Abstract
Museum genomics provide an opportunity to investigate population demographics of extinct species, especially valuable when research prior to extinction was minimal. The Bachman’s warbler (Vermivora bachmanii) is hypothesized to have gone extinct due to loss of its specialized habitat. However, little is known about other potential contributing factors such as natural rarity or changes to connectivity following habitat fragmentation. We examined mitochondrial DNA (mtDNA) and genome-wide SNPs using specimens collected from breeding and migration sites across the range of the Bachman’s warbler. We found no signals of strong population structuring across the breeding range of Bachman’s warblers in both mtDNA and genome-wide SNPs. Thus, long-term population isolation did not appear to be a significant contributor to the extinction of the Bachman’s warbler. Instead, our findings support the theory that Bachman’s warblers underwent a rapid decline likely driven by habitat destruction, which may have been exacerbated by the natural rarity, habitat specificity and low genetic diversity of the species.
Similar content being viewed by others
Introduction
Nine avian species have gone extinct in North America since the nineteenth century1. While the factors leading to these extinctions were complex and synergistic, many involved overexploitation and habitat loss1, among the primary drivers of biodiversity loss today2. Such proximate causes of extinction are well-documented; however, the specific factors that make some species at higher risk for extinction are still not well understood3,4. For example, habitat fragmentation can inhibit dispersal among populations, leading to isolation and, potentially, inbreeding, loss of adaptive potential, and decrease of long-term population fitness5,6,7. In the extreme, then, isolation can contribute to heightened extinction risk, especially in species which may have already experienced declines or are naturally uncommon7,8,9. However, many species persist at low densities in patchy and rare habitat10, and it is unclear why some disappear while others remain. Investigating demographic histories and parameters such as range-wide connectivity of extinct species may therefore shed insight on intrinsic factors contributing to a species’ extinction6. Museum collections provide invaluable opportunities to reconstruct population dynamics of extinct species. As data collection was minimal for many of these species before their extinction, genomic analyses of museum or other historical materials often represent the only means of unlocking past dynamics of extinct species.
Here, we conducted genomic analysis of the extinct Bachman’s warbler (Vermivora bachmanii) to investigate the hypothesis that long-term population isolation may have contributed to the species’ decline and ultimate extinction. The Bachman’s warbler was a Neotropical migrant that bred in the southeastern United States and overwintered in Cuba11,12,13. The last Bachman’s warbler sighting occurred in 198811, and the species was proposed for Endangered Species Act (ESA) delisting in 2021, effectively declaring it extinct14. Contemporaneous records of the Bachman’s warbler are sparse, but they were known to occur primarily on ephemeral canebrake (Arundinaria gigantea) stands in flooded forest on their breeding grounds13. The high soil fertility of this habitat type led to widespread conversion of canebrakes and other flood-plain forest habitat to agricultural land in the nineteenth and twentieth centuries15,16. During this period, habitat in the Bachman’s warblers’ narrow wintering range in Cuba was also in serious decline due to hurricanes and extensive agricultural activities17,18. The wide-scale destruction of these specialist habitats has led to speculation that habitat loss and fragmentation across both the wintering and breeding grounds were the main drivers of the Bachman’s warbler’s extinction11,12,13. For Bachman’s warblers, such a rapid and large-scale habitat loss would have been exacerbated by the fact that cane species are semelparous, undergoing synchronized die-offs on 20–30 year cycles, that make canebrakes a spatially and temporally variable habitat19,20.
Habitat loss poses the greatest risk for specialist species3,21, particularly when their required habitat is rare and/or patchily available. However, not all habitat-restricted species respond the same to threatening processes22, and some are able to persist following large-scale habitat loss23. It has also been suggested that human persecution in the form of overharvest, including by museum institutions, may have contributed to population declines13, rendering the Bachman’s warbler even more sensitive to environmental perturbations, but this hypothesis has not been investigated. Thus, it remains unclear how potential characteristics of Bachman’s warblers such as restricted dispersal, rarity, or inbreeding may have interacted with habitat fragmentation and other threatening factors such as overharvest to contribute to their decline.
Previous genomic work comparing extant and extinct species, including the Bachman’s warbler, estimated lower genetic diversity and smaller effective population sizes in extinct species, but found largely similar demographic histories reflecting population expansions following late Pleistocene climate fluctuations in both groups4. This same study found no evidence for population structuring within samples of Bachman’s warblers, although the sample size and sampling breadth was largely confined to one breeding region, and was not adequate to investigate range-wide population structure4. Another study found higher mean runs of homozygosity (ROH) in the Bachman’s warbler versus its extant congeners, suggesting increased levels of inbreeding could have contributed to, or been a by-product of, its extinction24. The narrow habitat specialization of Bachman’s warblers fits with simulations showing that small populations may be more susceptible to genetic drift and inbreeding if they are restricted to specialized habitat patches isolated within a large breeding range7,25. Critically, we still lack a strong understanding of the pre-decline population connectivity of Bachman’s warblers, which could shed light on factors that made them vulnerable to extinction.
The decline of Bachman’s warblers was rapid12; however, it is unclear whether the species was naturally rare or whether populations fluctuated temporally or spatially11,12,13. Anecdotal accounts of historical abundance in the breeding range vary from locally common to sporadic11. The large breeding distribution of Bachman’s warblers (Fig. 1a) and patchy nature of their canebreak breeding habitat does indicate the potential for lack of connectivity and differentiation of isolated populations. Notably, however, most extant New World warbler species (Parulidae) show weak population structuring within major geographic regions26,27. When structuring is seen, it is hypothesized to be driven by local adaptation28 or geographic isolation29. It is unlikely that disjunct Bachman’s warbler populations developed any local adaptive differentiation based on habitat differences, given that their specialized breeding habitat was likely consistent across their range30. Thus, if Bachman’s warbler populations were genetically structured, this likely would result from isolation, perhaps as a consequence of anthropogenic habitat fragmentation.
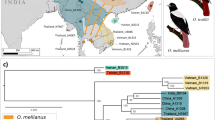
(a) Hypothesized breeding (blue) and wintering (orange) range of the Bachman’s warbler (Vermivora bachmanii) prior to extinction. Circles show collection localities of specimens examined in this study with color corresponding to the three mainland sampling regions—Eastern (n = 10; light green), Migratory (n = 17; brown), and Western (n = 19; dark green). Specimens in the Eastern and Western regions collected outside of the hypothesized breeding range (blue) likely represent an expansion of the known breeding range. Bachman’s warbler image by Louis Agassiz Fuertes, 1907, Warblers of North America, New York, Appleton. (b) Unrooted network of five unique mitochondrial DNA (mtDNA) haplotypes derived from a concatenated alignment of domain I of the control region, NADH dehydrogenase subunit 2, and cytochrome b observed in individuals sampled from the migratory route (FL and LA) and breeding ranges (all other states). See Table 1 for key to state abbreviations. (c) Bayesian skyline estimates of historical population size (Ne) based on mtDNA (dark middle line represents the median estimate of Ne, while the gray outer lines represent the upper and lower 95% HPD confidence intervals).
Without reliable census data, the only method of assessing historical demography of Bachman’s warbler populations is through genetic analysis of museum specimens. Here, we use mtDNA and nuclear SNPs to investigate population connectivity and estimate the effective population size (Ne) of Bachman’s warblers using historical museum specimens collected between 1888 and 1924, a period during which the species became known as one of the rarest warblers in North America12.
Results
Mitochondrial DNA
For all analyses, samples were designated as “Eastern'' (Atlantic coastal plain), “Western” (interior U.S.; Fig. 1a), and “Migratory” (south of breeding range) based on collection location (Table S1). Concatenated alignments were up to 117 bp for the control region, 100 bp for cytochrome b, and 126 bp for ND2. Out of n = 48 individuals, we recovered five haplotypes (Fig. 1b; Table 1; Table S4). The majority of individuals shared a single common haplotype, and three out of the four other haplotypes were sampled from individuals in the migratory route (Fig. 1b). θ was estimated to be 0.014 with a 95% credible interval of 0.007–0.0261, which corresponds to a Nef of approximately 574 (95% CI, 316–1060; Fig. 1c).
Nuclear population structuring
Of the n = 46 individuals sequenced, we retained n = 32 samples after filtering for missingness in the restricted dataset (Table 2). From 761,336,806 aligned reads with a 58% alignment rate resulting in 22,853,493 sites, we retained 6,436 SNPs with a mean missingness of 0.16% and mean depth of 54.4 × for the full dataset (Table S1) and 12,509 SNPs with a mean missingness of 0.05% and mean depth of 46.9 × for the restricted dataset.
Estimates of genetic diversity were comparable between the Eastern, Migratory, and Western sampling regions (Table 2). Although differences between regions were minor, genetic diversity estimates were slightly higher for samples collected on the migratory grounds (Table 2), potentially reflecting either higher genetic diversity or unknown population structuring in unsampled breeding locations represented by these migrants. We found greater estimates of FIS for the Western samples (Table 2); however, this may have been an artifact of sampling error since samples from this region had greater mean frequency of missing data and lower depth (Table S1, Fig. S2), FST was low between all pairs of sampling locations, despite being statistically significant for Eastern-Western and Migratory-Western pairings (Table S5). However, the test for IBD was significant for both the entire dataset (r = 0.24; p < 0.0001) and the dataset of breeding birds only (r = 0.26; p = 0.002), indicating a correlation between geographic and genetic distances (Fig. S3a, b).
STRUCTURE identified K = 1 as the likeliest number of clusters (Fig. 2a), with STRUCTURE plots showing partitions evenly split between samples for all iterations of K (Fig. 2). Results were identical for the subsampling test (Fig. S4). Geographic regions were largely overlapping in PCA, with the likeliest number of clusters as K = 1 (Fig. 2b). However, while PCA showed overlap between the Eastern and Migratory regions, there was some differentiation of the Western region, with some samples from Kentucky spreading away from the Eastern and other Western samples on PC1 (Fig. S5).
Estimates of genetic clustering among the Eastern, Western and Migratory regions based on the full SNP set. (a) Bayesian analysis of population structure and admixture estimated in STRUCTURE, with bars along the x-axis representing individuals within each region and the y-axis representing their proportion of group membership under models estimated for 2, 3, and 4 populations (K). (b) Principal components analysis (PCA) plotted on the first two PCs, with points representing individual samples, colors representing sampling region, and ellipses representing clustering of each region.
Plumage genes
We observed a total of 1769 candidate SNPs [of which 716 with a minor allele count (MAC) ≥ 2] within 1 kb of the baited plumage genes. 199 of these candidate SNPs (93 MAC ≥ 2) overlapped the baited plumage gene exons. Further functional analyses are required to elucidate the possible roles these variants may play in warbler plumage.
Overharvest by museum collections
We determined that 332 specimens of Bachman’s warblers were collected in the eighteenth and nineteenth centuries, with over 200 of these being collected in a 5-year period (1888–1893, Ornis Data Portal). When compared to the six other extant warbler species with similar distributions, this number was found to be on the lower end of the number of specimens collected between 1820 and 1940, suggesting Bachman’s warbler was likely not impacted by overharvesting by museum collections more than other contemporaneous species. Relative proportions of historical collection totals were roughly correlated to modern population sizes for extant species (Fig. 3).
Comparison of modern population size (as reported by BirdLife International, http://datazone.birdlife.org/species/) and number of specimens collected between the years 1820–1940. Collections numbers were obtained by searching for the species name using the advanced search option on the VertNet online database (http://portal.vertnet.org/search) with Basis of Record noted as Preserved Specimen and with all entries corresponding to nest and eggs and without a recorded collection year removed.
Discussion
Genomics is increasingly being used to help guide the conservation management of threatened species31. Museum genomic approaches offer a unique avenue for enhancing genomics-guided conservation perspectives—e.g., via exploring temporal changes in genetic diversity24,32,33, testing species boundaries between extinct and extant species6,24,30, and exploring historical demographic changes4. Here, we investigated the historical population structuring of the Bachman’s warbler, a narrow habitat specialist that went extinct in the Anthropocene. As in situ data are sparse for most species that went extinct prior to the twenty-first century, our study showcases how invaluable museum collections are as a repository of species’ responses to past threatening processes, often representing the only means of unlocking the historical demographics of extinct species.
We found no signals of strong population structuring across the breeding range of Bachman’s warblers in both mtDNA and genome-wide SNPs. Nuclear SNPs showed little differentiation, however, significant isolation-by-distance was detected, indicating minor geographic variation between populations. We also found some subtle east–west partitioning, with some samples from Kentucky as differentiated from the rest of the region, although it is not clear why these specific samples exhibited more variation. MtDNA haplotypes were not geographically structured, and all but one sampling site shared a common haplotype. This suggests either ongoing connectivity between populations or recent common ancestry following late Pleistocene population expansions. Signals from the nuclear and mtDNA genomes suggest that Bachman’s warblers did not experience long periods of isolation within fragmented habitat patches prior to their extinction, which is consistent with anecdotal reports of rapid population declines beginning in the 1920s12. Estimates of genetic diversity were equivalent between breeding populations, indicating similar demographic trajectories between sites across the Bachman’s warbler’s breeding range. These results provide the first evidence of range-wide population connectivity for this extinct Parulid species, and are consistent with the weak population structuring typically found in other, extant New World warbler species26,27.
Our sequence data lends further support to the hypothesis that Bachman’s warblers were not a common species, which may have contributed to their decline. Our mtDNA findings of low Ne are consistent with results from prior findings of low heterozygosity in Bachman’s warblers24, and may most accurately reflect the state of the Bachman’s warblers’ population demographics at the time of its extinction. Results from the Bayesian skyline estimates indicate a relatively stable long-term Ne, which supports the theory that Bachman’s warbler was a naturally rare species, likely due to the ephemeral nature of their primary breeding habitat. This rarity, combined with ecological traits, may have made the Bachman’s warbler more vulnerable to ecological disturbances. Swainson’s warblers (Limnothlypis swainsonii) are habitat specialists with a similar breeding distribution to Bachman’s warblers, and have also been found to have a low population-level Ne, with an estimated Ne of < 200 individuals per breeding population33. Swainson’s warblers are also considered an uncommon species; however, although they experienced the same loss of flooded forest habitat as the Bachman’s warbler, the species persisted and is currently not a species of conservation concern. Although similar ecologically, Swainson’s warblers have a broader overwintering range than Bachman’s warblers, which may have contributed to greater population stability in the face of breeding habitat loss33. This example highlights the complex nature of the various intrinsic and extrinsic traits that work together to contribute to heightened extinction risk.
Additional factors could have been responsible for the Bachman’s warblers’ decline. It has been suggested that human persecution in the form of overharvest by museum institutions may have contributed to population declines13. At the lower range of our population estimates, specimen collection could have represented additive mortality that may have contributed to population instability34. However, we found the rate of Bachman’s warbler collecting to be comparable to or lower than that of other extant warbler species with similar restricted ranges and population sizes (Fig. 3). Based on these findings it is possible that collection did not significantly impact the Bachman’s warbler populations; however, without contemporaneous population estimates or other forms of data, it is difficult to speculate on how such external factors such as harvest, disease, or parasitism may have contributed to population declines in the species.
Our analyses provide further evidence that the Bachman’s warblers’ story is a cautionary tale of extinction resulting from habitat destruction. The Bachman’s warblers’ breeding grounds historically hosted 56% of bottomland forest in the United States, a habitat type that currently occupies less than 2% of its former range35. Although rates of loss have slowed in modern times, wetland destruction continues in the southeastern United States, and wetlands-dependent taxa continue to accordingly decline36,37. The same habitat destruction that devastated the Bachman’s warbler also led to the extinction of the Carolina parakeet and the likely extinction of the ivory-billed woodpecker, two species which also relied on the same flood-plain forests38. Although remnants of that habitat type persisted and remain today, it was not enough to support viable populations of these habitat specialists. The rapid decline and extinction of these species serve as an example of the importance of habitat conservation and a reminder that wildlife extinctions will continue as habitat destruction persists.
Methods
We sampled toe pad tissue samples from n = 55 museum specimens from 7 institutions (Table S1) from across the known Bachman’s warbler range (Fig. 1a). Total genomic DNA was extracted from tissue samples at a specialized facility using an organic DNA extraction method39. From the 55 specimens, we performed mitochondrial PCR and sequence analysis on n = 48 specimens and obtained genome-wide SNPs via sequence capture on n = 46 individuals.
Mitochondrial DNA
Based on sequence we obtained from the Bachman’s warbler’s closest relatives, blue-winged (Vermivora cyanoptera) and golden-winged (V. chrysoptera) warblers40, we designed amplification and sequencing primers targeting ~ 100 bp fragments of the mitochondrial genome in domain I of the control region, NADH dehydrogenase subunit 2 (ND2) and cytochrome b. We then performed amplifications using these primers in n = 48 samples (Eastern: n = 11; Migratory, n = 17; Western, n = 13). To minimize contamination risk, all PCRs were prepared in the ancient DNA facility at the Smithsonian’s Center for Conservation Genomics (CCG) before transfer to PCR thermocyclers in the general genetics lab for amplification under the conditions reported in39. Raw mtDNA sequence data was aligned using GENEIOUS PRO 5.1.741, and converted to analysis formats using FABOX 1.3542. We combined all sequences and used LAMARC 2.043 to estimate Bayesian skyline estimates of historical population size (Ne) using a mutation rate of 2% per million years, based on the average reported for Passerines4. Generation time was defined as 1.8 years based on the estimated generation time for the Yellow-Rumped Warbler (Dendroica coronata)44. We defined θ as θ = 2Nefμ, where Nef equals the effective female population size and μ equals the mutation rate per sequence, per generation. We then quantified θ with the Bayesian module in LAMARC 2.043, using ten initial chains with a sampling interval of 100 steps and with 50,000 trees sampled with a burn-in of 10,000 and with two replicates per run for two runs. We used TRACER v 1.545 to determine run length.
Bait design and genome capture methods
We designed a custom myBaits (Arbor Biosciences, Ann Arbor, MI) in-solution capture baits set from three primary sources of genetic variation—shotgun genomes of Bachman’s warblers and their two congeners, Ultra-conserved Element (UCE) raw reads from Bachman’s warblers4, and candidate plumage genes known from other warblers28,46,47,48,49,50. We selected the chromosome-level genome assembly of a close relative, the myrtle warbler (Setophaga coronata coronata) (Assembly mywa_2.1; GenBank Accession GCA_001746935.2)46, as the reference genome for this study. To prepare the reference genome, we used RepeatMasker 4.0.951 (using RMBlast 2.9.0+) to annotate repeats using the Aves repeat database (options: --gccalc --nolow -species Aves). Next, we used RepeatModeler 2.0.152 on the initial repeat-masked genome to build a custom myrtle warbler repeat database. We produced a final repeat-masked genome by rerunning RepeatMasker on the myrtle warbler assembly using the custom myrtle warbler repeat database.
Genome-wide baits
We shotgun sequenced two Bachman’s warblers (AMNH 380148 and CAS 53742) on a test MiSeq and then a full a HiSeq lane, obtaining a total of 308,014,040 and 365,719,262 reads, respectively. We also downloaded raw genome resequencing reads for the golden-winged warbler (Vermivora chrysoptera; SRR4017514) and blue-winged warbler (Vermivora cyanoptera; SRR4017516) from NCBI SRA47. AdapterRemoval 2.3.153 was used to remove adapter sequences, trim Ns and low quality reads, discard reads shorter than 25 bp, and merge paired end reads. BWA 0.7.1754 was used to align reads to the repeat-masked myrtle warbler reference genome using the backtrack algorithm and a minimum quality score of 15. Within PALEOMIX, we also used mapDamage 2.2.155 to rescale quality scores of the two Bachman’s warbler shotgun genomes derived from historical museum specimens. SNPs were called on each alignment using GATK 4.1.3.0 HaplotypeCaller56 with default settings, followed by combineGVCF and GenotypeGVCFs to perform joint genotyping across all four genomes and between just the two Bachman’s warbler genomes, and VariantFiltration to perform initial hard filtering based on the following settings—QD < 2.0, FS > 40.0, MQ < 30.0, MQRankSum < − 12.5, ReadPosRankSum < − 8.0). We then used VCFtools 0.1.1657 and BCFtools 1.7.258 to filter SNPs based on minimum quality < 30, depth < 5 and > 20, N_ALT = 1, removing invariants and restricting SNPs to those mapped to chromosomes.
A total of 30,486 SNPs were called across a dataset containing only Bachman’s warblers and 8089 SNPs for the dataset containing all three warbler species. We retained SNPs localized on chromosomes using VCFtools. We generated 120 bp (option -L120) candidate baits using BaitsTools 1.7.2 vcf2baits59. Each SNP was covered by one candidate bait with the SNP placed at the 61st base of the bait (options -b60 -k1). We requested up to 1500 sites that segregated between Bachman’s warblers and the other warbler species and 13,500 sites that were variable within Bachman’s warblers (options --taxacount 0,1500,13500 --popcategories 13500,0). We required SNP sites to be a minimum 10,000 bp apart (option -d10000) and scaled the number of selected SNPs per chromosome by the chromosome lengths (option -j). We excluded candidate baits that included gaps, Ns or were less than 120 bp long (options -c -N -G exclude). We required baits to have GC contents between 30 and 50% (options -n30.0 -x50.0) and have a linguistic complexity at least 0.9 (option -y0.9). We removed baits that had homopolymer runs longer than 4 bp (option -J4) or overlapped repeat-masked regions by more than 25% (option -K25). After filtration, we retained 4340 candidate baits (representing 4340 SNPs at 1× coverage).
Ultra-conserved element baits
We obtained NCBI SRA raw sequence reads for ten Bachman’s warblers previously generated using sequence capture of UCE baits from Tilston Smith et al.4 (Table S2). These were aligned to the myrtle warbler genome using a custom pipeline (derived from60) incorporating Trim Galore! 0.6.461 (using Cutadapt 2.462 and FastQC 0.11.863) for read trimming and adapter removal, BWA-MEM 0.7.1764 for read mapping, Picard Tools 2.20.6 for marking PCR duplicates65 and GATK 3.8.1.0 for read re-alignment56. SNPs were called using GATK 4.1.3.0 and filtered as per the shotgun genomes above. Using BaitsTools 1.7.2 vcf2baits59, we then generated complementary UCE SNP baits to the genome-wide SNP baits (option --previousbaits). We requested baits to cover up to 30,000 SNPs. Otherwise, we generated baits under the same parameters as the genome-wide SNP baits. After filtration, we retained 7814 candidate baits (representing 7814 SNPs at 1× coverage).
Plumage gene baits
We performed a literature search for plumage pigmentation genes involved in carotenoid and melatonin production, and this list was narrowed down to seven candidate genes discovered between golden-winged and blue-winged warblers and other warblers28,46,47,48,49,50 (Table S2). We then used Ensembl version 102 (accessed November 2020) to extract sequences for each gene using the zebra finch genome (Taeniopygia guttata; bTaeGut1_v166). These sequences were then aligned to our repeat-masked myrtle warbler genome and we extracted their locations in BED format. We used BaitsTools 1.6.8 bed2baits to generate 120 bp baits (option -L120), padding 60 bp upstream and downstream of the gene coordinates (option -P60), and tiling every 15 bp (option -O15). We retained baits with gaps (option -G include). We excluded candidate baits less than 120 bp (option -c) and unresolved bases (Ns, option -N). We required candidate baits to have GC contents between 30 and 60% (options -n30.0 -x60.0). We removed baits that had homopolymer runs longer than 4 bp (option -J4) or overlapped repeat-masked regions by more than 25% (option -K25). After filtration, we generated 175 baits, covering the target sequences at a mean depth of 2.0×.
Final bait set
The three candidate bait sets were submitted to Daicel Arbor Biosciences for BLAST (Basic Local Alignment Search Tool67) analysis for building a myBaits in-solution capture assay. 283 bait candidates (5 plumage baits, 56 genome baits, and 222 UCE baits) were removed for having more than one BLAST hit. To improve the likelihood of capturing damaged DNA molecules, each of the surviving 120 bp candidate bait sequences was converted into two tiled 80 bp baits (overlapping by 40 bp), generating a total of 24,092 candidate baits (340 plumage gene baits, 8,568 genome SNP baits, and 15,184 UCE baits). We refiltered the 80 bp bait sequences using BaitsTools 1.7.4 checkbaits using the appropriate filtration parameters noted above. After final filtration, we retained 21,581 candidate baits (317 plumage gene baits, 7637 genome SNP baits, and 13,627 UCE baits). Using a custom script (random_downsample.rb), we then randomly removed 1581 UCE baits (leaving 12,046 UCE baits) to fit into a 20,000-bait myBaits kit (Fig. S1). The final bait set and custom scripts are available in the bait-development repository (https://github.com/campanam/bait-development/tree/main/BAWA).
Genomic library construction and capture
We prepared genomic libraries using SRSLY PicoPlus Uracil + kits (Claret Bioscience, Santa Cruz, CA, USA). We prepared and double-indexed libraries with unique P5 and P7 barcodes in a PCR-free ancient laboratory before transferring them to a separate laboratory facility for PCR. We quantified PCR product concentration using a Qubit Fluorometer 3.0 (Invitrogen, Carlsbad, CA, USA) and visualized mean library insert sizes with an Agilent 2200 TapeStation (Agilent Technologies, Santa Clara, CA, USA). We performed capture by first combining libraries into equimolar pools of three samples per pool with a targeted amount of ~ 300 ng DNA per pool. We then followed the MyBaits v5.0 protocol (Arbor Biosciences, Ann Arbor, MI, USA) for capture, with a modified 65 °C hybridization temperature, a 48-h hybridization time, and a 16 cycle post-capture PCR. Enriched pools were combined at equimolar ratios. We sequenced captures as paired-end 150 bp reads on a single lane of an Illumina HiSeq X by Admera Health (South Plainfield, NJ, USA).
Genotyping
We demultiplexed sequenced genomic reads with Bcl2fastq 1.8.4 (Illumina, Inc., San Diego, CA, USA). Following demultiplexing, we cleaned raw sequence files by trimming adaptors and removing low-quality bases with Trimmomatic 0.3968 within the illumiprocessor 2.10 wrapper69. We then aligned reads from individual samples to our myrtle warbler reference genome using BWA-MEM 0.7.1764. Damage to museum DNA can lead to false identification of SNPs, and we therefore rescaled quality scores for each sample using mapDamage 2.055. Following rescaling, we removed duplicates with Picard 2.20.665 and called SNPs for individual sample GVCF files using default settings in the program HaplotypeCaller in GATK 4.1.3.0. We then combined individual GVCF files into a single file using the GATK command CombineGVCFs, indexed the merged GCVF file, and genotyped all samples in the combined file with GenotypeGVCFs, resulting in our final file of raw SNPs. We filtered SNPs in the final VCF file within VCFtools 0.1.1657 by removing indels and all loci below a Phred-scaled minimum genotype quality of 30. We then filtered samples to a minimum per sample site depth of 5×, total maximum site depth of 100×, and minor allele frequency of 0.01. For the first set, hereby referred to as the “full” set, we removed SNPs with > 20% missing data across all samples. For the second set, hereby referred to as the “restricted” set, we removed SNPs with > 10% missing data across all samples and individual samples with > 60% missing data. Most of the specimens removed due to low quality had a small initial library fragment size (≤ 170 bp) before capture, which may have resulted from degradation of either the skin or the DNA sample during storage or handling. Finally, for both sets, we thinned SNPs within 1000 bp of each other to remove loci potentially in linkage disequilibrium.
Data analysis
Spatial analyses of genetic structure were conducted using the three a priori regional groups. High rates of missing data in individual samples can bias inferences of population genomic parameters such as genetic diversity70, and population genetic parameters were therefore estimated using the restricted SNP set. We quantified population genetic parameters and pairwise FST using the basic.stats function in the R v4.2.271 program hierfstat v.0.5-1172. We estimated pairwise FST between populations with the genet.dist function in hierfstat and calculated 95% confidence intervals for both F statistics with 104 permutations. We tested for isolation-by-distance (IBD) using the full SNP set for all birds and breeding birds only by constructing an identity-by-state (IBS) distance matrix (1 − pairwise proportion of shared alleles) in SNPRelate 1.14.073. We converted Euclidean geographical distance between sample sites to geodesic distance (in kilometers) using the R package ‘geodist’ (Padgham 2021), and used this geographic metric and the IBS distance metric to conduct a Mantel test with 104 permutations in ADE4 1.7.1174.
We investigated the likeliest number of genetic clusters using STRUCTURE 2.3.475. Because of potential for sampling bias given the large difference in number of samples between regions in the full set76, we also ran STRUCTURE with samples from the breeding grounds only, subsampling Western birds to approximate the Eastern sample size by removing Western samples based on missingness. We ran STRUCTURE simulations for 1–5 possible clusters (K) for 10 iterations each with 104 MCMC repetitions after a burn-in of 104 generations using the no admixture model with correlated allele frequencies and with location information included as a prior. We determined the likeliest number of genetic clusters via the mean log likelihood from each iteration of K [LnP(K)]. We then aligned clusters, merged STRUCTURE runs by K value, and visualized output using the R v4.2.2 package Pophelper 2.3.077.
We also explored sampling clustering using Principal Components Analysis (PCA) in the R package ‘adegenet’78. In addition to using PCA to visualize data clustering a priori, we used the K-means clustering via the find.clusters function to identify the most likely number of genetic clusters in our dataset without prior population information, retaining all PCs and selecting the number of clusters based on the lowest Bayesian Information Criterion (BIC) value.
Museum specimen analysis
We used the Ornis Data Portal and the VertNet online database (http://portal.vertnet.org/search) to search for all recorded Bachman’s warbler specimens collected in the eighteenth and nineteenth centuries and compiled records by year and sampling location. To compare between species, we used the same portals to search for records of seven warbler species (Fig. 3) using the advanced search function and with Basis of Record noted as “Preserved Specimen.” For all species, we restricted years of collection to between 1820 and 1940 (corresponding to the earliest and latest recorded Bachman’s warbler specimens), and removed all entries that corresponded to nests and eggs or lacked a collection year. We then obtained modern population sizes for all extant species using data compiled by BirdLife International, http://datazone.birdlife.org/species/).
Data availability
Genomic data from this project are available on Genbank under project number PRJNA994867. Bait set and custom bait design scripts are available at https://github.com/campanam/bait-development/tree/main/BAWA. Data analysis scripts and SNP VCF files are available at https://github.com/pabyerly/BAWA.
References
Elphick, C. S., Roberts, D. L. & Michael Reed, J. Estimated dates of recent extinctions for North American and Hawaiian birds. Biol. Conserv. 143, 617–624 (2010).
Ducatez, S. & Shine, R. Drivers of extinction risk in terrestrial vertebrates. Conserv. Lett. 10, 186–194 https://doi.org/10.1111/conl.12258 (preprint) (2017).
Owens, I. P. F. & Bennett, P. M. Ecological basis of extinction risk in birds: Habitat loss versus human persecution and introduced predators. Proc. Natl. Acad. Sci. USA 97, 12144–12148 (2000).
Smith, B. T., Gehara, M. & Harvey, M. G. The demography of extinction in eastern North American birds. Proc. R. Soc. B Biol. Sci. 288 (2021).
Johnson, J. A., Toepfer, J. E. & Dunn, P. O. Contrasting patterns of mitochondrial and microsatellite population structure in fragmented populations of greater prairie-chickens. Mol. Ecol. 12, 3335–3347 (2003).
Kearns, A. M. et al. Conservation genomics and systematics of a near-extinct island radiation. Mol. Ecol. 31, 1995–2012 (2022).
Mathur, S., Tome, J. M., Tarango-arámbula, L. A., Perez, R. M. & Dewoody, J. A. An evolutionary perspective on genetic load in small, isolated populations as informed by whole genome resequencing and forward-time simulations. Evolution 77, 1–15 (2023).
O’Grady, J. J. et al. Realistic levels of inbreeding depression strongly affect extinction risk in wild populations. Biol. Conserv. 133, 42–51 (2006).
Palstra, F. P. & Ruzzante, D. E. Genetic estimates of contemporary effective population size: What can they tell us about the importance of genetic stochasticity for wild population persistence?. Mol. Ecol. 17, 3428–3447 (2008).
Femerling, G. et al. Genetic load and adaptive potential of a recovered avian species that narrowly avoided extinction. Biol. Sci. https://doi.org/10.1101/2022.12.20.521169 (2022).
Lawrence, G. N. The rediscovery of Bachman’s Warbler, Helminthophila bachmani (Aud.), in the United States. Auk 4, 35–37 (1887).
Stevenson, H. M. The recent history of Bachman’s Warbler. Willson Bull. 84, 344–347 (1972).
Hamel, P. B. Bachman’s Warbler. Audubon Wildl. Rep. 1988(1989), 624–635 (1988).
U.S. Fish and Wildlife Service. Endangered and Threatened Wildlife and Plants; Removal of 23 Extinct Species from the Lists of Endangered and Threatened Wildlife and Plants. Vol. 86. 54298–54338 (2021).
Knopf, F., Johnson, R., Rich, T., Samson, F. & Szaro, R. Conservation of riparian ecosystems in the United States. Wilson Bull. 100, 272–284 (1988).
Steinberg, M. K. The importance of cultural ecological landscapes to the survival of the Bachman’s Warbler (Vermivora bachmanii) in the Southeastern United States. Southeast. Geogr. 50, 272–281 (2010).
Terborgh, J. Preservation of natural diversity: The problem of extinction prone species. Bioscience 24, 715–722 (1974).
Rappole, J. H. Analysis of plumage variation in the Canada Warbler. J. Field Ornithol. 54, 152–159 (1983).
Hughes, R. H. Observations of cane (Arundinaria) flowers, seed, and seedlings in the North Carolina coastal plain. Bull. Torrey Bot. Club 78, 113 (1951).
Platt, S. G., Brantley, C. G., Platt, S. G. & Brantley, C. G. Canebrakes: An ecological and historical perspective. South. Appalachian Bot. Soc. 62, 8–21 (2019).
Andrén, H. Effects of habitat fragmentation on birds and mammals in landscapes with different proportions of suitable habitat: a review. NCASI Tech. Bull. 12–13 (1999).
Magrach, A., Larrinaga, A. R. & Santamaría, L. Changes in patch features may exacerbate or compensate for the effect of habitat loss on forest bird populations. PLoS One 6 (2011).
Lindsay, D. L. et al. Habitat fragmentation and genetic diversity of an endangered, migratory songbird, the golden-cheeked warbler (Dendroica chrysoparia). Mol. Ecol. 17, 2122–2133 (2008).
Wood, A. W., Szpiech, Z. A., Lovette, I. J., Smith, B. T. & Toews, D. P. L. Genomes of the extinct Bachman’s warbler show high divergence and no evidence of admixture with other extant Vermivora warblers. Curr. Biol. 33, 2823-2829.e4 (2023).
Frankham, R. Relationship of genetic variation to population size in wildlife. Conserv. Biol. 10, 1500–1508 (1996).
Bubac, C. M. & Spellman, G. M. How connectivity shapes genetic structure during range expansion: Insights from the Virginia’s Warbler. Auk 133, 213–230 (2016).
DeSaix, M. G. et al. Population assignment reveals low migratory connectivity in a weakly structured songbird. Mol. Ecol. 28, 2122–2135 (2019).
Carpenter, J. P. et al. Genomic variation in the Black-throated Green Warbler (Setophaga virens) suggests divergence in a disjunct Atlantic coastal plain population (S. v. waynei). Ornithology 139, 1–11 (2022).
Milot, E., Lisle Gibbs, H. & Hobson, K. A. Phylogeography and genetic structure of northern populations of the yellow warbler (Dendroica petechia). Mol. Ecol. 9, 667–681 (2000).
Baveja, P. et al. Using historical genome-wide DNA to unravel the confused taxonomy in a songbird lineage that is extinct in the wild. Evol. Appl. 14, 698–709 (2021).
Kershaw, F. et al. The coalition for conservation genetics: Working across organizations to build capacity and achieve change in policy and practice. Conserv. Sci. Pract. 4, e12635 (2022).
DeGraaf, R. M. Neotropical Migratory Birds: Natural History, Distribution, and Population Change. (Comstock Publishing Associates, 1995).
Winker, K. & Graves, G. R. Genetic structure of breeding and wintering populations of Swainson’s Warbler. Wils 120, 433–445 (2008).
Winker, K. et al. The importance, Effects, and ethics of bird collecting. Auk 127, 690–695 (2010).
Noss, R. F., LaRoe, E. T. & Scott, J. M. Endangered ecosystems of the United States: A preliminary assessment of loss and degradation. In Biological Report 20, U.S. Department of the Interior Technical Report Series, Washington DC, USA (1995).
Rosenberg, K. V. et al. Decline of the North American avifauna. Science 366, 120–124 (2019).
Schipper, A. M. et al. Contrasting changes in the abundance and diversity of North American bird assemblages from 1971 to 2010. Glob. Chang. Biol. 22, 3948–3959 (2016).
Askins, R. A. Restoring North America’s Birds. (Yale University Press, 2000).
Fleischer, R. C., Olson, S. L., James, H. F. & Cooper, A. C. Identification of the extinct Hawaiian eagle (Haliaeetus) by mtDNA sequence analysis. Auk 117, 1051–1056 (2000).
Lovette, I. J. et al. A comprehensive multilocus phylogeny for the wood-warblers and a revised classification of the Parulidae (Aves). Mol. Phylogenet. Evol. 57, 753–770 (2010).
Drummond, A. J. A. B. Geneious v5. 3. http://www.geneious.com (2010).
Villesen, P. FaBox: An online toolbox for FASTA sequences. Mol. Ecol. Notes 7, 965–968 (2007).
Kuhner, M. K. LAMARC 2.0: Maximum likelihood and Bayesian estimation of population parameters. Bioinformatics 22, 768–770 (2006).
Milá, B., Smith, T. B. & Wayne, R. K. Speciation and rapid phenotypic differentiation in the yellow-rumped warbler Dendroica coronata complex. Mol. Ecol. 16, 159–173 (2007).
Rambaut, A., Drummond, A. J., Xie, D., Baele, G. & Suchard, M. A. Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Syst. Biol. 67, 901–904 (2018).
Baiz, M. D., Wood, A. W., Brelsford, A., Lovette, I. J. & Toews, D. P. L. Pigmentation genes show evidence of repeated divergence and multiple bouts of introgression in setophaga warblers. Curr. Biol. 31, 643-649.e3 (2021).
Toews, D. P. L. et al. Plumage genes and little else distinguish the genomes of hybridizing warblers. Curr. Biol. 26, 2313–2318 (2016).
Baiz, M. D. et al. Genomic and plumage variation in Vermivora hybrids. Auk 137, 1–14 (2020).
Wang, S. et al. Selection on a small genomic region underpins differentiation in multiple color traits between two warbler species. Evol. Lett. 4, 502–515 (2020).
Brelsford, A., Toews, D. P. L. & Irwin, D. E. Admixture mapping in a hybrid zone reveals loci associated with avian feather coloration. Proc. R. Soc. B Biol. Sci. https://doi.org/10.1098/RSPB.2017.1106 (2017).
Smit, A. F. A., Hubley, R. & Green, P. RepeatMasker Open-4.0.2013-2015. http://www.repeatmasker.org (2010).
Flynn, J. M. et al. RepeatModeler2 for automated genomic discovery of transposable element families. Proc. Natl. Acad. Sci. USA 117, 9451–9457 (2020).
Schubert, M., Lindgreen, S. & Orlando, L. AdapterRemoval v2: Rapid adapter trimming, identification, and read merging. BMC Res. Notes 9 (2016).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
Jónsson, H., Ginolhac, A., Schubert, M., Johnson, P. L. F. & Orlando, L. MapDamage2.0: Fast approximate Bayesian estimates of ancient DNA damage parameters. In Bioinformatics. Vol. 29. 1682–1684 (Oxford Academic, 2013).
McKenna, A. et al. The genome analysis toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 (2010).
Danecek, P. et al. The variant call format and VCFtools. Bioinformatics 27, 2156–2158 (2011).
Danecek, P. et al. Twelve years of SAMtools and BCFtools. Gigascience 10, 1–4 (2021).
Campana, M. G. BaitsTools: Software for hybridization capture bait design. Mol. Ecol. Resour. 18, 356–361 (2018).
Cassin-Sackett, L. et al. Genetic structure and population history in two critically endangered Kaua‘i honeycreepers. Conserv. Genet. 22, 601–614 (2021).
Krueger, F. Trim Galore! Version 0.6.4. https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/ (2019).
Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17, 10 (2011).
Simon, A. FastQC Version 0.11.8. https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (2018).
Li, H. Aligning Sequence Reads, Clone Sequences and Assembly Contigs with BWA-MEM (2013).
Institute, B. Picard Tools Version 2.20.6. https://broadinstitute.github.io/picard/ (2019).
Warren, W. C. et al. The genome of a songbird. Nature 464, 757–762 (2010).
Camacho, C. et al. BLAST+: Architecture and applications. BMC Bioinform. 10, 1–9 (2009).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Faircloth, B. C. Illumiprocessor: A Trimmomatic Wrapper for Parallel Adapter and Quality Trimming (2013).
Gautier, M. et al. The effect of RAD allele dropout on the estimation of genetic variation within and between populations. Mol. Ecol. 22, 3165–3178 (2013).
R Core Team. R: A Language and Environment for Statistical Computing. ISBN 3-900051-07-0. https://doi.org/10.1038/sj.hdy.6800737 (R Foundation for Statistical Computing, 2018).
Goudet, A. J. & Goudet, M. J. Package ‘hierfstat’ (2014).
Zheng, X. et al. A high-performance computing toolset for relatedness and principal component analysis of SNP data. Bioinformatics 28, 3326–3328 (2012).
Chessel, D., Dufour, A. B. & Thioulouse, J. The Ade4 Package-I: One-Table Methods. Vol. 4 (2004).
Pritchard, J. K. Inference of population structure using multi-locus genotypes. Genetics 155, 945–959 (2000).
Puechmaille, S. J. The program structure does not reliably recover the correct population structure when sampling is uneven: Subsampling and new estimators alleviate the problem. Mol. Ecol. Resour. 16, 608–627 (2016).
Francis, R. M. pophelper: An R package and web app to analyse and visualize population structure. Mol. Ecol. Resour. 17, 27–32 (2017).
Jombart, T. & Bateman, A. adegenet: A R package for the multivariate analysis of genetic markers. Bioinformatics 24, 1403–1405 (2008).
Acknowledgements
We are grateful to staff of the following institutions for providing historical samples: the American Museum of Natural History (Paul Sweet), Cornell University Museum of Vertebrates (Irby Lovette), Field Museum of Natural History (John Bates), California Academy of Sciences (Jack Dumbacher), National Museum of Natural History (Gary Graves), the Harvard Museum of Comparative Zoology (Scott Edwards), the Academy of Natural Sciences Philadelphia (Nate Rice), and the University of Michigan Museum of Zoology (Janet Hinshaw). We thank Brian Brunelle and Zachary Hanf for their assistance with the bait design. Computing was performed on the Smithsonian High Performance Cluster ‘Hydra’ (SI/HPC), Smithsonian Institution (https://doi.org/10.25572/SIHPC). The authors have no competing interests to declare. This research was funded by a Smithsonian Postdoctoral Fellowship (AW), the Center for Conservation Genomics, and James Bond Fund grants to RCF, AMK and PPM.
Author information
Authors and Affiliations
Contributions
This study was designed by AMK, AW, PPM, AW, MGC, and RCF. Museum samples were collected by PMM, RCF and AW. Laboratory work was performed by PAB, AMK, AW, MEO under RCF supervision. Data analysis was performed by PAB, AMK, AW, AW, and MGC. PAB, AMK, and AW wrote the manuscript. All authors contributed to and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Byerly, P.A., Kearns, A.M., Welch, A. et al. Museum genomics provide insight into the extinction of a specialist North American warbler species. Sci Rep 14, 17047 (2024). https://doi.org/10.1038/s41598-024-67595-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-67595-5
- Springer Nature Limited