Abstract
The effective identification of fruit tree leaf disease is of great practical significance to reduce pesticide spraying, improve fruit yield and realize ecological agriculture. Computer vision technology can be effectively identifying and prevent plant diseases and insect pests. However, the lack of consideration of disease diversity and accuracy of existing detection models hinders their application and development in the field of plant pest detection. This paper proposes an efficient detection model of apple leaf disease spot through the improvement of the traditional Yolov5 detection network called A-Net. In order to significantly increase the A-Net's detection speed and accuracy, the A-Net model applies the loss function Wise-IoU, which includes the attention mechanism and the dynamic focusing mechanism, to the Yolov5 network model. The RepVGG module is then used to replace the original model's convolution module. The experimental results show that the improved model effectively suppresses the growth of some error weights. Compared with several object detection models, the improved A-Net model has a Mean Average Precision across IoU threshold 0.5 and an accuracy of 92.7%, which fully proves that the improved A-Net model has more advantages in detecting apple leaf diseases.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Introduction
Apples are a great source of vitamins, minerals, and organic acids that provide excellent material protection for human health. The apple business is also one of the most frequently produced fruit industries in the world1. Because the apple tree is prone to apple leaf disease, branch wheel grain disease and other invasion, which leads to the serious deterioration and production reduction of apple, which brings great economic losses to the fruit farmers. One kind of traditional diagnostic approach for apple leaf disease involves using fruit farmers' knowledge to appraise and detect illnesses and insect pests. This approach of subjective perception is inefficient, labor-intensive, and prone to inaccuracy2. The other is the traditional machine learning method to identify apple leaf disease, which has been a satisfactory alternative to the former method in the past.
For example, Liu et al.3 focused their efforts on detecting four common types of diseases that affect apple leaves: Mosaic, Rust, Brown spot, and Alternaria leaf spot. They acknowledged the significance of early and precise disease identification in order to prevent the spread of infections and ensure the healthy growth of the apple industry. The authors emphasized the drawbacks of existing methods, which often involve complex image preprocessing and yield inconsistent recognition rates. To overcome these challenges, the researchers proposed a new deep convolutional neural network (DCNN) architecture based on AlexNet. They compiled a comprehensive dataset consisting of 13,689 images depicting the four target diseases and trained their DCNN model to accurately identify these diseases. The model exhibited an impressive overall accuracy of 97.62% on a separate test set, surpassing the performance of the standard AlexNet while significantly reducing the number of model parameters by over 51 million. One notable contribution of this study was the utilization of image generation techniques to enhance the resilience of the DCNN model. By generating additional pathological images, they achieved a 10.83% improvement in accuracy compared to the standard model. The research underscored the potential of deep learning models, particularly DCNNs, in offering a faster and more precise solution for controlling apple leaf diseases. However, it is important to note that the study had certain limitations. The model was initially trained and evaluated on a specific dataset without exploring its performance on other apple leaf diseases or in different environmental conditions. Additionally, the computational requirements and inference speed of the model were not addressed, which are crucial for real-world deployment. Furthermore, the authors did not compare their proposed architecture with other state-of-the-art DCNN models, leaving room for further exploration of more advanced architectures. While this study provided a strong foundation for using DCNNs in apple leaf disease detection, our research seeks to expand on these findings by overcoming limitations and enhancing the model's applicability and accuracy. We suggest improvements to the YOLOv5 architecture, an advanced object detection model, and assess its performance on a diverse dataset that includes various natural backgrounds to enhance the A-Net's generalization capabilities. Our work also concentrates on enhancing detection speed and accuracy, addressing the shortcomings of the previous study, and making our model more suitable for practical agricultural applications.
Zhong and Zhao4 concentrated on the detection of apple leaf diseases by employing a DenseNet-121 deep convolution network. They devised and evaluated three approaches, namely regression, multi-label classification, and focus loss function, using a dataset comprising 2462 images of six apple leaf diseases. These methods exhibited remarkable accuracies ranging from 93.31% to 93.71%, surpassing conventional techniques reliant on cross-entropy loss function. Nevertheless, the study encountered constraints in relation to the dataset's limited size and diversity, as well as the applicability of the proposed methods in real-world scenarios.
Yan et al.5 focused on the identification of common apple leaf diseases such as scab, frogeye spot, and cedar rust, which are crucial for the advancement of apple production. The study introduced an enhanced model based on VGG16, utilizing a global average pooling layer to decrease parameters and incorporating a batch normalization layer to enhance convergence speed. Additionally, a transfer learning approach was employed to expedite training. The experimental outcomes demonstrated an impressive 99.01% accuracy in apple leaf disease classification using the proposed model. Notably, compared to the conventional VGG16, the model parameters were reduced by 89%, while achieving a 6.3% accuracy improvement and significantly reducing training time. Consequently, the deep convolutional neural network model presented in this study offers a superior solution for apple leaf disease identification with heightened accuracy and accelerated convergence speed. Limitations of the previous study include the lack of diversity in the types of apple leaf diseases considered, potentially limiting the generalizability of the model to a broader range of diseases. Additionally, the study did not thoroughly explore the robustness of the model under varying environmental conditions or leaf image qualities, which are critical factors for real-world applicability. Furthermore, the evaluation was based on a specific dataset, raising questions about the A-Net's performance when confronted with new and unseen data. These limitations underscore the need for further research to address these concerns and enhance the practical utility of the proposed model in real-world settings.
Bi et al.6 concentrated on detecting common diseases that affect apple leaves, such as Alternaria leaf blotch and rust. These diseases have a significant impact on the yield of apple crops. The traditional method of inspecting diseases by experts is time-consuming and not very accurate. To tackle this problem, the study suggested a low-cost, stable, and highly accurate identification method that utilizes the MobileNet model. The MobileNet model was chosen because it is cost-effective and can be easily used on mobile devices, making disease detection more accessible to everyone. By replacing the need for expert inspection with the algorithm, the method ensures consistent and reliable disease identification. The precision of the MobileNet model was found to be similar to more complex deep learning models like ResNet152 and InceptionV3. The researchers conducted experiments using apple disease datasets collected by agriculture experts in Shaanxi Province, China. However, there are some limitations to the study. Firstly, it only focused on two specific types of apple leaf diseases, which may restrict its applicability to a wider range of diseases. Secondly, the study did not investigate how the model performs under different environmental conditions or image qualities, which are important factors for real-world implementation. Lastly, the evaluation was based on datasets from a specific region, which raises concerns about whether the model can be applied to other regions with different disease patterns. These limitations emphasize the need for further research to address these gaps and improve the reliability and practicality of the proposed method in various settings.
Khan et al.7 tackled the significant economic losses caused by apple diseases and pests. This was done by detecting and identifying apple diseases at an early stage. The researchers started by collecting a large dataset of high-quality RGB images of apple diseases, which included approximately 9000 images. These images were then used to train a deep learning-based detection system. The proposed system followed a two-stage approach. In the first stage, a lightweight classification model was used to categorize the images as diseased, healthy, or damaged. In the second stage, the system performed the actual detection and localization of each symptom from the diseased leaf images. The results of the study showed promising classification accuracy of around 88% and a mean average precision (mAP) of 42% for disease detection. This highlighted the practical usefulness of the system for farmers and apple growers, as it could effectively detect various types of apple diseases. However, it is important to note that the study had some limitations. Firstly, the focus was solely on apple diseases, which means that the proposed system may not be applicable to other fruits and vegetables. Additionally, the researchers did not thoroughly investigate how the system would perform under different environmental conditions or with varying image qualities. These factors are crucial for real-world implementation. Furthermore, the mAP of 42% indicates that there is room for improvement in the accuracy of symptom detection. These limitations emphasize the need for further research to enhance the system’s performance and make it applicable across different agricultural settings.
In the realm of pattern recognition, convolutional neural networks, or CNNs, and deep learning have shown promising results recently8. CNN becomes a target detection research hotspot because it can extract features directly from the input pictures, avoiding the need for laborious image preparation9. Inspired by it, the research and application of CNN in crop disease detection have also appeared, and have achieved some good results1. In addition to establishing a novel model for apple leaf disease detection utilizing the mean block and apple initial model, H. Sun et al. developed a lightweight CNN model that can detect apple leaf disease in real time15. With an average accuracy of 82.48% for apple leaf disease under natural settings, Li et al. suggested an enhanced and faster model for RCNN apple leaf disease detection16. YADAV et al. presented a unique CNN-based apple leaf disease detection and classification model that used the fuzzy c-mean (FCM) clustering technique and contrast stretch-based preprocessing technology to identify plant illnesses. In 400 picture samples, its accuracy was 98%10. Mohanty et al. utilized GoogleNet to detect 26 illnesses of 14 crops based on the publicly available dataset PlantVillage. The findings demonstrated that the GoogleNet pre-trained model's pest identification accuracy is high after parameter fine-tuning17. Sun Jun et al. enhanced the AlexNet network to achieve a high identification rate by examining the PlantVillage dataset and the disease leaf photos18. To increase the accuracy of target identification, Linbai Wang et al. suggested the CenterNet-SPP model, which introduces the SPP spatial pyramid pooling model19. Although there are still some shortcomings, the work of the aforementioned researchers has greatly advanced the use and advancement of CNN in the fields of plant leaf disease and insect pest identification. It mostly shows itself in two ways: first, there is a need to increase the accuracy of model identification11. Secondly, there are insufficient training data for the model and poor generalization capability. Consequently, in order to discover a practical approach that can take into account the model's accuracy as well as its adaptability and be extensively used in the diagnosis of several common apple leaf diseases, it is imperative that the current algorithms be optimized and improved14. In order to address the shortcomings of the current model in the identification of diseases and insect pests in apple leaves, an enhanced YOLOv5 apple leaf disease identification model is proposed in this paper12. The underlying model, YOLOv5, was selected due to its small target size and subtle features. We built the leaf detection dataset using five apple leaf diseases that had a static backdrop and a complex background in the natural environment, in an effort to increase the YOLOv5's capacity for generalization and identification accuracy13. The YOLOv5 network model is enhanced by the model by using Wise-IoU, and by substituting the RepVGG module for the original YOLOv5's convolution module, the model's detection speed and accuracy are significantly increased.
Apple leaf disease dataset
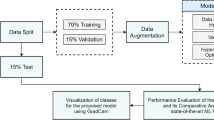
Due to a shortage of appropriate datasets for the experiment, we used a combination of artificially created apple blade photos from real-world settings and apple blade images from Baidu Flying Paddle's image collection. We augment the dataset using image processing techniques including filtering, rotation, and translation in order to increase its size and improve the generalization performance20. We primarily chose five frequent diseases—high incidence, rust, mosaic, black rot, brown spot disease, and gray spot—in order to test the model's efficacy. Figure 1 displays a photo sample of each ailment. It is easy to see how different the five apple leaf diseases are from one another in Fig. 1. Comparative research reveals that, above all, rust spots are made up of rust yellow patches with brown needle-sized patches in the center. Rust may be differentiated from other illnesses more readily because of this notable distinction. Second, under comparable natural circumstances, lesions resulting from the same illness exhibit certain similarities. Third, the mosaic yellow lesion covers the whole leaf, in contrast to other areas. Lastly, the gray areas exhibit geometric feature similarities, making detection more difficult.
The collected dataset contained 12,000 images. Among these, including healthy and five diseased leaves, 1500 copies of each leaf were selected as the training set and 500 images as the test set. In order to verify the generalization performance of the model, the collected dataset has three characteristics: First, multiple diseases may occur simultaneously on the same disease image. The second is that most images are taken from natural conditions and contain complex backgrounds. Third, all the images of lesions in the dataset are manually annotated and labeled for different diseases, as shown in Table 1. Label numbers 0–5 represents healthy leaves, rust, mosaic, black rot, brown spot, and gray spot diseases separately.
Improved network model
Training model
In this paper, YOLOv5-6.0 is used as the training model. The YOLOv5-6.0 model is improved over the earlier YOLOv5-5.0 model, using a convolution kernel with size 666,666, step size 2 and padding 2 instead of the Focus module. In version YOLOv5-6.0, Focus was replaced with a 66 convolution layer to improve the convolution efficiency, as shown in Fig. 2. Meanwhile, the YOLOv5-6.0 model adjusts the SPP in the original Backbone module to use SPPF. SPPF uses the input feature layer in parallel through multiple MaxPool2d fusion output modules in different formats. As parallel operation requires simultaneous Concat fusion after 3 × 3, 9 × 9, 13 × 13 convolution, the calculation amount will be greatly increased and the efficiency will be reduced, while SPPF combines the original input feature layer. Through serial operation, the calculation amount of each parallel MaxPool2d will be small, and the convolution time will be relatively reduced, and the efficiency will be greatly improved. As shown in Fig. 3.
Wise-IoU loss function
Considering that there are many overlapping phenomena of various disease spots in the leaves, and this phenomenon will have an impact on the identification, so the prediction of the positioning anchor frame of disease spots anchor is very critical. In the YOLOv5 model, the CIOU_Loss loss function detects the boundary box scale. Although the threshold of the boundary box length and width is increased, the difference between the prediction box and the real box position of some low-quality targets is ignored, and the convergence rate is slower. In view of this problem, this paper adopts Wise-IoU loss function, which adds attention mechanism and dynamic focusing mechanism. By decreasing the detrimental gradient produced by low-quality instances, this optimization not only lessens the competitiveness of high-quality anchor boxes but also improves the generalization impact of the model. Wise-IoU loss function is as follows:
The parameters Wg and Hg represent the length and width of the prediction box, x and y represent the corresponding coordinates of the target. In order to effectively eliminate the hindering factors, the convergence is prevented by width and coordinate, while RWIoU and LWIoUv1 suppress the difference between high quality and low quality anchor. For different categories of Wise-IoU simultaneously set FM for cross-entropy, the classification can be optimized for samples, such as Eq. 2.
On the basis of \(L_{{{\text{WIoUv1}}}}\), the \(L_{{{\text{IoU}}}}^{r*}\) is set to the focusing coefficient of \(L_{{{\text{WIoU1}}}}\) monotone clustering. \(L_{{{\text{IoU}}}}^{ - }\) represents the dynamic average. As the coefficient increases, the gradient will gradually decrease, making the convergence speed also greatly reduced, so that the efficiency and accuracy of training will also be reduced.\(L_{{{\text{IoU}}}}^{ - }\) normalize dynamic vectors to alleviate the problem of slow convergence of coefficients and improve the accuracy of the training model.
Detection model
The RepVGG network model's single channel layout allows for reasonably high parallelism and a modest memory footprint in the area of plant leaf disease identification. Therefore, it is more efficient with the same computational amount compared with other algorithms, and better identification results are obtained by pre-training the model. In this paper, the RepVGG model is introduced instead of the original convolution model, and the improved model is shown in Fig. 4.
RepVGG reparameterization process, as shown in Fig. 5, in the structural reparameterization process, (1) module needs to be converted into (2) module. The winning green module combines the convolution layer of 3 × 3 and BN to form a new 3 × 3 convolutional layer. The standard blue module first converts the 1 × 1 convolution layer of 3 × 3 and BN to form a new 3 × 3 convolution layer, while the standard orange module only has a BN branch. First, it constructs a 3 × 3 convolution layer to get a new 3 × 3 convolution layer. Finally, the three new 3 × 3 convolution layers are fused into a 3 × 3 convolution layer. The reparameterized parameters part is combined with two groups of 3 × 3 convs and two groups of 1 × 1 convs and the mean value of BN module and standard deviation to form an operator for multiple refusion.
Regularized during the fusion of BN and Convs. As shown in Eq. 3, the process yi in the fusion convolution BN is the ith channel for the input feature graph, u represents the mean of the feature graph, σ is its variance is constantly determined by training, γ and β learn the parameters obtained through training, ε prevents the error of yi for the part of the target convolution channel.
The calculation process of bn is performed as described in Eq. 4.
i is the sequence of passage in Eq, \(b_{n} (M,u,\sigma ,\gamma ,\beta ):i,:,\) representing data parameters through the ith channel, \(M_{:,i,:,:}\) represents the value of the feature value corresponding to the i channel. \(W_{i,:,:,:}{\prime} ,b_{i}{\prime}\) indicates the new convolution layer weights and paranoia after fusion convolution. Through the above continuous accumulation of the mean, fusion convolution and other operations to further optimize the previous single model, making the model more lightweight and greatly improved in performance.
Experimental result
Experimental environment
In this paper, pytorch deep learning framework is adopted, and the hardware sets used in the experiment are shown in Table 2.
Model evaluation parameters
Precision, recall, F1-score, mAP @ [0.5:0.95], mAP @ 0.5, and speed were the six characteristics from which the optimized model performance was evaluated in order to validate the effectiveness of the suggested network model for apple leaf disease spot identification. Accuracy The fraction of accurately predicted samples in all anticipated positive samples. As shown in formula 5, TP is True Positive, which indicates the IoU calculation with the annotation box. The calculated IoU is greater than the IoU set previously, and the prediction label is the same parameter as the calibration label obtained after the IoU calculation. FP is False Positive, which is the parameter in which the IoU calculation in the sample does not reach the set IoU threshold, this parameter can be used to reflect the correct prediction accuracy in the sample.
Recall, also known as recall rate, is a measure of coverage in the sample. The proportion of correctly predicted positive examples is mainly used to evaluate the coverage of the target detection in the evaluation detector. As shown in Eq. 6, FN is the positive case parameter that misjudged negative cases in this model, and a higher recall rate in the trained model indicates a higher recognition rate in the detection process.
mAP @ [0.5:0.95] is the mAP averaged across the different IoU thresholds, with the Iou threshold (0.5–0.95) with a step size of 0.5. As shown in Eq. 7, AP, Average Precision is the individual class average accuracy. The greater the accuracy of the general MAP in the model, the better the detection of its target.
Model testing
To improve the accuracy of identifying apple leaf diseased spots, this paper uses a lightweight YOLOv5s deep learning framework. The model was trained on 9000 training images and 3000 test images. The model was updated every ten rounds, the input picture's size was 640 × 640, the batch size was set to 4, the number of training rounds was set to 100, and the initial learning rate was set to 0.01. The Adam optimization technique was used21. and set the weight decay coefficient to 0.0005. After 75 rounds, the loss reached a stable minimum. After 75 rounds, the loss reached a stable minimum. Figure 6 shows the change in training losses. From the figure, we can see that the loss function eventually tends to zero, which proves that the proposed model works well, and the various performance indicators of the model can be evaluated next.
Ablation experiment
The optimized model was first evaluated by ablation experiments. The detection of diseased spots in apple tree leaves is shown in Fig. 7. By randomly selecting an apple tree leaf with an infected spot in the dataset, the red target frames the infected spot of the apple tree leaves detected after training and their confidence level. The higher the confidence during the detection process, the better the detection effect of the model is. (a) Apple tree leaves detected in the original YOLOv5s model; (b) Optimize the YOLOv5s model loss function with the disease spot detected by Wise-IoU; (c) Optimize convolution RepVGG detection for YOLOv5s; (d) shows the disease spots detected by the YOLOv5s model combining Wise-IoU with the RepVGG module. From the identification result diagram, it can be clearly seen that the detection accuracy and effect from (a) to (b) have been greatly improved. Part of the disease spots that could not be detected in the original leaves can be accurately identified through loss function modification and RepVGG optimization, and the overall confidence has been greatly improved. This detection effect allows for an earlier and faster detection of smaller and more obscure disease spots in the leaves.
The ablation experiment can see the parameters improved during the model optimization process more accurately, as shown in Table 3. The optimized model has a certain improvement in Precision, Recall and other parameters. In the model, the loss function is replaced by Wise-IoU, and the convolution part uses the RepVGG module, and both Precision and Recall are also greatly improved. After the model is introduced into RepVGG, the model in the training stage is equally fused to become a single branch structure, which greatly saves the training time. The optimized model's mean is smooth and the loss mean is low, as seen in Fig. 8 by the loss of the bounding box and the classified loss mean before and after model optimization. This suggests that the prediction box and improved model's detection are more accurate.
Data augmentation comparison experiments
We use data augmentation methods to address the occurrence of overfitting due to insufficient datasets and to further improve the robustness of the model. First, we collected suspected diseased apple leaves in different weather conditions in natural environments and apple leaves in laboratory environments. The aim is to ensure the generalization of the model by choosing different shooting backgrounds, thus reducing the occurrence of overfitting. Second, to further enhance the model's generalization performance and avoid overfitting, the variety and quantity of training photos are augmented by rotating, transforming, and mirroring the gathered images. We performed two sets of experiments to validate the model performance and trained before and after the dataset expansion. As shown in Fig. 9. Without increasing the data, the training process had high loss and low accuracy, finally reaching 73.12% mAP. However, 79.05% mAP was achieved after passing the data-enhanced model, a 5.93% improvement in detection accuracy over the dataset with unenhanced data.
Detection visualization and failure analysis
Our proposed detection model can detect the leaves of a single disease spot, a single disease spot and a multiple disease spot, as shown in Fig. 10a–c, respectively. Therefore, it shows that our proposed detection model has a relatively powerful detection performance.
Performance comparison between the different network models
As shown in Table 4, we contrasted the enhanced model with three sophisticated detector models: YOLOv5, SSD, and Faster-RCNN. As can be seen, the accuracy of the enhanced model is 90.7%, up 3.9% from the YOLOv5 model; the recall rate is 93.8%, up 4.2%, and the mAP@0.5 value is 94.8%, up 5.2%. The enhanced model's assessment index has also significantly improved when compared to the other models.
Conclusions
This work provides an improved real-time detection technique of apple leaf disease based on YOLOv5 model to efficiently identify insect pests and diseases, minimize pesticide spraying, enhance fruit production, and achieve ecological agriculture. By applying the loss function Wise _ IoU, which includes the attention mechanism and the dynamic focusing mechanism, to the Yolov5 network model, the conventional YOLOv5 model is adjusted to increase the detection efficiency of the model. The datasets were produced using data augmentation technology and gathered from both the actual forest environment with complicated situations and the laboratory setting in order to validate the generalization performance of the proposed model. Furthermore, by substituting the original model's convolutional module with the RepVGG module, the model's performance and degree of lightweighting are significantly enhanced. The findings of the experiment demonstrate that the suggested model is more effective at identifying five different types of apple leaf illnesses spots that are likely to appear in real time and offers a workable method for apple leaf disease detection in real time. Long training time cycles are a consequence of the model in this article's failure to properly account for model parameter optimization throughout the training phase. In subsequent studies, we'll concentrate on two areas: (1) Reduce the model's size and computational complexity while making sure the accuracy value stays constant. (2) Use the enhanced model to identify pests and illnesses on fruit trees more easily by deploying it on mobile devices like drones.
Data availability
All data generated or analysed during this study are included in this published article.
References
Jiang, P., Chen, Y. H., Liu, B., He, D. J. & Liang, C. Q. Real-time detection of apple leaf diseases using deep learning approach based on improved convolutional neural networks. IEEE Access 7, 59069–59080 (2019).
Dutot, M., Nelson, L. M. & Tyson, R. C. Predicting the spread of postharvest disease in stored fruit, with application to apples. Postharvest. Biol. Tec. 85, 45–56 (2013).
Liu, B., Zhang, Y., He, DongJian & Li, Y. Identification of apple leaf diseases based on deep convolutional neural networks. Symmetry 10(1), 11 (2017).
Zhong, Y. & Zhao, M. Research on deep learning in apple leaf disease recognition. Comput. Electron. Agricult. 168, 105146 (2020).
Yan, Q. et al. Apple leaf diseases recognition based on an improved convolutional neural network. Sensors 20(12), 3535 (2020).
Bi, C., Wang, J., Duan, Y., Fu, B., Kang, J. R., Shi, Y. MobileNet based apple leaf diseases identification. Mob. Netw. Appl. 1-9 (2022).
Khan, A. I., Quadri, S. M. K., Banday, S. & Shah, J. L. Deep diagnosis: A real-time apple leaf disease detection system based on deep learning. Comput. Electron. Agricult. 198, 107093 (2022).
Wu, H. et al. A CNN-SVM combined model for pattern recognition of knee motion using mechanomyography signals. J. Electromyogr. Kinesiol. 42, 136–142 (2018).
Melchiorre, J., D’Amato, L., Agostini, F. & Rizzo, A. M. Acoustic emission onset time detection for structural monitoring with U-Net neural network architecture. Dev. Built Environ. 18, 100449 (2024).
Yadav, D., Akanksha, Yadav, A. K. A novel convolutional neural network based model for recognition and classification of apple leaf diseases. Trait Signal 37(6), 1093–1101 (2020).
Mao, X., Hijazi, S., Casas, R., Kaul, P., Kumar, R., Rowen, C. Hierarchical CNN for traffic sign recognition. In: 2016 IEEE intelligent vehicles symposium (IV), 130–135 (2016).
Zhang, B., Quan, C., Ren, F. Study on CNN in the recognition of emotion in audio and images. In: 2016 IEEE/ACIS 15th International Conference on Computer and Information Science (ICIS), 26, 1–5 (2016).
Mathew, M. P. & Mahesh, T. Y. Leaf-based disease detection in bell pepper plant using YOLO v5. Signal Image Video 16(3), 841–847 (2022).
Zhang, Y., Yang, G. P., Liu, Y. K., Wang, C. & Yin, Y. L. An improved YOLO network for unopened cotton boll detection in the field. J. Intell. Fuzzy Syst. 42(3), 2193–2206 (2022).
Sun, H. N. et al. MEAN-SSD: A novel real-time detector for apple leaf diseases using improved light-weight convolutional neural networks. Comput. Electron. Agr. 189, 106379 (2021).
Lie, W. L., Chen, P., Wang, B. & Xie, C. J. Automatic localization and count of agricultural crop pests based on an improved deep learning pipeline. Sci. Rep.-Uk 9, 1–11 (2019).
Mohanty, S. P., Hughes, D. P., Salathé, M. Using deep learning for image-based plant disease detection. Front. Plant Sci. 7 (2016).
Sun, J. et al. Recognition of multiple plant leaf diseases based on improved convolutional neural network. Trans. Chin. Soc. Agricult. Eng. 33(19), 209–215 (2017).
Linbai, W. et al. Potato leaf disease recognition and potato leaf disease spot detection based on Convolutional Neural Network. J. Chin. Agricult. Mech. 42(11), 122 (2021).
Putra, Y. C., Wijayanto, A. W. & Chulafak, G. A. Oil palm trees detection and counting on Microsoft Bing Maps Very High Resolution (VHR) satellite imagery and Unmanned Aerial Vehicles (UAV) data using image processing thresholding approach. Ecol. Inf. 72, 101878 (2022).
Reddy, V. K. & Ravikumar, A. V. Multi-channel neuro signal classification using Adam-based coyote optimization enabled deep belief network. Biomed. Signal Process. Control 77, 103774 (2022).
Acknowledgements
This work was supported by Natural Science Research Project of Anhui Province (Grant No. 2022AH051325, KJ2021A0662, KJ2021A0682, KJ2020A0539) , Natural Science Research Project of Fuyang Normal University (Grant No.2021FSKJ02ZD).
Author information
Authors and Affiliations
Contributions
All authors wrote the main manuscript text. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Liu, Z., Li, X. An improved YOLOv5-based apple leaf disease detection method. Sci Rep 14, 17508 (2024). https://doi.org/10.1038/s41598-024-67924-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-67924-8
- Springer Nature Limited