Abstract
Mammalian dentition exhibits distinct heterodonty, with more simple teeth located in the anterior area of the jaw and more complex teeth situated posteriorly. While some region-specific differences in signalling have been described previously, here we performed a comprehensive analysis of gene expression at the early stages of odontogenesis to obtain complete knowledge of the signalling pathways involved in early jaw patterning. Gene expression was analysed separately on anterior and posterior areas of the lower jaw at two early stages (E11.5 and E12.5) of odontogenesis. Gene expression profiling revealed distinct region-specific expression patterns in mouse mandibles, including several known BMP and FGF signalling members and we also identified several new molecules exhibiting significant differences in expression along the anterior–posterior axis, which potentially can play the role during incisor and molar specification. Next, we followed one of the anterior molecules, SATB2, which was expressed not only in the anterior mesenchyme where incisor germs are initiated, however, we uncovered a distinct SATB2-positive region in the mesenchyme closely surrounding molars. Satb2-deficient animals demonstrated defective incisor development confirming a crucial role of SATB2 in formation of anterior teeth. On the other hand, ectopic tooth germs were observed in the molar area indicating differential effect of Satb2-deficiency in individual jaw regions. In conclusion, our data provide a rich source of fundamental information, which can be used to determine molecular regulation driving early embryonic jaw patterning and serve for a deeper understanding of molecular signalling directed towards incisor and molar development.
Similar content being viewed by others
Introduction
Tooth development is a complex process involving close interaction between the oral mesenchyme and epithelium during prenatal stages. Odontogenesis starts at about the 11th day of embryonic development by a thickening of the oral epithelium followed by epithelial protrusion into the mesenchyme and the formation of buds surrounded by condensed mesenchyme originating from the neural crest1. Tooth development is initiated by a set of reciprocal and precisely synchronised interchanges of a large number of signalling molecules from several gene families, the most explored being BMP, WNT, FGF and HH2.
In nearly all mammalian species, there are differences in the development and final morphology of the anterior and posterior teeth. Mammalian molars and premolars are multicuspidal teeth with several roots, while incisors and canines are typically single-rooted teeth. Previously, several homeobox genes have been found to display specific expression in the anterior (area of forthcoming incisors and surrounding tissue) and posterior (area of forthcoming molars including surrounding tissues) jaw development in mice. Epithelial Bmp4 together with mesenchymal Msx1 and Msx2 are expressed in the anterior region of the jaw, the future incisor area. On the other hand, epithelial Fgf8 together with mesenchymal Barx1, Dlx1 or Dlx2 are localised in the posterior jaw region corresponding to the future molar area. The changes in expression of these homeobox genes cause shape deformities of teeth or hypodontia3,4,5,6,7 and loss of function of these genes leads to developmental arrest of odontogenesis8,9. Misexpression of Barx1 in the presumptive incisor mesenchyme at E10 results in the transformation of tooth identity from incisor to molar10.
Also in humans, there are differences in tooth shape along the anterior–posterior jaw axis with more complex teeth located posteriorly. However, in some patients, abnormalities in the shape of teeth can occur, which includes an anomaly called the talon cusp, a developmental dental disruption in which an accessory cusp arises from the cingulum or cementoenamel junction of the anterior teeth in the maxilla or mandible11. The aetiology of the talon cusps is not totally clear. Talon cusp arises during the morphodifferentiation stage of tooth development. It usually occurs as an isolated anomaly rather than being part of any disorder12. Nevertheless, the anomaly has been reported in patients with Mohr syndrome12 (Xp22 gene), Rubinstein–Taybi syndrome13 (CREBBP or EP300 gene), Sturge–Weber syndrome14 (GNAQ gene) and Ellis–van Creveld syndrome15 (EVC or EVC2 gene). Here, we ask which genes underlay anterior–posterior dental patterning along the jaw. Especially, we focus on the anterior genes as more severe dental and craniofacial defects have been described in human patients in this area16.
Until now, gene expression in orofacial structures has been well studied by several methods17,18; however, genome-wide analysis is a necessary tool to obtain information about molecules not yet associated with a certain area or domain. It is also considered an important tool for clinical medicine and gene expression profiling, which can help to identify diagnostic or prognostic biomarkers, classify diseases, monitor the response to therapy and understand the mechanisms involved in the genesis of disease processes19,20. This method has also been used to study the development of craniofacial structures17,21,22. However, the global gene expression differences between the anterior and posterior lower jaw areas separately have not yet been evaluated. This approach can help us uncover new genes already contributing to the specification of anterior and posterior jaw areas early during development and enable us to reveal new players involved in complex tooth patterning along the anterior–posterior axis.
Here, we compared the gene expression between the anterior (future incisor) and posterior (future molar) parts of the mouse mandible in the initial stages of dental development with the aim to expand recent knowledge of molecular patterning in the area-specific matter. We first introduced and reviewed the expression of top genes revealed by microarray screen, which are known for their participation during incisor and molar development and can serve us to validate our profiling. Moreover, we collected facts about their expression during development, their influence on cell functions and the impact of gene mutations on embryo development. We also paid attention to genes which have not been yet disclosed as having a role in tooth development.
To further follow the role of uncovered genes in odontogenesis, we selected SATB2 for a more detailed protein expression analysis and also evaluated the effect of Satb2-deficiency on incisor and molars development. SATB2 is a member of the special AT-rich binding proteins family, which are associated with nuclear MARs (matrix attachment regions) mediating higher-order chromatin integration and therefore modulating target gene expression. SATB2 participates in the process of regulating several transcription factors both during early embryogenesis and later in the process of cell differentiation23, therefore, we followed the role of this protein as a potential molecule involved in regulation of early odontogenesis.
Results and discussion
Distinct expression profiles of anterior and posterior areas during the tooth initiation stages
Whole-genome gene expression profiling was performed in the anterior (forthcoming incisor) and posterior (forthcoming molar) areas of the mouse jaw (Fig. 1). We selected the mouse mandible for our study as there are only two types of teeth—incisors and molars with a large diastema between them, which help us to easily separate the two areas from each other (Fig. 1A and B). We collected three samples for each area at two different stages, E11.5 and E12.5, when tooth development is initiated and distinct tooth domains are established.
Mandibular dissection and gene expression analysis of dissected zones. (A) Lower jaw of an adult mouse contains one incisor (I) and three molars (M1–M3) in each half with a large diastema between anterior and posterior part.(B) Dental germs of the first molar (M1) and incisor (I) are present in the mandible at E11.5 and E12.5 day old mouse embryos, when dissection for gene expression analyses were performed. These lower jaws were dissected into two segments—anterior part including incisors and posterior zone with molars. (C) Heat map of the genes that were differentially expressed in at least one comparison. (D) Principal component analysis revealed clear separation between E11.5 and E12.5 embryos along the first principal component (PC1) and between the molar and incisor parts along the second principal component (PC2).
The samples from individual stages/areas clustered well together (Fig. 1). We observed; more significant variability in samples collected at stage E11.5 in contrast to E12.5 most probably because it is more difficult to collect material from exactly the same areas at earlier stages of development. Interestingly, samples cluster together based on the developmental stage, i.e., the anterior at E11.5 is more similar to the posterior area at this stage than to the anterior area at stage E12.5 (Fig. 1C). This indicates that gene expression profiles changed very significantly during one day. However, we detected 146 genes (350 transcripts) significantly differentially expressed between posterior and anterior areas at E11.5 and 252 genes (631 transcripts) at E12.5 (Tables S1, S2, S3 and S4).
To validate our findings, we performed an extensive literature survey for 20 highly expressed genes from our data sets for each collection group (Tables S5, S6, S7 and S8). Large number of top genes from microarray analyses were previously associated with craniofacial development or odontogenesis. Moreover, we validated several differentially expressed genes using RT-qPCR (Fig. S1). Gene expression pattern was same as observed in microarray analyses for the gene Dlx5 (significantly upregulated in the molar area compared to the incisor region) as well as for genes downregulated in the molar area, such as Dlg2, Dpep1 and Satb2. RT-qPCR together with the literature screen confirmed strong agreement with our array data and indicated high credibility obtained data.
Expression of neural system-associated molecules is enhanced in the posterior (molar) region
Our microarray analysis determined the expression of genes previously described in posterior areas, such as Barx1 and Barx2 (Figs. 2, S2, Table S3). These nuclear mesenchymal genes are important for craniofacial development including odontogenesis24,25. Barx1 expression in the head region is detected in spatially restricted areas of the first and second branchial arches, before any apparent cellular or morphological differentiation at embryonic day 10.5. Later in development, BARX1 is involved in the development of the primary and secondary palate, the molar tooth papillae and the stroma of the submandibular gland26. Barx1−/− mouse embryos display mid-gestational lethality and a spectrum of anomalies27; mutations of Barx1 lead to cleft lip palate28. Barx1−/− embryos die around embryonic day E13.5, so the effect of Barx1 deficiency on the differentiation of dental tissues could not be analysed. We observed higher expression in the posterior region in both stages analysed; this difference was greater and statistically significant at E12.5 (Figs. 2 and S2). Interestingly, this gene was previously associated with anterior–posterior patterning in the jaw. The inhibition of BMP signalling early during mandible development by the action of exogenous Noggin protein results in ectopic Barx1 expression in the distal, presumptive incisor mesenchyme and a transformation of tooth identity from incisor to molar24.
The distal-less (DLX) family of nuclear homeodomain transcription factors were also found to be expressed in the molar mesenchyme. Our analyses uncovered higher expressions of Dlx1, Dlx4, Dlx5 and Dlx6 (Figs. 2, S2 and S3). Differences in expression of these Dlx genes were observed in the molar compared to the incisor region by the earlier E11.5 stage but increased at the later stage analysed, E12.5, where they reached statistical significance (Tables S1 and S3). Regionally restricted expression of Dlx homeobox-containing genes was also found previously to be responsible for generating early polarity in the first branchial arch29. Reciprocal signalling interactions between the ectoderm and the underlying ectomesenchymal cell populations control the odontogenic developmental program, from early patterning of the future dental axis to the initiation of tooth development at specific sites within the ectoderm29. Loss-of-function mutation in DLX genes results in abnormalities of the first branchial arch derivatives and the disruption of molar development6,30. A crucial role of Dlx1 and Dlx2 in patterning of the jaw and the induction of a specific type of dentition has been proven in mice with targeted null mutations of Dlx1 and Dlx2, which do not develop maxillary molars, but whose incisors and mandibular molars are normal7. Dlx1 and Dlx2 genes have been proposed to participate in the patterning of dentition through an “odontogenic homeobox gene code” by specifying the molar area. Rescue of the development of lower molars has been proposed to occur by compensation of their function by other Dlx genes in this area.
The gene Nr2f1 is also potentially important for jaw patterning as its expression profile was similar to that of Barx1 and Dlx1 genes (Figs. 2, S2 and S3). The expression of all three genes was higher posteriorly than anteriorly in both stages. At the same time, there was a decrease in expression within a day, as the expression in E12.5 was lower than in E11.5 for all three genes, especially in the anterior area (Fig. 2, Tables S1 and S3). The protein encoded by the gene Nr2f1 is a nuclear hormone receptor and transcriptional regulator. During development, its expression has been detected in telencephalon neurons31,32. Upregulation of the Nr2f1 gene and its antisense long non-coding RNA in murine neural crest cells results in neurocristopathy33. Mutation of Nr2f1 is associated with Bosch–Boonstra–Schaaf optic atrophy syndrome (BBSOAS), the common features of which include developmental delay, intellectual disability, hypotonia, optic nerve atrophy, attention deficit disorder, autism spectrum disorder, seizures, hearing defects, spasticity, thinning of the corpus callosum and the presence of micro- and retrognathia with large-appearing teeth34. The loss of Nr2f1 significantly impacts craniofacial structures and disrupts lower jaw development, as has been shown for zebrafish35; however, the role of Nr2f1 in odontogenesis has not yet been evaluated.
Sim2 also appears to have the potential to be involved in jaw patterning as its expression profile resembles that of Barx1 and Dlx1 genes (Figs. 2 and S3). Sim2 encodes a transcription factor that is a master regulator of neurogenesis. It has been detected in the brains of mouse embryos31 and pharyngeal arches36. Overexpression of the Sim2 gene brings about some of the phenotypic deformities characteristic of Down syndrome. SIM2 mRNA is detected in multiple brain regions and has been linked to deformities in individuals with Down syndrome. It is also expressed in the epithelia of the palate, oral cavity, and tongue, as well as the mandibular and hyoid bones37. Knockout of this gene causes malformations, particularly in the craniofacial area. Sim2−/− individuals display either a full or partial secondary palate cleft and malformations in the tongue and pterygoid processes of the sphenoid bone37. On other hand, tooth development in Sim2−/− mice was not affected37.
Genes with the highest level of expression in the posterior region at E11.5 were associated with neurogenesis. Neurod1 and Neurod4 (neuronal differentiation 1 and 4) are transcription factors contributing to the regulation of several cell differentiation pathways in the neural system (Fig. 2). NEUROD1 is, together with PAX6 or SIX3, required for the regulation of amacrine cell fate specification or dendrite morphogenesis. Moreover, there is expression of genes such as Fabp7 (fatty acid binding protein 7, brain; or brain lipid binding protein, Blbp), Nefl (neurofilament light polypeptide), Neurog1 (neurogenin 1), Gbx2 (gastrulation brain homeobox 2) and Elavl4 (ELAV-like neuron-specific RNA binding protein 4) (Fig. 2, Tables S1 and S3). Also, genes playing a role in microtubule dynamics and signal transduction such as Nefm (neurofilament medium polypeptide), Ina (internexin neuronal intermediate filament protein, alpha), Snap25 (synaptosomal-associated protein 25) or Stmn2 (Stathmin 2) are highly expressed in the caudal region associated with the posterior area. Neurog1 null mice fail to form trigeminal ganglia38; however, tooth development exhibits only subtle changes, indicating that the innervation is not driving early tooth development39. Neurogenesis and axonogenesis in the mandibular prominence are boosted during these early stages and nerves progressively extend from the caudal region to the more rostral area, as demonstrated by our expression data uncovering very low expression in the anterior area. Therefore, the differential neuronal marker profile reflects the progression of jaw development in the rostral direction.
Members of the BMP pathway are enriched in the anterior (incisor) area of the jaw together with Wnt inhibitors
The gene with the highest level of expression in the anterior region at E11.5 was aristaless-like homeobox 3 (Alx3), a nuclear protein with a homeobox DNA-binding domain that functions as a transcriptional regulator40 (Fig. 3; Table S2). Alx3 exhibited higher levels of expression in the incisor area when compared to the posterior jaw domain also at a later analysed stage at E12.5 (Figs. 3, S3 and S4). The gene Alx3 has previously been found to play a role in neural crest-derived mesenchyme, frontonasal head mesenchyme, the development of the first and second pharyngeal arches and their derivatives40 and in the formation of the nose and jaws41. In humans, the Alx3 mutation is associated with frontonasal dysplasia42, which is characterised by frontonasal malformation, a wide nasal bridge, a short nasal ridge and a midline notch in the upper lip, which confirms the significance of this gene for anterior structure development. However, no phenotypic abnormalities in the lower jaw have yet been reported in Alx3-deficient mice43.
Gene expression analysis also uncovered that several members of the TGF-beta family are expressed in the anterior area at E11.5, such as Bmp4 (2.17-fold change), Msx1 (1.98-fold change), Msx2 (1.73-fold change) and Bambi (1.63-fold change), and they were also upregulated at E12.5 in the anterior area (Figs. 3, S5; Tables S2 and S4). High expression of these mesenchymal transcription factors is consistent with the results of previous studies44. They have also been associated with craniofacial development, particularly odontogenesis in the anterior part of the jaw41,45. Defects in these genes cause distinct malformations in tooth development, where Msx1 failure leads to ectodermal dysplasia, tooth agenesis and orofacial clefting46. Homozygous Msx1-deficient mice exhibit complete secondary cleft palate, failure of incisor development and bud-stage arrest of molar development8. In humans, heterozygous loss-of-function mutations in MSX1 cause selective tooth agenesis, which often preferentially affects the posterior molars and second premolars3,4,5. Although mice heterozygous for Msx1 do not exhibit tooth defects, many Msx1+/−compound heterozygous mice lack third molars47.
The Msx2 expression pattern closely resembled that of Msx1 with higher expression located in the anterior area (Figs. 3 and S4). Mutations in Msx2 lead to specific abnormalities in teeth, but mainly affecting cell differentiation in the enamel organ48. In Msx2−/− mice, epithelial cells exhibit divergent alterations in the crown and root. Reduced expression of enamel matrix proteins in the crown is associated with the disappearance of ameloblasts48 resulting in the degeneration of the ameloblast and ultimately the enamel organ49.
Cited1 (Cbp/p300-interacting transactivator with Glu/Asp-rich carboxy-terminal domain 1), which functions as a transcriptional coactivator, was also upregulated in the rostral area (Fig. 3; Tables S2 and S4). CITED1 was previously found to interact with BMPR2 and SMAD4 to activate the CITED1-BMPR2-SMAD1/5/8 axis50 and play a role in activating the BMP signalling pathway. Moreover, it is closely related to transcriptional regulation by the AP-2 (TFAP2) family of transcription factors, and Tfap2c was also upregulated in the anterior area of the mandible (Fig. 3).
In the anterior area, numerous Wnt signalling members were found to be highly expressed (Fig. 3; Tables S2 and S4) and regulation of canonical Wnt signalling is known to be key for numerous processes during odontogenesis51. Inhibitors of the canonical Wnt signalling pathway were found to be highly expressed in this area, such as Dickkopf (Dkk1 and Dkk4). DKKs play a vital role in the anterior–posterior axial patterning as they locally inhibit canonical Wnt signalling52. Dkk1 deficiency leads to the development of smaller craniofacial structures and induction of clefts53. Dkk4 is a Wnt/β-catenin target gene54 and it is expressed in the enamel knots in the dental epithelium 55. Dkk1 exhibited the same pattern of expression differences between the two areas as other Wnt inhibitors: Wif1, Sfrp4, Lef1 or Sostdc1 (also known as Ectodin and Wise) (Fig. 3, Tables S1, S2, S3 and S4). WIF1 (Wnt inhibitory factor 1) is a secreted Wnt antagonist, which localises to the dental epithelium and mesenchyme during early tooth development; knockdown of Wif1 leads to promotion of apoptosis, resulting in delayed enamel nodule formation, during early tooth development56. LEF1 (lymphoid enhancer binding factor 1) is a transcription factor which mediates the Wnt signalling pathway by association with its coactivator beta-catenin. Loss of Lef1 leads to developmental arrest of the tooth at an early stage, as it is critical for mediating the epithelial–mesenchymal interaction during tooth morphogenesis57.
Moreover, members of non-canonical Wnt signalling pathway were upregulated in the anterior area, such as Wnt5a (wingless-type MMTV integration site family, member 5A), Wnt4 and Rspo2 (Fig. 3; Tables S2 and S4). Mutating the Rspo2 gene in mice leads to supernumerary lower molars58, inactivation of Rspo3 causes the loss of lower incisors and simultaneous deletion of Rspo2 and Rspo3 induces disruption of all tooth development59.
From the above-mentioned genes, Dkk4, Lef1 and Sfrp4 showed no differences at the earlier stage analysed (E11.5), but exhibited significant differences at the later stage (E12.5) (Fig. 3). On the other hand, significant differences between regions were found in case of Rspo2 just for the earlier stage, while differences were only small at E12.5. Our array therefore uncovered time-dependent dynamic recruitment of Wnt pathway members during anterior–posterior patterning of the lower jaw.
We also observed differences in Pax3 expression between incisor and molar areas (Fig. 3, S6; Tables S2 and S4). PAX3 is a downstream factor of β-catenin and a member of the paired box (PAX) family of transcription factors60. Mutations of PAX3 are associated with Waardenburg syndrome in humans, the features of which include teeth with a conical appearance, oligodontia involving both the lower lateral incisors and taurodontism61,62.
Our analysis also discovered numerous new genes potentially involved in tooth patterning that have not previously been associated with incisor development yet and it will be necessary to evaluate their role in tooth patterning in future. The expression pattern of some newly uncovered genes at the anterior area, such as Dlg2, exhibited similarities to that of Bmp4 and Pax3, where the expression of these genes was stronger in the anterior area in contrast to the posterior area (Figs. 3, S4; Tables S2 and S4) suggesting possibly comparable function in jaw patterning (Fig. 3). Dlg2 encodes a member of the membrane-associated guanylate kinase (MAGUK) family and its protein forms a heterodimer with a related family member that may interact at postsynaptic sites63. Its expression was previously detected in the mouse mandible, upper jaw and neural ganglia in embryonic stage 14.564; however, it has not been analysed in an odontogenesis context yet.
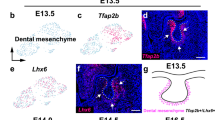
Satb2 expression is enriched in the anterior area at the mRNA as well as protein level
One of genes exhibiting strong expression in the anterior jaw area in contrast to the posterior area at both analysed stages was Satb2 (SATB homeobox 2) (Fig. 3; Tables S2 and S4).
SATB2 protein is a transcription factor and chromatin modulator. Its function at the molecular level is to attach itself to chromatin and change its 3D arrangement, most often into loop shapes65. This way, SATB2 is able to bring otherwise distant genes closer together, thus ensuring their joint expression. Its second function is to bind various proteins into complexes, which it directs and binds to specific sites on the DNA. These are most commonly gene promoters and enhancers66,67. This way, SATB2 is involved in regulating numerous genes during early embryogenesis and plays a crucial role in cell differentiation23. Expression of this gene during early development has previously been found to be restricted to the medial portion of the first to fourth pharyngeal arches23. It also plays a role in later craniofacial patterning and development, development of hard tissues, primarily bones, palate formation, osteoblast differentiation, and maturation68,69. Likewise, it has been associated with several morphological alterations during tooth development. Moreover, nonsense mutation of SATB2 has been found in patients with multiple craniofacial defects, cleft palate, osteoporosis and cognitive defects23,70,71. SATB2 is also associated with 2q32q33 microdeletion syndrome, including tooth abnormalities72 such as abnormally sized or shaped teeth, tooth crowding, supernumerary teeth or oligodontia73. SATB2 enhances migration and invasion in osteosarcoma by regulating genes involved in cytoskeletal organisation74.
Here, we first confirmed differences in Satb2 expression level found between anterior and posterior areas observed in gene expression profiling while using RT-qPCR and Satb2 expression was found to change with the progress of development (Fig. S1).
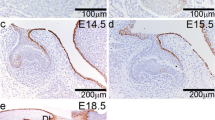
Also, at the protein level, SATB2 exhibited distinct differences in craniofacial areas on sagittal sections in an anterior–posterior direction, with stronger expression in the anterior area (Fig. 4). At epithelial thickening of the oral epithelium (E11-E12), SATB2-positive cells were located in the mesenchyme surrounding developing incisors and the rostral part of palatal shelves (Fig. 4). Only a few positive cells were found in the posterior area surrounding molars (Fig. S7). A transversal section revealed SATB2-positive cells located medially in the mesenchyme from the future molars and in forming palatal shelves (Fig. S7). Similarly, at later stages (E13-E15), areas of high SATB2 positivity were evident in the mesenchyme around teeth, with stronger expression around incisors and asymmetrical expression around molar germs, and the intensity of the signal increased at later developmental stages (Figs. 4 and S7). Observed protein expression pattern of SATB2 described here corresponds to previous findings in the palatal shelves where positive cells appeared at stage E10.5 and the signal intensity increased until E13.568. While the influence of Satb2-deficiency on skeletogenesis has already been elucidated23; the impact on developmental processes during odontogenesis remains incompletely understood therefore we focused on its role in incisor and molar development.
SATB2 protein expression during odontogenesis in the incisor area. Immunohistochemical analyses of SATB2 protein expression in the anterior area of the jaw. Sagittal sections of head at stage from E11 up to E15. SATB2-negative nuclei are stained by Hematoxylin (blue), SATB2-positive nuclei are visualized by DAB (brown). Scale bar = 100 μm.
deficiency causes abnormalities of the incisor and also molar development
To investigate the potential contribution of SATB2 in teeth development, we examined Satb2-/- mice (Figs. 5, S8, S9, S10 and S11). Therefore, our study endeavours to unveil potential morphological variations in the developing incisor and molar regions of Satb2-/- animals. Loss of Satb2 led to an abnormal craniofacial pattern in our animals, most notably associated with a shorter mandible at all analyzed stages and cleft palate (Fig. 5) similarly as was previously described23. Such phenotype has also been found in human patients with SATB2-associated syndrome (SAS, Glass syndrome). When considering intraoral dental concerns, individuals with SAS frequently manifest issues such as crowding, enlarged upper incisors, irregular dental morphology, hypodontia and, on a less frequent basis, delayed tooth eruption, tooth fusion, and supernumerary teeth75.
Abnormalities in odontogenesis in Satb2-deficient animals in micro-CT. The tooth germs (pink color) and the mesenchymal dental papilla reflecting the shape of the future teeth (white) were manually segmented in the micro- CT data. In the Satb2-deficient mice of both analyzed stages, the ectopic molar teeth are clearly distinguishable in the maxillary area. The incisor teeth in both mandible and maxilla are reduced in size in the Satb2-deficient mice compared to the WT mice. Scale bars: lower resolution = 1 mm, higher resolution = 0.5 mm.
Therefore, here we proposed a differential effect of this deficiency on tooth development in anterior and posterior jaw segments based on strong distinction of expression pattern in these areas. We used micro-CT analyses, which enabled us to follow soft and hard tissues on the same samples (Fig. S8). Segmentation analysis of incisor and molar epithelium while using micro-CT images allowed us to perform 3D analysis of volumes or distances between structures. Incisors were found to be present in Satb2-/- mice, albeit as extremely reduced structures in both the upper and lower jaws (Fig. 5). Furthermore, we observed molar germs exhibiting altered morphology. The epithelial compartments of the first molars were also significantly diminished in Satb2-/- mice (Fig. 5). However, the most unexpected finding from this analysis is the occurrence of ectopic teeth (Fig. 5) at both analysed stages in Satb2-/- mice.
Next, we performed Wall thickness analysis (software fits spheres of different diameters in the segmented tooth), revealing distinct changes in tooth thickness in Satb2−/− mice (Fig. 6) compared to Satb2+/+. Differences were observed in both tooth types, with the more striking decrease of wall thickness observed in the incisor area (Fig. 6).
Wall thickness analyses of teeth. Wall thickness analysis of the tooth germs in the E16.5 and E18.5 stages displays that the molars in the Satb2-deficient mice is similar in shape to the wt mice, but the thickness of walls in both stages are greatly reduced in the Satb2-deficient mice. The largest difference is visible in the incisors, which exhibit arrested development in Satb2-deficient mice and thus the quantified wall thickness is incomparably smaller. Scale bar = 650 µm.
We also evaluated the volume and surface of segmented teeth (Figs. S12, S13 and S14). While Satb2−/− animals exhibited a smaller volume as well as the surface area of incisors, the volume and surface area of mutant molars were only slightly decreased compared to Satb2+/+ animals (Figs. S13 and S14). Length of the molar area in Satb2-/- mice was reduced in both jaws compared to WT individuals at the E18.5 stage (Figs. S15 and S16) however when we included ectopic teeth, the size of the molar area was almost the same as WT animals (Fig. 7).
Measurements of molar area length and distances between incisor and molar area including ectopic tooth. (A–C) In the micro CT data, the lengths of the molars were measured at stage E18.5. (A) The length of the molar was measured in the 3D view between the most anterior and posterior points of the tooth germ in Satb2+/+ mice. (B) In the Satb2-deficient mice, the length of the molar was measured including the ectopic tooth. (D–E) The distance between the molar and the incisor was defined by the length between the most anterior part of the molar and the most anterior part of the incisor in Satb2+/+ mice (D). E In case of the Satb2-deficient mice, the ectopic tooth was included in measurements. Scale bars: 500 µm.
Based on these results, our analysis provides evidence that neglected molar area is severely affected in Satb2-/- mice. A question, which remains, is whether SATB2 is controlling the size of these structures or whether this change is caused secondary by space restriction in the jaws or limited cellular resources because of ectopic teeth induction.
Ectopic teeth are located in the diastema region in Satb2 -/- animals
Our analysis further revealed that the absence of Satb2 disrupts the development of the mouse diastema, the toothless space between the first molar and incisor, which was associated with two phenomena. Firstly, the absolute lengths of diastemas (distance between incisor and molar) were shorter in Satb2-/- mice than in WT animals in both the upper and lower jaws (Fig. S17), indicating an overall reduction of diastema length in the mutants. Secondly, a comparison between the lower and upper jaws within individual mice indicates that Satb2−/− mice exhibited a shorter diastema in the lower jaw than in the upper jaw (Fig. S17).
Segmentation of both the tooth germ epithelium and dental pulp mesenchyme on micro-CT scans revealed that the molar epithelium displayed modified morphology in Satb2−/− mice compared to Satb2+/+ mice (Fig. 5), along with the presence of ectopic teeth anteriorly before the first molar. These structures were identified in both the upper and lower jaw but were more prominent in the upper jaw (Fig. 5).
While the mouse diastema in adults is toothless, primordial remnants of the evolutionary reduced teeth can be observed during development in the maxilla76,77,78,79. However, later in development, apoptosis reduces and assimilates these structures. These sites are commonly referred to as R1 (anterior) and R2 (posterior) in the maxilla and MS and R2 in the mandible. Based on our results, we propose that the absence of Satb2 prompts the development of ectopic teeth in the areas of previously described rudimental teeth corresponding to premolars79. Moreover, it is necessary to mention that also in human patients with SATB2 mutations, macrodontia, accessory cusps were found in a permanent maxillary first premolar or the first and second deciduous molars80,81,82. Also teeth in the anterior area were found to be disrupted in human patients with SAS such as missing lower permanent incisors or large upper central incisors indicating the role of SATB2 in development of replacement teeth in the incisor area83.
The evaluation of the ectopic tooth position in the jaw in relation to the regular teeth was conducted by measuring tooth lengths and distances between teeth in 3D space (Fig. 7). Morphometric analysis indicated that the molar area is reduced in Satb2−/− animals in contrast to the epithelial area in Satb2+/+ mice (Fig. 7). However, the molar area with the inclusion of ectopic teeth in the analyses, particularly in the upper jaw, exhibits a closer resemblance to WT dental epithelium (Fig. 7). Also, length of diastema in the upper jaw in Satb2−/− animals resembled those in WT animals when ectopic teeth were included. However, the distances were still shorter in the lower jaw (Fig. 7). Ectopic teeth were initiated in the regions, which at least partially belongs to the first molar area of WT animals. This suggests that the presence of ectopic teeth is not the main reason for the shift of the molar epithelium from the original position.
To confirm that the observed ectopic teeth in Satb2−/− animals were not just epithelial protrusions but real teeth was initiated, we evaluated the proliferation, stem cell marker analyses while using immunohistochemistry and enamel knot markers expression in an ectopic tooth and the first molar (m1) using RNAScope (Fig. 8). The first molar and ectopic tooth of Satb2−/− animals exhibited high proliferation in the epithelial as well as mesenchymal compartments at E16.5 (Fig. 8A–C). This was in contrast to proliferation inincisor area, whhere only few Ki67-positive cells were detected in Satb2−/− animals (Fig. 9A, B). SOX2 expression, which is typical for the lingual side of the dental stalk in the molars as well as incisors84, was preserved in Satb2-deficient animals (Fig. 8D–F and 9C, D). In ectopic epithelial structures, Shh- and Fgf4-positive areas were detected in Satb2−/− animals, indicating a prolonged existence of rudimental germ and new tooth formation. Both Shh and Fgf4 expressions were however stronger in Satb2−/− first molars in contrast to ectopic teeth. Moreover, Shh signal was downregulated in Satb2-deficient animals in insisors in agreement with their smaller phenotype (Fig. 9E, F). In addition, the Shh signal was already propagating along the cervical loop extending beyond the area of the enamel knot in WT animals and Satb2−/− first molars. This was in contrast to the expression of both markers in the ectopic teeth, where Shh expression was still restricted just to the enamel knot area corresponding to simpler morphology of ectopic teeth. Similarly, the diastemal region was previously found to express numerous genes crucial for early tooth development, enabling the transformation of epithelium and ectomesenchyme into the tooth germ85. Moreover, the epithelium thickening can persist in several locations along the dental lamina and can be associated with Shh expression, which is characteristic for early-stage dental organ induction85.
Cell proliferation and gene expression of enamel knot markers in molar area in Satb2-deficient animals. A–C Proliferation of cells detected by immunohistochemistry while labelling Ki67-positive cells in the first molar (m1) and ectopic tooth (Et) of Satb2+/+ mice (A) and Satb2-/- mouse at E16.5 (B, C). D–F SOX2 expression visualized by immunohistochemistry is located in the labial part of the dental stalk of Satb2+/+ mice (D) and Satb2-/- mouse at E16.5 (E, F). G, J Expression of two enamel knot markers Shh and Fgf4 detected by RNAScope in the first molars of Satb2+/+ mice. H, K The signal of both genes is stronger in the Satb2-/- mouse first molar. I, L The expression of Shh and Fgf4 is also visible in the ectopic tooth of Satb2-/- mouse indicating that this structure is not just a protrusion of epithelial tissues, but it possesses a tooth organizer-enamel knot. Detected protein (A–F) or gene expression (G–L) is labelled by magenta colour. Erythrocytes, which are naturally autofluorescent, are labelled by green, yellow or red colour. Scale bar = 100 µm.
Cell proliferation and gene expression of enamel knot markers in incisor area in Satb2-deficient animals. Proliferation of cells detected by immunohistochemistry while labelling Ki67-positive cells in the incisor of Satb2+/+ mice (A) and Satb2-/- mouse at E16.5 (B). SOX2 expression in the vestibule surrounding the incisor of Satb2+/+ mice (C) and Satb2-/- mouse (D). E Expression of Shh is localized in the inner enamel epithelium detected by RNAScope in the incisor of Satb2+/+ mice. F No expression of Shh was detected in Satb2-/- mouse. Detected protein (A–D) or gene expression (E, F) is labelled by magenta colour. Naturally autofluorescent erythrocytes are labelled by green, yellow or red colour. Scale bar = 100 µm.
The presence of ectopic teeth in the diastema region was uncovered in a wide array of mouse models, such as Gas1−/−86, Tg737orpk hypomorphic87, Spry2/Spry4-deficient animals78, Wnt1-Cre;Polarisflox/flox86; and Lrp4−/−(Megf7) hypomorph88, all exhibiting extra teeth at the R2 position in both jaws89. All these genes are associated with primary cilia dysfunction, which however have not been described in Satb2-deficient mice yet, including no connection of SATB2 protein with ciliary signalling. As SATB2 can enhance migration of cells in some pathologies through regulation of genes involved in cytoskeletal organisation74, we propose similar molecular mechanisms occur during ectopic tooth induction in our model organism.
Nevertheless, more information about the involvement of SATB2 in regulating the main signalling pathways during odontogenesis and their possible alterations in Satb2-deficient animals will be necessary, including follow up study of later processes driving differentiation of hard dental tissue producing cells.
Conclusions
In this study, gene expression profiling was carried out to uncover differences between the anterior and posterior regions of the early embryonic lower jaw in the mouse, at stages when incisor and molar areas are established. Our data provide a rich source of fundamental information which can be used for further studies focused on early embryonic jaw patterning and serve to enhance understanding of the initial molecular signalling driving incisor and molar development. For more detailed analysis of odontogenesis, we selected the transcriptional factor SATB2, which displayed stronger expression in the anterior area on RNA level. SATB2 protein was also expressed in the anterior mesenchyme where incisor germs are forming, however, we uncovered a distinct SATB2-positive region in the mesenchyme closely surrounding molars. Satb2-deficient mice exhibited defects in incisor morphology confirming a crucial role of SATB2 in formation of anterior craniofacial structures. On the other hand, ectopic tooth germs were observed in the molar area indicating differential effect of Satb2-deficiency in individual jaw regions, which will be necessary to follow further with focus on the role of SATB2 in cytoskeletal components organisation during odontogenesis.
Material and methods
Mouse strains, genotyping and tissue collection
The housing of mice and in vivo experiments were performed in compliance with the European Communities Council Directive of 24 November 1986 (86/609/EEC) and national and institutional guidelines. Animal care and experimental procedures were approved by the Animal Care Committee of Masaryk University (no. MSMT 8337/2019-3) and the study is reported in accordance with ARRIVE guidelines.
Satb2 gene targeting in mice was performed by a b-galactosidase/pGKneo-pA cassette insertion in frame with the ATG codon of the Satb2 gene as described previously28. Animals were genotyped by PCR using a set of three primers: Satb2-specific reverse primer detecting the wildtype (WT) allele 5’ GCC ACC CTC TGG GTA AAC CAC 3’, Satb2-LACZ-specific reverse primer 5’ CGG GCC TCT TCG CTA TTA CG3’ and the common forward primer 5’ CGG TGG GAA CTT TGT CTC CA 3’ annealing to the 5’ untranslated region (UTR) of the Satb2 gene. PCR (35 cycles; denaturation: 94 °C, 30 s; annealing: 58 °C, 30 s; extension: 72 °C, 30 s) was performed on genomic DNA isolated from the tail tips.
RNA isolation and microarray analysis
We collected the anterior (forthcoming incisor) and posterior (forthcoming molar) areas of lower jaws for gene expression profiling from mouse embryos at stages E11.5 and E12.5. Three biological replicates were used for every stage and area. Anterior and posterior samples were separated in the future diastemal area (Fig. 1) and both epithelium and surrounding mesenchyme were included in samples. The posterior dissection was performed just behind the molar germ.
RNA was extracted using a commercial RNeasy Mini Kit (cat. no. 74106, Qiagen, Germany). The quantity and quality of RNA were ascertained using a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies LLC, USA) and Agilent 2100 Bioanalyzer (Agilent Technologies, CA, USA). All samples had RIN above 9 and were used for the microarray analysis on the GeneChip Mouse Genome 430A 2.0. Array (cat. no. 900499, Affymetrix, USA) following the GeneChip 3’IVT express protocol: total RNA (100 ng) was amplified and fragmented using the provided kit and 10 μg of the resulting fragmented complementary deoxyribonucleic acid (cDNA) was hybridised on the chips according to the manufacturer’s protocol. This whole-genome chip probes 39,059 well-characterised mouse targets.
The resulting CEL files were processed using the packages oligo90 and limma91 of the Bioconductor92 within the R environment (R Core Team (2023). R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria. https://www.R-project.org). Probe set targets were annotated using the BrainArray annotation package93; version 24.0.0, ENST at the level of Ensembl mouse transcripts94, version 97, GRCm38). Next, robust multiarray analysis95 was used to standardise the data. All samples passed technical quality controls. Moderated t-test was used to detect differentially expressed transcripts after fitting linear model I ~ stage * area within limma. False discovery rate less than 0.0596 and a minimally two-fold change in expression intensity were required to consider genes differentially transcribed. The minimum information about a microarray experiment (MIAME) compliant data was deposited to the ArrayExpress database97, E-MTAB-13604).
Correlation analyses
Correlation analyses were performed while using GenespringGX version 7.3 (Agilent Technologies) for the identification of co-regulated genes while using pair-wise correlations. As statistical methods for computing correlation coefficients we selected Pearson.
Validation of the microarray data with qRT-PCR
Tissues from the mandibles of mouse embryos at stage E12 were collected separately from the posterior (forthcoming molar) and the anterior (forthcoming incisor) areas. Three biological replicates were analysed. Each replicate for each area was generated as a pool of eight collected embryos to obtain enough RNA for further analyses. RNA was extracted using an RNeasy Mini Kit (cat. no. 74106, Qiagen, Germany).
The cDNA samples were preamplified using a gb Reverse Transcription Kit (cat. no. 3012, Generi Biotech, Czech Republic). Pre-amplified cDNA was used for q-PCR in a Vapo. Protect Cycler system (Eppendorf, USA). The light cycler programme consisted of three main steps: Preincubation at 95 °C for 10 min, denaturation at 95 °C for 15 s and annealing at 62 °C for 60 s. Data were analysed using LightCycler 96 software 1.1 to obtain Ct values. Three technical replicates were performed for each sample. Gene expression values were expressed regarding the threshold cycle normalised to the expression of beta-actin (Actb; TaqMan® Gene Expression Assay, ID: Mm00607939_s1, Thermo Fisher Scientific, USA). The following TaqMan® Gene Expression Assays (cat. No. 4351372, Applied Biosystems, USA) were used: Dio2 (ID: Mm00515664_m1), Dlg2 (ID: Mm01318472_m1), Dlx5 (ID: Mm00438430_m1), Dpep1 (ID: Mm00514599_g1) and Satb2 (ID: Mm00507331_m1), Rerg (ID: Mm07305356_m1).
Fold-change differences relative to incisors (at E12) were calculated using the ∆∆Ct method as described previously33. Standard errors of the means were calculated as described previously33. Statistical analysis consisted of t-tests comparing dCt values between anterior and posterior areas with p < 0.05 considered to be significant.
Immunohistochemical analysis of protein expression
For SATB2 protein expression, deparaffinizated and rehydrate sections of mouse heads at embryonic days E11.5–E16.5 were processed and antigen retrieval was performed in a boiling water bath (97 °C) in Dako Target Retrieval Solution, pH 9 (cat. No. S2367, Dako Agilent, USA) for 20 min. A non-specific secondary antibody was bound on the antigen by incubation in horse serum for 20 min. Primary antibody (anti-SATB2, 1 : 100, cat. no. ab51502, Abcam, UK) was applied for 1 h. The secondary biotinylated anti-mouse antibody (1 : 500, ABC kit, cat. no. PK-4002, Vectastain, USA) and avidin–biotin complex (cat. no. PK-4002, ABC kit, Vectastain, USA) were applied for 30 min each. Products were visualised with 3,3′-diaminobenzidine tetrachloride chromogenic substrate (DAB, cat. no. K3466, Dako, USA), and slides were counterstained with Hematoxylin. Samples were analysed and photographed using a Leica microscope (DMLB2) with a Leica camera (DFC480) attached (Leica Microsystems, Wetzlar, Germany).
For Ki-67 and SOX2 expression, alternative slides were deparaffinized and rehydrated through a series of ethanol. Water bath (97 °C) with DAKO solution (pH = 6) was used for 5—20 min for antigen retrieval. Blocking serum was applied on the samples for 30 min to prevent nonspecific binding of antibodies. Next, slides were incubated with primary antibodies SOX2 (cat. No. 2748, 1:100, Cell Signaling, Danvers, USA) or Ki-67 (1:200, cat. no. 275-R-16, Cell Marque, USA) for 1 h or overnight, alternatively. The secondary antibody with fluorophore was Alexa Fluor 594 both foth staining (1:200, A11037, Invitrogen, USA). The sections were mounted in a histologically mounted medium called Fluoroshield with DAPI (F6059, Sigma-Aldrich, USA). This mounting medium counterstained cell nuclei in blue.
Micro-CT analysis
Micro-CT analysis was conducted to acquire comprehensive 3D structural data of the incisor and molars germs, as well as the measurement of diastema length. Additionally, the 3D structure of Meckel's cartilage was also incorporated into the analysis. This technique facilitated the assessment of various parameters, such as the shape, size, volume, area and spatial arrangement of the dental elements.
Wildtype and knockout SATB2 mouse embryos in stages E16.5 and E18.5 were dehydrated in ethanol gradient solutions (30%, 50%, 70%, 80%, 90%). To visualise the soft tissues, the E16.5 embryos were stained in 1% I2 in 90% MeOH solution for 48 h; the E18.5 embryos were stained for 4 days in 1% I2 in 90% MeOH (after 48 h of staining, the solution was changed to a fresh one). To prevent the motion of the sample during the measurement, embryos were placed in a 15 mL falcon tube and embedded in 1% agarose gel.
Micro-CT measurement was performed on a GE Phoenix v|tome|x L 240 laboratory system equipped with a 180 kV/15 W nanofocus tube with the scanning parameters described in Table S9.
The tooth germs and Meckel's cartilage were manually segmented using Avizo software. All measurements of lengths were performed in VG Studio MAX 2022 software. The length of the dental area was measured from the frontal protrusion in the anterior part of the Meckel's cartilage by placing the points of the polyline tool in the centre of the transversal cross section of the cartilage; in the posterior part of the Meckel's cartilage, the measurement was ended at the point where the cartilage started to branch. The cartilage length was then defined by the length of the placed polyline.
3D morphometrics of the tooth germs: lengths of the individual tooth germs and the dimensions between the tooth germs were measured in 3D space by placing landmarks on the most distal and proximal parts of the tooth germ and then measuring the distance between those defined points.
For the wall thickness analysis of the tooth germs, all of the incisors in one animal were grouped together and all of the molars were grouped together. Then for each category—incisors and molars—wall thickness analysis was applied. The analysis is based on a function where the software fits spheres of different diameters in the segmented model in the 3D space of the tooth germ, resulting in a colour map of wall thickness distribution. The obtained data were subsequently exported into the graph of the quantifications of the wall thicknesses for each sample.
Messenger RNA detection in tissues
The expression of Fgf4 and Shh was determined by RNAScope (Advanced Cell Diagnostics, USA) in mice at E16.5. We analysed at least three mouse embryos for each studied genotype, WT, HET and mutant. Mouse embryos were euthanized by decapitation and fixed in 4–10% PFA for a maximum of 48 h. After fixation, samples were decalcified in 10% EDTA for 3 days at 4 °C, then dehydrated by standard ethanol series followed by xylene. The samples were embedded in paraffin and sectioned at 5 µm. Tissues were processed using anRNAScope Multiplex Fluorescent v2 assay (cat. no. 323 110, Advanced Cell Diagnostics, USA). Samples were incubated in hydrogen peroxide (cat. no. 322 335, Advanced Cell Diagnostics, USA) at RT for 10 min before they were boiled in the target retrieval reagent (cat. no. 322 001, Advanced Cell Diagnostics, USA) and pretreated with Protease Plus (cat. no. 322 331, Advanced Cell Diagnostics, USA). Probes (RNAScope® Probe Mm-Fgf4, cat. no. 514311; Mm-Shh, cat. no. 314361, Advanced Cell Diagnostics, USA) were used to detect transcripts, and DAPI (cat. no. 323 108, Advanced Cell Diagnostics, USA) was used for nucleus staining. Pictures were taken under a Leica DM LB2 microscope (Leica Microsystems, Germany) and processed by Adobe Photoshop 7.0.
Fundings
Research was supported by the Czech Science Foundation (original microarray analyses by 304/08/P289, Satb2-deficient mice evaluation by 22-02794S) and LIH by the Ministry of Health, Czech Republic (preliminary results by NU20-06–00189/2020, revisions by NW24-10–002024). This work was supported by ELIXIR CZ research infrastructure (MEYS Grant No: LM2023055). We acknowledge the core facility CELLIM supported by the Czech-BioImaging large RI project (LM2018129 funded by MEYS CR) for their support with obtaining scientific data presented in this paper.
Data availability
Miroarray data was deposited to the ArrayExpress database (E-MTAB-13604). Other supporting datasets and pictures are available as Supplementary Information files that are freely accessible on nature.com upon publication.
References
Rossant, J. & Tam, P. Mouse development: patterning, morphogenesis, and organogenesis. 1st edn. (Academic Press, 2002).
Thesleff, I. & Tummers, M. Tooth organogenesis and regeneration. Stembook (2009) https://doi.org/10.3824/STEMBOOK.1.37.1.
Stockton, D. W., Das, P., Goldenberg, M., D’Souza, R. N. & Patel, P. I. Mutation of PAX9 is associated with oligodontia. Nat. Genet. 24, 18–19 (2000).
Vastardis, H., Karimbux, N., Guthua, S. W., Seidman, J. G. & Seidman, C. E. A human MSX1 homeodomain missense mutation causes selective tooth agenesis. Nat. Genet. 13, 417–421 (1996).
Mostowska, A., Biedziak, B. & Jagodzinski, P. P. Novel MSX1 mutation in a family with autosomal-dominant hypodontia of second premolars and third molars. Arch. Oral Biol. 57, 790–795 (2012).
Thomas, B. L., Liu, J. K., Rubenstein, J. L. R. & Sharpe, P. T. Independent regulation of Dlx2 expression in the epithelium and mesenchyme of the first branchial arch. Development 127, 217–224 (2000).
Thomas, B. L. et al. Role of Dlx-1 and Dlx-2 genes in patterning of the murine dentition. Development 124, 4811–4818 (1997).
Satokata, I. & Maas, R. Msx1 deficient mice exhibit cleft palate and abnormalities of craniofacial and tooth development. Nat. Genet. 6, 348–356 (1994).
Du, W., Du, W. & Yu, H. The role of fibroblast growth factors in tooth development and incisor renewal. Stem Cells Int. 2018, 7549160 (2018).
Miletich, I., Buchner, G. & Sharpe, P. T. Barx1 and evolutionary changes in feeding. J. Anat. 207, 619 (2005).
Elmubarak, N. A. R. Genetic risk of talon cusp: Talon cusp in five siblings. Case Rep. Dent. https://doi.org/10.1155/2019/3080769 (2019).
Segura-Egea, J. J., Jiménez-Rubio, A., Ríos-Santos, J. V. & Velasco-Ortega, E. Dens evaginatus of anterior teeth (talon cusp): Report of five cases. Quintessence Int. 34, 272–277 (2003).
Gardner, D. G. & Girgis, S. S. Talon cusps: A dental anomaly in the Rubinstein-Taybi syndrome. Oral Surg Oral Med. Oral Pathol. 47, 519–521 (1979).
Chen, R. J. & Chen, H. S. Talon cusp in primary dentition. Oral Surg Oral Med. Oral Pathol. 62, 67–72 (1986).
Hattab, F. N., Yassin, O. M. & Sasa, I. S. Oral manifestations of Ellis-van Creveld syndrome: Report of two siblings with unusual dental anomalies. J. Clin. Pediatr. Dent. 22, 159–165 (1998).
Kjær, I. Dental approach to craniofacial syndromes: How can developmental fields show us a new way to understand pathogenesis?. Int. J. Dent. https://doi.org/10.1155/2012/145749 (2012).
Buchtová, M. et al. Whole genome microarray analysis of chicken embryo facial prominences. Dev. Dyn. 239, 574–591 (2010).
Sehic, A., Risnes, S., Khan, Q. E. S., Khuu, C. & Osmundsen, H. Gene expression and dental enamel structure in developing mouse incisor. Eur. J. Oral Sci. 118, 118–130 (2010).
Tarca, A. L., Romero, R. & Draghici, S. Analysis of microarray experiments of gene expression profiling. Am. J. Obstet. Gynecol. 195, 373 (2006).
van IJzendoorn, D. G. P. et al. Machine learning analysis of gene expression data reveals novel diagnostic and prognostic biomarkers and identifies therapeutic targets for soft tissue sarcomas. PLoS Comput. Biol. 15, e1006826 (2019).
Soldatov, R. et al. Spatiotemporal structure of cell fate decisions in murine neural crest. Science 364, eaas9536–eaas9536 (2019).
Nimmagadda, S. et al. Identification and functional analysis of novel facial patterning genes in the duplicated beak chicken embryo. Dev. Biol. 407, 275–288 (2015).
Dobreva, G. et al. SATB2 is a multifunctional determinant of craniofacial patterning and osteoblast differentiation. Cell 125, 971–986 (2006).
Tucker, A. S., Matthews, K. L. & Sharpe, P. T. Transformation of tooth type induced by inhibition of BMP signaling. Science 1979(282), 1136–1138 (1998).
Mitsiadis, T. A. et al. Expression of the transcription factors Otlx2, Barx1 and Sox9 during mouse odontogenesis. Eur. J. Oral Sci. 106(Suppl 1), 112–116 (1998).
Tissier-Seta, J. P. et al. Barx1, a new mouse homeodomain transcription factor expressed in cranio-facial ectomesenchyme and the stomach. Mech. Dev. 51, 3–15 (1995).
Kim, B. M. et al. Independent functions and mechanisms for homeobox gene Barx1 in patterning mouse stomach and spleen. Development 134, 3603–3613 (2007).
Krivicka-Uzkurele, B., Pilmane, M. & Akota, I. Barx1, growth factors and apoptosis in facial tissue of children with clefts (2008).
Cobourne, M. T. & Sharpe, P. T. Tooth and jaw: Molecular mechanisms of patterning in the first branchial arch. Arch. Oral Biol. 48, 1–14 (2003).
Suryadeva, S. & Begum, M. Role of homeobox genes in tooth morphogenesis: A review. J. Clin. Diagn. Res. 9, ZE09 (2015).
Gray, P. A. et al. Mouse brain organization revealed through direct genome-scale TF expression analysis. Science 306, 2255–2257 (2004).
Tripodi, M., Filosa, A., Armentano, M. & Studer, M. The COUP-TF nuclear receptors regulate cell migration in the mammalian basal forebrain. Development 131, 6119–6129 (2004).
Bergeron, K. F. et al. Upregulation of the Nr2f1-A830082K12Rik gene pair in murine neural crest cells results in a complex phenotype reminiscent of Waardenburg syndrome type 4. Dis. Model Mech. 9, 1283–1293 (2016).
Kaiwar, C. et al. Novel NR2F1 variants likely disrupt DNA binding: molecular modeling in two cases, review of published cases, genotype-phenotype correlation, and phenotypic expansion of the Bosch-Boonstra-Schaaf optic atrophy syndrome. Cold Spring Harb. Mol. Case Stud. 3, a002162 (2017).
Barske, L. et al. Essential role of Nr2f nuclear receptors in patterning the vertebrate upper jaw. Dev. Cell 44, 337-347.e5 (2018).
Reymond, A. et al. Human chromosome 21 gene expression atlas in the mouse. Nature 420, 582–586 (2002).
Shamblott, M. J., Bugg, E. M., Lawler, A. M. & Gearhart, J. D. Craniofacial abnormalities resulting from targeted disruption of the murine Sim2 gene. Dev. Dyn. 224, 373–380 (2002).
Ma, Q., Anderson, D. J. & Fritzsch, B. Neurogenin 1 null mutant ears develop fewer, morphologically normal hair cells in smaller sensory epithelia devoid of innervation. J. Assoc. Res. Otolaryngol. 1, 129–143 (2000).
Sudiwala, S. & Knox, S. M. The emerging role of cranial nerves in shaping craniofacial development. Genesis https://doi.org/10.1002/dvg.23282 (2019).
Ten Berge, D. et al. Mouse Alx3: An aristaless-like homeobox gene expressed during embryogenesis in ectomesenchyme and lateral plate mesoderm. Dev. Biol. 199, 11–25 (1998).
Sharpe, P. T. Homeobox genes and orofacial development. Connect. Tissue Res. 32, 17–25 (1995).
Twigg, S. R. F. et al. Frontorhiny, a distinctive presentation of frontonasal dysplasia caused by recessive mutations in the ALX3 homeobox gene. Am. J. Hum. Genet. 84, 698 (2009).
Beverdam, A., Brouwer, A., Reijnen, M., Korving, J. & Meijlink, F. Severe nasal clefting and abnormal embryonic apoptosis in Alx3/Alx4 double mutant mice. Development 128, 3975–3986 (2001).
Jowett, A. K., Vainio, S., Ferguson, M. W. J., Sharpe, P. T. & Thesleff, I. Epithelial-mesenchymal interactions are required for msx 1 and msx 2 gene expression in the developing murine molar tooth. Development 117, 461–470 (1993).
Liang, J. et al. MSX1 mutations and associated disease phenotypes: genotype-phenotype relations. Eur. J. Hum. Genet. 24, 1663–1670 (2016).
Milet, C. & Monsoro-Burq, A. H. Neural crest induction at the neural plate border in vertebrates. Dev. Biol. 366, 22–33 (2012).
Nakatomi, M. et al. Genetic interactions between Pax9 and Msx1 regulate lip development and several stages of tooth morphogenesis. Dev. Biol. 340, 438–449 (2010).
Molla, M. et al. Enamel protein regulation and dental and periodontal physiopathology in MSX2 mutant mice. Am. J. Pathol. 177, 2516–2526 (2010).
Alappat, S., Zhang, Z. Y. & Chen, Y. P. Msx homeobox gene family and craniofacial development. Cell Res. 13, 429–442 (2003).
Xu, Y. et al. Transcription coactivator Cited1 acts as an inducer of trophoblast-like state from mouse embryonic stem cells through the activation of BMP signaling. Cell Death Dis. 9, 1–17 (2018).
Tamura, M. & Nemoto, E. Role of the Wnt signaling molecules in the tooth. Jpn. Dent. Sci. Rev. 52, 75 (2016).
Niida, A. et al. DKK1, a negative regulator of Wnt signaling, is a target of the β-catenin/TCF pathway. Oncogene 23, 8520–8526 (2004).
Brugmann, S. A. et al. Wnt signaling mediates regional specification in the vertebrate face. Development 134, 3283–3295 (2007).
Zhang, Y. et al. Reciprocal requirements for EDA/EDAR/NF-kappaB and Wnt/beta-catenin signaling pathways in hair follicle induction. Dev. Cell 17, 49–61 (2009).
Järvinen, E., Shimomura-Kuroki, J., Balic, A., Jussila, M. & Thesleff, I. Mesenchymal Wnt/β-catenin signaling limits tooth number. Development https://doi.org/10.1242/dev.158048 (2018).
Lee, M. J., Kim, E. J., Li, L. & Jung, H. S. Roles of Wnt inhibitory factor 1 during tooth morphogenesis. Cell Tissue Res. 362, 61–68 (2015).
Sasaki, T. et al. LEF1 is a critical epithelial survival factor during tooth morphogenesis. Dev. Biol. 278, 130–143 (2005).
Yamada, W. et al. Craniofacial malformation in R-spondin2 knockout mice. Biochem. Biophys. Res. Commun. 381, 453–458 (2009).
Dasgupta, K. et al. R-spondin 3 regulates mammalian dental and craniofacial development. J. Dev. Biol. 9, 31 (2021).
Palmer, A. J., Savery, D., Massa, V., Copp, A. J. & Greene, N. D. E. Genetic interaction of Pax3 mutation and canonical Wnt signaling modulates neural tube defects and neural crest abnormalities. Genesis 59, e23445 (2021).
Bandyopadhyay, S., Prasad, S. & Singhania, P. K. Partial anodontia in a case of Waardenburg’s syndrome. J. Laryngol. Otol. 113, 672–674 (1999).
Sólia-Nasser, L. et al. Waardenburg syndrome type I: Dental phenotypes and genetic analysis of an extended family. Med. Oral. Patol. Oral Cir. Bucal. 21, e321 (2016).
Krüger, J. M. et al. Differential roles of postsynaptic density-93 isoforms in regulating synaptic transmission. J. Neurosci. 33, 15504–15517 (2013).
Diez-Roux, G. et al. A high-resolution anatomical atlas of the transcriptome in the mouse embryo. PLoS Biol. 9, e1000582 (2011).
Bell, R. A. V. et al. Chromatin reorganization during myoblast differentiation involves the caspase-dependent removal of SATB2. Cells 11, 966 (2022).
Gyorgy, A. B., Szemes, M., De Juan Romero, C., Tarabykin, V. & Agoston, D. V. SATB2 interacts with chromatin-remodeling molecules in differentiating cortical neurons. Eur. J. Neurosci. 27, 865–873 (2008).
Dobreva, G., Dambacher, J. & Grosschedl, R. SUMO modification of a novel MAR-binding protein, SATB2, modulates immunoglobulin μ gene expression. Genes Dev. 17, 3048–3061 (2003).
Zhao, X. et al. The role of SATB2 in skeletogenesis and human disease. Cytokine Growth Factor Rev. 25, 35–44 (2014).
He, L. et al. Expression and localization of special AT-rich sequence binding protein 2 in murine molar development and the pulp-dentin complex of human healthy teeth and teeth with pulpitis. Exp. Ther. Med. 14, 3507–3512 (2017).
FitzPatrick, D. R. et al. Identification of SATB2 as the cleft palate gene on 2q32-q33. Hum. Mol. Genet. 12, 2491–2501 (2003).
Leoyklang, P. et al. Heterozygous nonsense mutation SATB2 associated with cleft palate, osteoporosis, and cognitive defects. Hum. Mutat. 28, 732–738 (2007).
Rosenfeld, J. A. et al. Small deletions of SATB2 cause some of the clinical features of the 2q33.1 microdeletion syndrome. PLoS One 4, 6568 (2009).
Döcker, D. et al. Further delineation of the SATB2 phenotype. Eur. J. Hum. Genet. 22, 1034 (2014).
Seong, B. K. A. et al. SATB2 enhances migration and invasion in osteosarcoma by regulating genes involved in cytoskeletal organization. Oncogene 34, 3582–3592 (2015).
Scott, J. et al. Dental radiographic findings in 18 individuals with SATB2-associated syndrome. Clin. Oral Investig. 22, 2947–2951 (2018).
Peterková, R. et al. Different morphotypes of the tabby (EDA) dentition in the mouse mandible result from a defect in the mesio-distal segmentation of dental epithelium. Orthod. Craniofac. Res. 5, 215–226 (2002).
Peterkova, R., Lesot, H. & Peterka, M. Phylogenetic memory of developing mammalian dentition. J. Exp. Zool. B Mol. Dev. Evol. 306B, 234–250 (2006).
Klein, O. D. et al. Sprouty genes control diastema tooth development via bidirectional antagonism of epithelial-mesenchymal FGF signaling. Dev. Cell 11, 181–190 (2006).
Klein, O. D. et al. Developmental disorders of the dentition: An update. Am. J. Med. Genet. C Semin. Med. Genet. 163, 318–332 (2013).
Kurosaka, H. et al. Craniofacial and dental characteristics of three Japanese individuals with genetically diagnosed SATB2-associated syndrome. Am. J. Med. Genet. A 191(7), 1984–1989 (2023).
Li, X., Ye, X. & Su, J. The dental phenotype of primary dentition in SATB2-associated syndrome: A report of three cases and literature review. BMC Oral Health 22(1), 522 (2022).
Mouillé, M. et al. SATB2-associated syndrome: Characterization of skeletal features and of bone fragility in a prospective cohort of 19 patients. Orphanet. J. Rare Dis. 17(1), 100 (2022).
Zarate, Y. A., Bosanko, K., Derar, N. & Fish, J. L. Abnormalities in pharyngeal arch-derived structures in SATB2-associated syndrome. Clin. Genet. https://doi.org/10.1111/cge.14540 (2024).
Dosedělová, H. et al. Fate of the molar dental lamina in the monophyodont mouse. PLoS One 10(5), e0127543 (2015).
Prochazka, J. et al. Patterning by heritage in mouse molar row development. Proc. Natl. Acad. Sci. U. S. A. 107, 15497–15502 (2010).
Ohazama, A. et al. Primary cilia regulate Shh activity in the control of molar tooth number. Development 136, 897–903 (2009).
Zhang, Q. et al. Loss of the Tg737 protein results in skeletal patterning defects. Dev. Dyn. 227, 78–90 (2003).
Ohazama, A. et al. Lrp4 modulates extracellular integration of cell signaling pathways in development. PLoS One 3, e4092 (2008).
Cobourne, M. T. & Sharpe, P. T. Making up the numbers: The molecular control of mammalian dental formula. Semin. Cell Dev. Biol. 21, 314–324 (2010).
Carvalho, B. S. & Irizarry, R. A. A framework for oligonucleotide microarray preprocessing. Bioinformatics 26, 2363–2367 (2010).
Ritchie, M. E. et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47–e47 (2015).
Huber, W. et al. Orchestrating high-throughput genomic analysis with Bioconductor. Nat. Methods 12, 115–121 (2015).
Dai, M. et al. Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. Nucleic Acids Res. 33, e175–e175 (2005).
Cunningham, F. et al. Ensembl 2022. Nucleic Acids Res. 50, D988–D995 (2022).
Bolstad, B. M. Low-level analysis of high-density oligonucleotide array data: Background, normalization and summarization (2004).
Benjamini, Y. & Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Stat. Methodol. 57, 289–300 (1995).
Sarkans, U. et al. From ArrayExpress to BioStudies. Nucl. Acids Res. 49, D1502–D1506 (2021).
Acknowledgements
We would like to thank prof. Rudolf Grosschedl from the Department of Cellular and Molecular Immunology, Max Planck Institute of Immunobiology and Epigenetics (Freiburg, Germany), for a kind gift and providing us with the Satb2-deficient mice line. We thank Veronika Klatovská and the Core facility of Genomics and Bioinformatics at Institute of Molecular Genetics for excellent support.
Author information
Authors and Affiliations
Contributions
Data analysis: PN, MS, MKa, DZ, DK, MKo, SMB, VJ, KP Visualization: PN, MKa, DZ, DK, KP, TZ Draft of manuscript writing: PN, MS, DZ, MKa, MB Final manuscript writing: MB, LIH, JK Grant support: MB, LIH.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Nevoránková, P., Šulcová, M., Kavková, M. et al. Region-specific gene expression profiling of early mouse mandible uncovered SATB2 as a key molecule for teeth patterning. Sci Rep 14, 18212 (2024). https://doi.org/10.1038/s41598-024-68016-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-68016-3
- Springer Nature Limited