed with methanol for 10 min and stained with crystal violet for 10 min. Invasive cells were photographed and counted under an inverted microscope at 100 × magnification.
Colony formation assay
Cells were digested, counted, and seeded at a confluency of 2000 cells/well into a 6-well plate (Corning). Cells were allowed to adhere and grow for 14–21 days. Once the colonies were visible (> 50 cells), the plates were harvested, and the cells were fixed with 4% paraformaldehyde for 10 min and stained with crystal violet for 10 min. Colonies were photographed and counted manually.
Cell proliferation assay
A 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide (MTT) assay was utilized to evaluate cell proliferation ability17. 4000 cells were suspended in 100 μl of DMEM containing 10% FBS and seeded into 96-well plates (Corning). The cells were allowed to adhere and grow for 4–5 days. 10 μl of MTT was added to each well, and the cells were incubated at 37 °C for 3 h. After incubation, the medium was discarded, and 100 μl of DMSO was added to each well to dissolve the formazan. The plate was wrapped in aluminum foil and incubated on a shaker for 15 min. The absorbance was measured at 490 nm.
Spheroid assay
Cells were digested, counted, and seeded at a confluency of 5000 cells/well (Ishikawa) or 10,000 cells/well (AN3-CA) into a low attachment 6-well plate (Corning). Complete DMEM supplied with B-27, 20 ng/ml EGF, and 10 ng/ml FGF was added to each well to induce spheroid formation. After 2 weeks, the plate was harvest and spheroids (diameter > 50 µm) were photographed and counted manually under microscope. The diameter of spheroids was measured using PAULA Cell Imager software.
Coculture assay
The CHD4 wild-type, CHD4 R975H and shCHD4 cell lines were cultured in complete DMEM supplemented with or without doxycycline for 2 days, after which the conditioned media was collected and filtered through a 0.45 µm sterile syringe filter (Corning). The THP-1 cell line was cultured in complete RPMI 1640 medium and activated by the addition of 100 ng/μl phorbol-12-myristate-13-acetate (PMA) for 24 h. After 24 h, the RPMI 1640 medium was discarded, and conditioned medium was added to the THP-1 cells for 2 days. THP-1 cells were then lysed with TRIzol and subjected to qRT‒PCR or flow cytometry analysis.
Immunoblotting
Cells were lysed in RIPA buffer (150 mM NaCl, 5 mM EDTA pH 8, 50 mM Tris pH 8, 1% CA-630, 0.5% sodium deoxycholate, 0.1% SDS) containing Pierce protease and phosphatase inhibitor (Thermo) for 30 min to obtain whole-cell lysates. The lysate was centrifuged at 13,000 rpm for 5 min. The supernatant was mixed well with 5 × Laemmli buffer (300 mM Tris–HCl (pH 6.8), 50% glycerol, 12.5% β-mercaptoethanol, 0.025% Blomophenol Blue, and 10% SDS) and subjected to heating at 100 °C for 15 min. After heating, the samples were cooled on ice and subjected to 10% SDS‒PAGE. The voltage is 80 V for 20 min and then 120 V for 70 min. The proteins were subsequently transferred to 0.45 μm polyvinylidene difluoride membranes (Merck Millipore) using Thermo Scientific Pierce G2 Fast Blotter (Thermo). The voltage is 25 V for 60 min. The membranes were then blocked in 5% bovine serum albumin for 30 min, incubated with primary antibody at 4 °C overnight. The membranes were cut before incubating with primary antibody because the antibody is not sufficient for full length membrane. After incubation, the membranes were washed with Tris-buffered saline (TBS) supplemented with 0.1% Tween-20 (TBST) for 5 min for three times and incubated with secondary antibody at room temperature for 1 h. After incubation, the membranes were washed with TBST for 5 min for three times, and the protein bands were visualized with the Alliance Q9 Advanced Chemiluminescence Imager System (USA). The antibody information is listed in Supplementary Table 3.
Flow cytometry
CHD4 R975H cells or THP-1 cells were digested, washed, and suspended in staining buffer (PBS with 1% BSA and 0.1% NaN3). Cells were incubated with Human TruStain FcX (Biolegend) for 10 min. Primary antibodies were then added into cell suspension and incubated for 20 min in the dark. After incubation, cells were washed twice and suspended with staining buffer. Cell suspension was subjected to BD FACSAriaIII (BD Biosciences) for flow cytometry. Results were analysed using BD FACSDiva Software. The antibody information is listed in Supplementary Table 4.
Mouse xenograft model
The study was reported in accordance with ARRIVE guidelines. The experiment protocol was approved by Committee on the Use of Live Animals in Teaching and Research (CULATR) of The University of Hong Kong (CULATR No. 5712-21). All treatment was performed under the guideline of Hong Kong SAR Government Department of Health and Centre for Comparative Medicine Research, The University of Hong Kong. CHD4 R975H cells with or without doxycycline induction were suspended in sterilized PBS at a concentration of 2 × 107 cells/ml, and 100ul of cell suspension was inoculated subcutaneously into the flank of 4–6 weeks female nude mice. The weight of mice and tumor volume were measured every 3–4 days. After 6 weeks of injection, Mice were anesthetized by intraperitoneal injection of 100 µl of 9.1 mg/ml pentobarbital and then sacrificed by dissection, and the tumor xenografts were weighed.
Immunohistochemistry
Tumor tissue formed in the mouse xenograft model was harvested, embedded in paraffin and sectioned. The slices were incubated with 5%BSA for 30 min, and then primary antibody of CD163 or CD206 for 1 h. The slices were washed with PBS and then incubated with secondary antibody for 30 min. After incubation, the slices were washed with PBS, stained with DAB staining kit (TCI) and then Hematoxylin. The slices were then photographed under microscope. The antibody information is listed in Supplementary Table 4.
RNA-seq analysis
Total RNA extracted from the CHD4 R975H cell line treated with or without doxycycline was used for cell line RNA-seq analysis by the Centre for PanorOmic Sciences (LKS Faculty of Medicine, HKU) in triplicate. RNA-seq data of EC patients was acquired from cbioportal [Uterine Corpus Endometrial Carcinoma (TCGA, PanCancer Atlas)]. The RNA-Seq results of cell line and EC patients are listed in Supplementary Tables 5 and 6. The cell line RNA-Seq result was subjected to GSEA with multiple gene sets and GSEA summary analysis. The EC patients RNA-Seq result was subjected to immune infiltration analysis and differential expression analysis.
Structure prediction and analysis
The structure of CHD4 WT and CHD4 R975H were predicted and analyzed using USCF ChimeraX. CHD4 WT structure was fetched by ID Q14839 in Alphafold database. CHD4 R975H structure was produced by the Rotamers tool. The atom–atom contacts were analyzed by the Contacts tool. The name and charge of atoms were added in the IDATM type.
Statistical analysis
Somatic mutation data of EC patients was acquired from cbioportal (Endometrial Cancer (MSK, 2018), Endometrial Cancer (MSK, Cancer Discovery 2023), Endometrial Carcinoma (CPTAC, Cell 2020), Uterine Corpus Endometrial Carcinoma (TCGA, Firehose Legacy), Uterine Corpus Endometrial Carcinoma (TCGA, Nature 2013), Uterine Corpus Endometrial Carcinoma (TCGA, PanCancer Atlas), Uterine Clear Cell Carcinoma (NIH, Cancer 2017)). For each of the database, data are listed in Supplementary Tables 7–13. Duplicate data have been eliminated during the statistical process. For the immune infiltration analysis, data analysis was performed by either the Wilcoxon signed-rank test or the Kruskal–Wallis test. For other assays, data was presented as the mean ± standard deviation (S.D.) of at least three independent experiments. Data analysis was performed by unpaired two-tailed t tests with GraphPad Prism 8. p < 0.05 (*), p < 0.01 (**), p < 0.001 (***) and p < 0.0001 (****) indicated statistical significance.
Results
CHD4 mutation occurs frequently in endometrial cancer
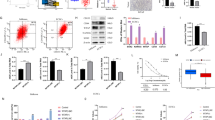
The frequency of CHD4 mutations in endometrial cancer was systematically investigated by analysing data from various databases associated with endometrial carcinoma in cBioPortal (https://www.cbioportal.org/). Across a comprehensive cohort of 2692 patients, 4.4% exhibited somatic mutations in CHD4 (119 out of 2692 patients) (Fig. 1A). Notably, within specific endometrial cancer datasets—the Uterine Corpus Endometrial Carcinoma (TCGA, PanCancer Atlas), Endometrial Carcinoma (CPTAC, Cell 2020), and Uterine Corpus Endometrial Carcinoma (TCGA, Nature 2013) datasets—the CHD4 mutation frequencies were 20.98% (111 out of 529 patients), 13.58% (11 out of 81 patients), and 9.43% (35 out of 371 patients), respectively (Fig. 1B). These collective findings underscore the high prevalence of CHD4 mutations among individuals with endometrial cancer.
The CHD4 R975H mutation occurs frequently in endometrial cancer. (A) Somatic mutation frequency of CHD4 in endometrial cancer patients in cBioPortal database. (B) Alteration frequency of CHD4 in various endometrial cancer datasets. (C) The corresponding number of samples with various CHD4 mutations. (D) Location of CHD4 R975H mutation on CHD4 schematic figure. (E,F) Prediction of the impact of the CHD4 R975H mutation on protein structure and function via two bioinformatics tools. (E) Polymorphism phenotyping v2. (F) Sorting intolerant from tolerant. (G) Prediction of CHD4 structural alteration induced by R975H mutation. Upper: Overall structure of CHD4 ATPase domain lobe 2. Middle: Detailed structure and interaction between 975R and 1182E in CHD4 WT. Lower: Detailed structure of 975H and 1182E in CHD4 R975H.
Upon closer examination of the mutation landscape, the missense mutation R975H emerged as the most common mutation, occurring 15 times within a pool of 271 total mutations (Fig. 1C). It locates in the lobe 2 of CHD4 ATPase domain, which is critical for DNA twisting and chromatin remodelling function of CHD4 (Fig. 1D)18. Consequently, our focus was on elucidating the impact of the CHD4 R975H mutation. Leveraging two bioinformatics analysis software—Polymorphism Phenotyping v2 (PolyPhen-2) and Sorting Intolerant from Tolerant (SIFT)—we assessed the predicted influence of the amino acid substitution 975aa R to H. Both analyses converged on a consensus: R975H is likely to be damaging to the structure and function of CHD4 (PolyPhen-2, Fig. 1E), with potential repercussions on CHD4 protein function (SIFT, Fig. 1F). We then compared the predicted structure of CHD4 wild type (WT) and CHD4 R975H using UCSF ChimeraX. In CHD4 WT, a salt bridge forms between the positively charged Nε in the side chain of 975R and the negatively charged Oε in the side chain of 1182E. The salt bridge could stabilize the α-helix structure of ATPase lobe 2. R975H disrupts the salt bridge and could cause instability of ATPase lobe 2 (Fig. 1G). In summary, our comprehensive analysis revealed a notable occurrence of CHD4 mutations in endometrial carcinoma, with particular emphasis on the recurrent CHD4 R975H mutation. Predictive analyses indicated that this mutation holds significance, potentially influencing the functional landscape of CHD4 in the context of endometrial cancer.
CHD4 R975H promotes EC cell proliferation in vitro and tumor growth in vivo
To further investigate the impact of the CHD4 R975H mutation on endometrial cancer, we engineered stable cell lines expressing CHD4 WT, CHD4 R975H, or short hairpin RNA targeting CHD4 (shCHD4). Given prior observations by Li et al., suggesting an increase in cell invasion and sphere formation capacity with both CHD4 R975H and shCHD4 in the EFE184 and SKUT-2 endometrial cancer cell lines15, we opted for the Ishikawa cell line, which is derived from an Asian EC patient. Inducible promoters within the SB vector (for CHD4 WT and CHD4 R975H) and the lentiviral vector (for shCHD4) were activated by doxycycline. The overexpression and knockdown efficacy of CHD4 WT and shCHD4 were validated through qRT–PCR (Fig. 2A), while the expression of the CHD4 R975H mutant was confirmed by western blot analysis (Fig. 4B).
The CHD4 R975H mutation promotes a cancer stem cell (CSC)-like phenotype in endometrial cancer cells. (A) CHD4 expression levels in the constructed CHD4 WT (left) and shCHD4 (right) cell lines determined via qRT‒PCR. (B–F) Transwell, colony formation and MTT assays of the CHD4 R975H, CHD4 WT and shCHD4 cell lines. (B) Stained invasive cells in the transwell assay. (C) Quantification of invasive cells. (D) Stained colonies in the colony formation assay. (E) Quantification of colonies. (F) Absorbance of the chromogenic product formazan produced by live cells at 490 nm. The number of live cells is linearly related to the absorbance. (G) Tumor weight and volume in the mouse experiment.
Subsequently, the CHD4 WT, CHD4 R975H, and shCHD4 cell lines were subjected to transwell, colony formation, and MTT assays. In transwell assay, CHD4 R975H demonstrated a proinvasive effect, in contrast with the inhibitory effects observed for CHD4 WT and shCHD4 (Fig. 2B,C). Colony formation assay revealed that CHD4 R975H promoted colony formation, while CHD4 WT and shCHD4 suppressed colony formation (Fig. 2D,E). Correspondingly, the MTT assay results indicated that CHD4 R975H enhanced cell proliferation, whereas CHD4 WT and shCHD4 repressed cell proliferation (Fig. 2F).
To further investigate the oncogenic role of the CHD4 R975H mutation, we conducted an in vivo experiment using a mouse xenograft model. Equal numbers of CHD4 R975H cells, either pre-induced with doxycycline or untreated, were subcutaneously injected into the flanks of nude mice. Six weeks post-injection, the results revealed that the expression of CHD4 R975H led to a significant increase in both tumor weight and volume compared to the control group (Fig. 2G). These comprehensive findings revealed that CHD4 R975H augments the CSC-mediated invasive and proliferative capabilities of endometrial cancer cells. In stark contrast, CHD4 WT and shCHD4 manifested opposing effects, collectively revealing the dynamic influence of CHD4 mutations on endometrial cancer cellular behaviors.
CHD4 R975H results in a cancerous transcriptome profile in EC cells
To unravel the underlying mechanisms through which CHD4 R975H enhances tumorigenesis in endometrial cancer (EC) cells, we conducted RNA-Seq analysis of total RNA extracted from the CHD4 R975H cell line cultured with or without doxycycline (Fig. 3A,B). The ensuing gene set enrichment analysis (GSEA) yielded insightful outcomes. A summary of the GSEA gene sets, spanning KEGG legacy gene sets, hallmark gene sets, and GO molecular function gene sets, provided a comprehensive overview (Fig. 3C). KEGG gene set analysis revealed that CHD4 R975H induced the expression of genes related to cancer-associated pathways. Hallmark gene sets further illuminated the spectrum of pathway activation, encompassing TNF-α signaling via NF-κB, KRAS signaling, the P53 pathway, Myc targets, and mTORC1 signaling—pathways commonly implicated in cancer19,20,21,22,23. GO functional gene sets confirmed the heightened growth factor activity induced by CHD4 R975H, a critical factor known to play a pivotal role in immune suppression within the tumor microenvironment24.
The CHD4 R975H mutant induces a cancer-specific transcriptome change in endometrial cancer cells. (A) Heatmap of gene expression profile of RNA-Seq result. (B) DEG volcano plot of RNA-Seq result. Differentially expressed genes (DEGs) are identified as | log2 fold change |> 0.5, p < 0.05. (C) Summary of GSEA results based on KEGG legacy gene sets, hallmark gene sets and GO molecular function gene sets. (D) GSEA results for Myc targets, mTOR signaling, TGF-β signaling, KRAS signaling, the p53 pathway, TNF-α signaling via NF-κB, growth factor signaling and EGFR signaling.
Individual gene set analyses for each pathway highlighted the robust activation induced by CHD4 R975H. Notably, enrichment of EGFR-associated gene sets revealed the activation of this driver of tumorigenesis25. Furthermore, consistent with the findings of Li et al., our GSEA confirmed the activation of TGF-β signaling by the CHD4 R975H mutant, a key contributor to endometrial tumorigenesis15 (Fig. 3D). Collectively, these results reveal that CHD4 R975H orchestrates the tumorigenic transcriptome in EC cells, unveiling the comprehensive landscape of activated pathways underlying its oncogenic influence.
CHD4 R975H stimulates oncogenic pathways and promotes cancer stemness
To corroborate the findings from our bioinformatics analysis, we conducted qRT–PCR assays to assess the expression levels of a set of genes in the CHD4 R975H, CHD4 WT, and shCHD4 cell lines. These genes, identified as upregulated by RNA-seq, encompass hallmarks of pathways implicated in our previous analyses. The pathways and their corresponding hallmark genes are detailed below: mTOR (SLC2A1, TRIB3, SLC7A5), KRAS (ALDH1A3, EPHB2, HBEGF), Myc (Myc), P53 (EPHA2, S100A10), TGF-β (PMEPA1, ITGB6, PDGFB), TNF-α signaling via NF-κB (CCND1), EGFR (SERPINE2, MICAL2), and growth factor (FGF18, MET). Additionally, SEMA3C, known for promoting cervical cancer growth through the activation of the p-ERK pathway26, was included due to its prominent variation in expression within the top 50 genes identified by RNA-seq analysis (Supplementary Table 4). The results unequivocally demonstrated a significant increase in the expression levels of all the aforementioned genes in the CHD4 R975H cell line. In contrast, limited upregulation was observed in the CHD4 WT and shCHD4 cell lines (Fig. 4A), suggesting that only CHD4 R975H effectively initiates the activation of tumorigenic pathways, while CHD4 WT and CHD4 depletion exert minimal influence on the oncogenic signaling network. The activation of mTOR and TGF-β signaling driven by CHD4 R975H was further validated through western blot analysis (Fig. 4B).
The CHD4 R975H mutant activates carcinogenic pathways and promotes the stemness of endometrial cancer cells. (A) Expression levels of marker genes in cancer-related pathways in the CHD4 R975H, CHD4 WT and shCHD4 cell lines measured by qRT‒PCR. (B) Expression levels of hallmark proteins involved in stemness (CD133), mTOR signaling (total S6K and pS6K) and TGF-βsignaling (β-catenin) in CHD4 R975H cells detected via western blotting. Original blots are presented in Supplementary Fig. 2. (C) Expression levels of stemness marker genes in the CHD4 R975H, CHD4 WT and shCHD4 cell lines measured by qRT‒PCR. (D) Flow cytometry analysis of CD133 staining in CHD4 R975H cell line. (E) Spheroids of Ishikawa and AN3-CA CHD4 R975H cell lines. (F) Number and diameter of spheroids.
In alignment with the findings of Li et al., Zhang et al., and Pratheeshkumar et al. on the role of CHD4 in cancer stemness in patients with endometrial cancer and other malignancies15,27,28, we sought to elucidate the impact of CHD4 R975H on the stemness of endometrial cells. We assessed the expression levels of a set of stemness marker genes (KLF4, NANOG, OCT4) in the CHD4 R975H, CHD4 WT, and shCHD4 cell lines. The results revealed a substantial increase in the expression levels of all stemness markers in the CHD4 R975H cell line, whereas no significant upregulation was observed in either the CHD4 WT or shCHD4 cell lines (Fig. 4C). This pronounced increase in stemness markers strongly suggested that CHD4 R975H actively promotes stemness in endometrial cancer cells. Intriguingly, CHD4 WT exhibited a suppressive effect on the expression of NANOG and OCT4, indicating an inhibitory influence on the stemness of endometrial cancer cells. Furthermore, the expression of CD133, a well-established marker of cancer stem cells, was elevated in the CHD4 R975H cell line, consistent with the qPCR results (Fig. 4B). Flow cytometry analysis of CD133 expression in CHD4 R975H cells revealed that, compared to the control group, the CHD4 R975H mutation significantly increased the proportion of CD133-positive cells (Fig. 4D). To further validate the promotive effect of CHD4 R975H on stemness traits in EC, we conducted spheroid formation assays in Ishikawa and AN3-CA CHD4 R975H cell lines. The results showed that CHD4 R975H expression markedly enhanced the growth of EC spheroids compared to the control group (Figs. 4E,F). Together, these findings provide compelling evidence that the CHD4 R975H mutation stimulates stemness in EC.
CHD4 R975H induces TAM polarization to M2-like macrophages
In light of the intricate composition of the tumor microenvironment (TME), encompassing diverse molecular constituents such as the extracellular matrix, growth factors, cytokines, and both nonimmune and immune cells29, we conducted an immune infiltration analysis focusing on multiple immune cell subgroups in endometrial cancer (EC) patients with the R975H mutation, other CHD4 mutations, or without any CHD4 mutation. The results revealed a discernible disparity in the distribution of various immune cells among the aforementioned patient categories (Supplementary Fig. S1). Given the multifaceted roles of tumor-associated macrophages (TAMs) and their ability to polarize towards either M1-like or M2-like macrophages, with M2-like macrophages well-established for their protumorigenic function in fostering immune suppression30, our attention turned to scrutinizing variations in the abundance of M2 macrophages in EC patients with or without CHD4 mutations. The findings indicated a significant enrichment of M2 macrophages in the tumor tissue of patients with CHD4 mutations compared to those without, suggesting a potential association between CHD4 mutation and M2-like macrophage polarization (Fig. 5A).
The CHD4 R975H mutation drives M2-like polarization in tumor-associated macrophages. (A) Abundance of M2 macrophages in tumor tissue of EC patients with or without CHD4 mutation. (B) Differential expression analysis of CCL and CXCL gene sets in tumor tissue of patients above. (C) Expression levels of M2 macrophage marker genes in the THP-1 cell line cocultured with the CHD4 R975H, CHD4 WT and shCHD4 cell lines measured by qRT‒PCR. (D) Flow cytometry analysis of CD68 + CD163/CD206 staining in cocultured THP-1 cell line. (E) Immunohistochemistry analysis of CD163/CD206 in tumor tissue of nude mice used in the mouse xenograft model. (F) Quantification of positive area of immunohistochemistry assay.
Recognizing the pivotal role of chemokines, secreted by cancer cells, as potent regulators influencing TAM polarization31, we further conducted a differential expression analysis of CCL and CXCL gene sets in EC patients with or without CHD4 mutations. The results unveiled upregulation of CCL4, CCL7, CCL8, CCL13, CXCL9, CXCL10, CXCL11, and CXCL13, coupled with downregulation of CCL14 in the tumor tissue of patients with CHD4 mutations in comparison to those without (Fig. 5B). This intriguing pattern aligns with the established functions of these chemokines: CCL7 activation promotes M2 macrophage enrichment in lung adenocarcinoma32; CCL14 induces M1 polarization and suppresses M2 polarization of TAMs in colon cancer33; in colorectal cancer, M2 macrophages release CXCL13, stimulating cancer metastasis34. The observed differential expression trends of these chemokines align cohesively with the heightened abundance of M2 macrophages in patients with CHD4 mutations. Collectively, these findings hint at the potential impact of CHD4 mutations within endometrial cancer cells on M2-like macrophages.
To explore whether CHD4 R975H could induce TAM polarization toward M2-like macrophages, we collected conditioned media from the CHD4 R975H, CHD4 WT, and shCHD4 cell lines. Subsequently, we separately cocultured THP-1 cells with conditioned medium from the aforementioned cell lines for 2 days and assessed the expression levels of CD23, CCL18, and CCL22, which are markers of M2 macrophages35,36. The results unequivocally demonstrated that CHD4 R975H significantly increased the expression levels of CD23, CCL18, and CCL22, whereas CHD4 WT and shCHD4 exhibited no discernible effect on their expression (Fig. 5C). This compelling evidence substantiates that CHD4 R975H actively stimulates TAM polarization toward an M2-like macrophage phenotype, whereas CHD4 WT and shCHD4 exert no discernable influence on TAM polarization. To further substantiate the inducible effect of CHD4 R975H on TAM M2-like polarization, we analysed the expression of CD163 and CD206 in cocultured THP-1 cells, both well-defined markers of M2 macrophages. The results revealed that, compared to the control group, CHD4 R975H expression significantly increased the ratio of CD68/CD163 and CD68/CD206 double-positive cells (Fig. 5D). To further confirm that CHD4 R975H triggers TAM M2-like polarization in vivo, we harvested tumor tissue from nude mice used in the mouse xenograft model and conducted immunohistochemistry of CD163 and CD206. The results showed that, compared to the control group, tumor tissue with CHD4 R975H expression exhibited higher expression of both CD163 and CD206. This demonstrates that CHD4 R975H expression in EC cells induces the M2-like polarization of infiltrated TAMs in vivo (Fig. 5E,F). Overall, these findings clearly demonstrate that the CHD4 R975H mutant induces TAM M2-like polarization in EC.
Discussion
Cancer initiation and progression hinge upon the accrual of somatic mutations, conferring cells with immortality and invasiveness37. In endometrial cancer, pivotal tumor suppressor genes, namely, PTEN and TP53, are the most frequently mutated genes, demonstrating their universality across various cancer types7,38,39. PTEN counteracts PI3K pathway activation by converting PIP2 to PIP3, while the somatic mutation of PTEN, which occurs in 69–80% of endometrioid tumors, underscores its critical role in endometrial cancer pathogenesis8. Similarly, TP53 mutations occur in more than 85% of serous endometrial cancers (SECs), a subtype of endometrial cancer8. As a transcription factor, p53 regulates the expression of a set of target genes and exerts function as a tumor suppressor. It has critical role in DNA repair, cell cycle arrest and apoptosis. In addition to losing tumor suppression function of wildtype p53, mutated p53 (mutp53) is known to have oncogenic functions. It forms complexes with NF-γ and p300 to transcriptionally activate NF-γ target genes, amplifying cell proliferation. Furthermore, mutp53 promotes metastasis, stemness, and immune suppression through diverse molecular mechanisms23.
Our analysis of endometrial cancer database data corroborates the frequent mutation of CHD4, prompting our investigation into the functional impact of CHD4 mutants in endometrial cancer8. Selecting CHD4 R975H due to its high prevalence in endometrial cancer patients, our study revealed a tumorigenic phenotype characterized by heightened proliferation, invasion, and colony formation capabilities compared to those of the CHD4 WT and shCHD4 cell lines. RNA-seq analysis revealed the activation of cancer-promoting pathways and the induction of a cancerous transcriptome in the CHD4 R975H cell line. Notably, this oncogenic function exhibited parallels with the known oncogenic traits of mutp53. Additionally, CHD4 R975H was found to promote the stemness and polarization of TAMs toward an M2-like phenotype, further corroborating its oncogenic effects.
The dearth of studies on CHD4 mutants, with Li et al.'s work being a notable exception15, motivated our comprehensive investigation. These findings, which emphasized that CHD4 mutants promote endometrial tumorigenesis via TGF-β signaling and increase cancer stem cell (CSC) traits, align with our results and conclusions. However, discrepancies exist, particularly regarding the similar functions of CHD4 knockdown in promoting cancer stemness and carcinogenesis. Our study, utilizing the Asian EC patient-derived Ishikawa cell line and White patient-derived AN3-CA cell line in endometrial cancer research40, contradicts this claim, suggesting potential context-dependent effects. While our study provides a multifaceted exploration of the oncogenic potential of CHD4 R975H, we acknowledge its limitations. Notably, we did not singularly focus on elucidating the mechanism of action for each pathway affected by CHD4 mutants, leaving these questions open for subsequent investigations.
Data availability
All data utilized for bioinformatical analysis are available in the public databases on cbioportal website (https://www.cbioportal.org/). All the data generated during this study are included in this article (and its Supplementary Information files).
References
Makker, V. et al. Endometrial cancer. Nat. Rev. Dis. Primers 7, 88. https://doi.org/10.1038/s41572-021-00324-8 (2021).
Katoh, M. & Katoh, M. WNT signaling and cancer stemness. Essays Biochem. 66, 319–331. https://doi.org/10.1042/EBC20220016 (2022).
Munoz-Galvan, S. et al. Downregulation of MYPT1 increases tumor resistance in ovarian cancer by targeting the Hippo pathway and increasing the stemness. Mol. Cancer 19, 7. https://doi.org/10.1186/s12943-020-1130-z (2020).
Genin, M., Clement, F., Fattaccioli, A., Raes, M. & Michiels, C. M1 and M2 macrophages derived from THP-1 cells differentially modulate the response of cancer cells to etoposide. BMC Cancer 15, 577. https://doi.org/10.1186/s12885-015-1546-9 (2015).
Sun, S. et al. 8-bromo-7-methoxychrysin reversed M2 polarization of tumor-associated macrophages induced by liver cancer stem-like cells. Anticancer Agents Med. Chem. 17, 286–293. https://doi.org/10.2174/1871520616666160204112556 (2017).
Rao, G. et al. Reciprocal interactions between tumor-associated macrophages and CD44-positive cancer cells via osteopontin/CD44 promote tumorigenicity in colorectal cancer. Clin. Cancer Res. 19, 785–797. https://doi.org/10.1158/1078-0432.CCR-12-2788 (2013).
Urick, M. E. & Bell, D. W. Clinical actionability of molecular targets in endometrial cancer. Nat. Rev. Cancer 19, 510–521. https://doi.org/10.1038/s41568-019-0177-x (2019).
Bell, D. W. & Ellenson, L. H. Molecular genetics of endometrial carcinoma. Annu. Rev. Pathol. 14, 339–367. https://doi.org/10.1146/annurev-pathol-020117-043609 (2019).
Sigal, A. & Rotter, V. Oncogenic mutations of the p53 tumor suppressor: The demons of the guardian of the genome. Cancer Res. 60, 6788–6793 (2000).
Tong, J. K., Hassig, C. A., Schnitzler, G. R., Kingston, R. E. & Schreiber, S. L. Chromatin deacetylation by an ATP-dependent nucleosome remodelling complex. Nature 395, 917–921. https://doi.org/10.1038/27699 (1998).
Polo, S. E., Kaidi, A., Baskcomb, L., Galanty, Y. & Jackson, S. P. Regulation of DNA-damage responses and cell-cycle progression by the chromatin remodelling factor CHD4. EMBO J. 29, 3130–3139. https://doi.org/10.1038/emboj.2010.188 (2010).
Wu, J. et al. CHD4 promotes acquired chemoresistance and tumor progression by activating the MEK/ERK axis. Drug Resist. Updat. 66, 100913. https://doi.org/10.1016/j.drup.2022.100913 (2023).
Ray Chaudhuri, A. et al. Replication fork stability confers chemoresistance in BRCA-deficient cells. Nature 535, 382–387. https://doi.org/10.1038/nature18325 (2016).
Xia, L. et al. CHD4 has oncogenic functions in initiating and maintaining epigenetic suppression of multiple tumor suppressor genes. Cancer Cell 31, 653-668 e657. https://doi.org/10.1016/j.ccell.2017.04.005 (2017).
Li, Y. et al. CHD4 mutations promote endometrial cancer stemness by activating TGF-beta signaling. Am. J. Cancer Res. 8, 903–914 (2018).
Kowarz, E., Loscher, D. & Marschalek, R. Optimized Sleeping Beauty transposons rapidly generate stable transgenic cell lines. Biotechnol. J. 10, 647–653. https://doi.org/10.1002/biot.201400821 (2015).
Kumar, P., Nagarajan, A. & Uchil, P. D. Analysis of cell viability by the MTT assay. Cold Spring. Harb. Protoc. https://doi.org/10.1101/pdb.prot095505 (2018).
Farnung, L., Ochmann, M. & Cramer, P. Nucleosome-CHD4 chromatin remodeler structure maps human disease mutations. Elife. https://doi.org/10.7554/eLife.56178 (2020).
Zou, Z., Tao, T., Li, H. & Zhu, X. mTOR signaling pathway and mTOR inhibitors in cancer: Progress and challenges. Cell Biosci. 10, 31. https://doi.org/10.1186/s13578-020-00396-1 (2020).
Madden, S. K., de Araujo, A. D., Gerhardt, M., Fairlie, D. P. & Mason, J. M. Taking the Myc out of cancer: Toward therapeutic strategies to directly inhibit c-Myc. Mol. Cancer 20, 3. https://doi.org/10.1186/s12943-020-01291-6 (2021).
Tang, D. et al. TNF-alpha promotes invasion and metastasis via NF-kappa B pathway in oral squamous cell carcinoma. Med. Sci. Monit. Basic Res. 23, 141–149. https://doi.org/10.12659/msmbr.903910 (2017).
Zhu, C. et al. Targeting KRAS mutant cancers: From druggable therapy to drug resistance. Mol. Cancer 21, 159. https://doi.org/10.1186/s12943-022-01629-2 (2022).
Zhang, C. et al. Gain-of-function mutant p53 in cancer progression and therapy. J. Mol. Cell Biol. 12, 674–687. https://doi.org/10.1093/jmcb/mjaa040 (2020).
Batlle, E. & Massague, J. Transforming growth factor-beta signaling in immunity and cancer. Immunity 50, 924–940. https://doi.org/10.1016/j.immuni.2019.03.024 (2019).
Sigismund, S., Avanzato, D. & Lanzetti, L. Emerging functions of the EGFR in cancer. Mol. Oncol. 12, 3–20. https://doi.org/10.1002/1878-0261.12155 (2018).
Liu, R., Shuai, Y., Luo, J. & Zhang, Z. SEMA3C promotes cervical cancer growth and is associated with poor prognosis. Front. Oncol. 9, 1035. https://doi.org/10.3389/fonc.2019.01035 (2019).
Zhang, J., Lv, X., Wei, B., Gong, X. & Chen, L. CHD4 mediates SOX2 transcription through TRPS1 in luminal breast cancer. Cell Signal 100, 110464. https://doi.org/10.1016/j.cellsig.2022.110464 (2022).
Pratheeshkumar, P. et al. CHD4 predicts aggressiveness in PTC patients and promotes cancer stemness and EMT in PTC cells. Int. J. Mol. Sci. https://doi.org/10.3390/ijms22020504 (2021).
Patel, H., Nilendu, P., Jahagirdar, D., Pal, J. K. & Sharma, N. K. Modulating secreted components of tumor microenvironment: A masterstroke in tumor therapeutics. Cancer Biol. Ther. 19, 3–12. https://doi.org/10.1080/15384047.2017.1394538 (2018).
Mehla, K. & Singh, P. K. Metabolic Regulation of macrophage polarization in cancer. Trends Cancer 5, 822–834. https://doi.org/10.1016/j.trecan.2019.10.007 (2019).
Qin, R. et al. Role of chemokines in the crosstalk between tumor and tumor-associated macrophages. Clin. Exp. Med. 23, 1359–1373. https://doi.org/10.1007/s10238-022-00888-z (2023).
Wu, Z., Bai, X., Lu, Z., Liu, S. & Jiang, H. LINC01094/SPI1/CCL7 axis promotes macrophage accumulation in lung adenocarcinoma and tumor cell dissemination. J. Immunol. Res. 2022, 6450721. https://doi.org/10.1155/2022/6450721 (2022).
Li, N. et al. C-C motif chemokine ligand 14 inhibited colon cancer cell proliferation and invasion through suppressing M2 polarization of tumor-associated macrophages. Histol. Histopathol. 36, 743–752. https://doi.org/10.14670/HH-18-348 (2021).
Zhao, S. et al. Tumor-derived exosomal miR-934 induces macrophage M2 polarization to promote liver metastasis of colorectal cancer. J. Hematol. Oncol. 13, 156. https://doi.org/10.1186/s13045-020-00991-2 (2020).
Murphy, B. S. et al. Azithromycin alters macrophage phenotype. J. Antimicrob. Chemother. 61, 554–560. https://doi.org/10.1093/jac/dkn007 (2008).
Arabpour, M., Saghazadeh, A. & Rezaei, N. Anti-inflammatory and M2 macrophage polarization-promoting effect of mesenchymal stem cell-derived exosomes. Int. Immunopharmacol. 97, 107823. https://doi.org/10.1016/j.intimp.2021.107823 (2021).
Stratton, M. R., Campbell, P. J. & Futreal, P. A. The cancer genome. Nature 458, 719–724. https://doi.org/10.1038/nature07943 (2009).
Chen, L., Liu, S. & Tao, Y. Regulating tumor suppressor genes: Post-translational modifications. Signal Transduct. Target Ther. 5, 90. https://doi.org/10.1038/s41392-020-0196-9 (2020).
Iranzo, J., Martincorena, I. & Koonin, E. V. Cancer-mutation network and the number and specificity of driver mutations. Proc. Natl. Acad. Sci. U. S. A. 115, E6010–E6019. https://doi.org/10.1073/pnas.1803155115 (2018).
Kozak, J., Wdowiak, P., Maciejewski, R. & Torres, A. A guide for endometrial cancer cell lines functional assays using the measurements of electronic impedance. Cytotechnology 70, 339–350. https://doi.org/10.1007/s10616-017-0149-5 (2018).
Acknowledgements
This work was supported by the National Natural Science Foundation of China (31970583 to JZ) and the Research Grants Council of Hong Kong China [HKU 17103920 General Research Fund (GRF) to JZ].
Author information
Authors and Affiliations
Contributions
J.Z. and Q.Z. designed the research; Q.Z. performed the bioinformatics tool analysis, structural analysis, functional assays, W.B. assays and qRT‒PCR assays of EC cells, conducted the data analysis except public databases and wrote the manuscript; F.Z. performed the RNA-seq analysis and data analysis of public databases. Y.T. performed the RNA-seq analysis of EC cell lines; D.S. conducted the qRT‒PCR analysis of THP-1 cells; J.Z. and Y.T. revised the manuscript. All the authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zhang, Q., Zhu, F., Tong, Y. et al. CHD4 R975H mutant activates tumorigenic pathways and promotes stemness and M2-like macrophage polarization in endometrial cancer. Sci Rep 14, 18617 (2024). https://doi.org/10.1038/s41598-024-69233-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-69233-6
- Springer Nature Limited