Abstract
The draft genome sequence of an agriculturally important actinobacterial species Amycolatopsis sp. BCA-696 was developed and characterized in this study. Amycolatopsis BCA-696 is known for its biocontrol properties against charcoal rot and also for plant growth-promotion (PGP) in several crop species. The next-generation sequencing (NGS)-based draft genome of Amycolatopsis sp. BCA-696 comprised of ~ 9.05 Mb linear chromosome with 68.75% GC content. In total, 8716 protein-coding sequences and 61 RNA-coding sequences were predicted in the genome. This newly developed genome sequence has been also characterized for biosynthetic gene clusters (BGCs) and biosynthetic pathways. Furthermore, we have also reported that the Amycolatopsis sp. BCA-696 produces the glycopeptide antibiotic vancomycin that inhibits the growth of pathogenic gram-positive bacteria. A comparative analysis of the BCA-696 genome with publicly available closely related genomes of 14 strains of Amycolatopsis has also been conducted. The comparative analysis has identified a total of 4733 core and 466 unique orthologous genes present in the BCA-696 genome The unique genes present in BCA-696 was enriched with antibiotic biosynthesis and resistance functions. Genome assembly of the BCA-696 has also provided genes involved in key pathways related to PGP and biocontrol traits such as siderophores, chitinase, and cellulase production.
Similar content being viewed by others
Introduction
The relationships of microbes with plants have been of diverse types, among which plant growth promotion (PGP) activities of microbes is one of the beneficial types to the plants. Beyond the lab or field-level characterizations of the PGP traits, there have been several recent studies where genomic approaches were used for the characterization of plant growth-promoting traits in rhizosphere bacteria. Some of the PGP traits studied include siderophores and indole acetic acid (IAA) production, anti-fungal properties, mineral solubilization, nitrogen fixation, activity of enzymes such as ACC deaminase, lipase, chitinase and cellulase1,2,3,4,5,6,7,8,9,10,11,12,13. Our group also reported draft genomes of sixteen PGP strains of Streptomyces and predicted genes underlying important PGP or biocontrol traits14. A typical approach used by these studies involves the decoding of genome sequence followed by identification of unique genomic islands, prediction of Biosynthetic Gene Clusters (BGC), comparison of BGCs as well as metabolic pathways across genomes, etc.
Amycolatopsis sp. BCA-696, one of the strains of an important genus of actinobacteria, was reported previously for its plant growth-promotion traits in sorghum and chickpea, and for its antagonistic potential against Macrophomina phaseolina that causes charcoal rot disease in sorghum in greenhouse and field screens15,16. Its PGP properties include the production of IAA, siderophore, lipase, cellulase, protease, chitinase, hydrocyanic acid (HCN), and β-1,3-glucanase. Besides these, it was also able to survive in moderate to extreme environmental conditions such as pH (5–11), salinity (0–6% NaCl), and temperature (20–40 °C)15.
Amycolatopsis was originally classified under the genus Nocardia17 but based on a combination of genotypic and phenotypic characters, this was reclassified under the genus Amycolatopsis18. It is a genus with high GC% and gram-positive bacteria belonging to the family Pseudonocardiaceae. These bacteria can produce different types of antibiotics and medicinally important metabolites due to this reason this genus is widely studied19,20,21. Due to the advanced and inexpensive genome sequencing techniques and growing clinical importance, 65 genome sequences of Amycolatopsis bacteria are currently publicly available in the NCBI genome database (https://www.ncbi.nlm.nih.gov/genome/). While Amycolatopsis genomes have been mined mainly for secondary metabolites22, not much information is available for novel genes/pathways underlying the PGP traits23. In the current investigation, the genome sequence of Amycolatopsis sp. BCA-696 was decoded to identify the BGCs and/or pathways associated not only with the PGP/biocontrol traits but also with some of the traits related to tolerance to pathogens. Besides, this strain was found to be distinct enough to be a separate species, and a pan-genome analysis of genomes of the Amycolatopsis genus was also conducted.
Results
Amycolatopsis sp. BCA-696 draft genome
In order to obtain the draft genome of this strain, sequencing was done using two types of libraries, i.e., paired-end (PE) and mate-pair (MP), yielding ~ 9.9 million PE reads of size 100 bases and ~ 6.2 million MP reads of size 250 bases, respectively, with approximate coverage of > 500× (assuming ~ 9.5 Mb as median assembly size of Amycolatopsis genomes available at NCBI).
The pre-processed reads were assembled de novo, initially generating 112 contigs, which were further ordered to obtain a single linear scaffolded genome sequence (Table 1, Supplementary Table S1). The genome sequence was of length 9,059,528 bp with GC content of 68.75% and a very nominal number of anonymous nucleotides (total size: ~ 6.5 Kb) (Fig. 1).
Visualization of different features on the Amycolatopsis sp. BCA-696 genome. Coding regions on forward and reverse strands are shown as 1st and 2nd concentric circular layers from outside(the colors differentiate the gene categories shown in Fig. 2), 3rd layer shows an average read depth in a 5 kb window along the genome, 4th layer shows GC skew (positive values in blue and negative values in brown color), 5th layer shows rRNAs (blue bands), tRNAs (red bands), and CRISPR spacers (black bands), and 6th layer shows similar repeats on the genome connected by lines (generated using Circos V.0.69.8).
To assess the completeness of the draft assembly, a reference set of 356 “Benchmarking Universal Single-Copy Orthologs” (BUSCOs) derived from 948 genomes from class actinobacteria lineage was created, and 355 (98.9%) BUSCOs were present in the Amycolatopsis sp. BCA-696 assembly. Out of the 355 BUSCOs identified, all of them were complete and just three of them were duplicated. Only one BUSCO was fragmented, indicating a high degree of completeness of the generated assembly.
Amycolatopsis sp. BCA-696 genome annotation
The annotation of the genome assembly provided a total of 8,716 protein-encoding genes (PEGs), with an average length of 939.02 bp, occupying 90.34% of the genome (Table 1). The genome also contained 61 RNA coding regions, 213 repeat regions, 42 CRISPR repeats, 40 CRISPR spacer, and 2 CRISPR array regions. Out of 61 RNAs, the number of rRNA and tRNA genes were 6 and 55, respectively (Table 1). Among the predicted PEGs, 5289 (60.66%) proteins were assigned with functions and 3427 (39.30%) were hypothetical proteins. The cofactors, vitamins, prosthetic groups, and pigments (344 genes) found to be the most abundant subsystem class, followed by amino acids and derivatives (247 genes), fatty acids, lipids, and isoprenoids (241 genes), stress response, defense, and virulence (208 genes), protein synthesis (204 genes), energy and precursor metabolites generation (167 genes), cell cycle, cell division, and death (117 genes), respiration (113 genes), and membrane transport (77 genes), etc. (Fig. 2).
Pan genome of Amycolatopsis species & genes specific to BCA-696
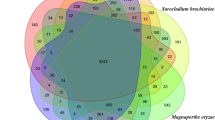
In order to infer the genomic similarity or variation among the Amycolatopsis species, a pan-genome analysis was conducted. A pan-genome, comprising all genomes of the Amycolatopsis genus with a scaffold or higher level assembly (n = 76) demonstrated huge diversity in terms of gene composition: out of a whopping 375,299 orthologous groups, core genes were a much smaller set (< 1%) compared to the cloud genes (> 95%) (Supplementary Fig. S1). Since our focus was on genomic regions unique to Amycolatopsis sp. BCA-696, a subset comprising this genome and the genomes of fourteen closely related species/strains, all from a single clade within the pan-genome-based tree, were re-analyzed. A total of 35,318 orthologous gene clusters could be divided into a core genome of 3,627 (41.6%) orthologous gene clusters, having more than 99% similarity present across all fifteen strains, and the unique genes which ranged from 654 (in A. keratiniphila) to 2557 genes (in Amycolatopsis coloradensis) (Supplementary Fig. S2). The genome of Amycolatopsis sp. BCA-696 has 1423 (16.3%) strain-specific genes. Since as many as 1/6th (n = 1423) of total genes were reported as unique, this required a rigorous evaluation by using an algorithm that establishes orthology very accurately. Orthology search using Orthofinder pipeline24 reported a core set of 4,733 (42.6%) genes and 466 (4.2%) unique genes in Amycolatopsis sp. BCA-696 genome (Fig. 3). Out of these 466 unique genes, only 53 genes could be functionally annotated(by RAST pipeline25 and Reciprocal Best Blast) (Supplementary Table S2).
Flower plot showing Core genes and unique genes between fifteen closely related Amycolatopsis strains based on Orthofinder results. The number of core genes, shared by all of them, is shown in the center of the flower plot, and the unique genes for each strain are as flower petals (generated using rstudio using plotrix V.3.8-4) (for the list of type strains refer to “Materials and methods”).
Among these 53 unique genes, one gene (Genbank ID: WYW18527.1) has been found to be involved in the biosynthesis of Bialaphos antibiotic (carboxyvinyl-carboxyphosphonate phosphorylase). Previously this antibiotic was reported only in Streptomyces species, possessing bactericidal, fungicidal, and herbicidal properties26. Other unique genes included drug antiporters, multiple transporter proteins, genes conferring resistance to antibiotic chloramphenicol and streptomycin, and endonucleases.
Phylogenetic relationships of BCA-696
To establish the taxonomic positioning of BCA-696 within the Amycolatopsis genus, we used methods based on the overall genome relatedness index, which showed that Amycolatopsis sp. BCA-696 genome was closer to the genomes of A. lurida strains than any other Amycolatopsis genomes (Fig. 4). But the bootstrap value for the branch leading to BCA-696 was poor (~ 50 out of 100) showing the uncertainty in the position of BCA-696. Since the previous taxonomic assignment of this strain was based on 16S rRNA sequence, the availability of draft genome information of the strain has positioned it in the phylogenetic tree equidistant to A. lurida and A. roodepoortensis without being closer to either of them, suggesting this strain might be a new species.
Phylogenetic tree obtained from TYGS server comparing genomes of Amycolatopsis BCA-696 with several closely related species/strains available in the online server (The type strains are indicated with a “T” in superscript). The TYGS analysis differed slightly from the results of Roary and Phylophlan3, showing A. roodepoortensis diverging from the common ancestors of A. lurida and Amycolatopsis sp. BCA-696. The bootstrap values (out of 100 iterations) are shown for each branch (generated from TYGS server https://tygs.dsmz.de/).
Biosynthetic gene clusters in Amycolatopsis sp. BCA-696 genome
In its genome of size ~ 9 Mb, 23 to 35 Biosynthetic Gene Clusters (BGCs) were predicted by two widely used BGC predictors (Fig. 5, Supplementary Table S3, Supplementary Fig. S3). The commonly observed BGC classes were ‘Resistance’, ‘Tailoring’, ‘Thiotemplated’, ‘Type II polyketide’, ‘ribosomally synthesized and post-translationally modified peptide product’ (RiPP), ‘phosphonate’, etc. Whether any of the genes unique to BCA-696 overlap with these BGCs, a comparison of their genomic coordinates did not show any overlap.
Physical maps of predicted biosynthetic gene clusters (BGCs) in the genome of Amycolatopsis sp. BCA-696. Arrows indicate the direction of gene transcription, while the chemical classes of products of the biosynthetic pathways encoded by the genes are indicated with different colors. The positions in the genome and the maps were generated from PRISM4 (https://prism.adapsyn.com/).
A detailed examination of core and additional biosynthetic genes showed two large clusters for the biosynthesis of two antibiotics- vancomycin and enediyne, spread within genomic coordinates 36,435—96,579 and 3,372,806—3,960,062, respectively (Supplementary Table S3). A full pathway for vancomycin biosynthesis was observed, however, for the Enediyne family of antibiotics, pathways were observed for the biosynthesis of its core molecule (neocarzinostatin) necessary for further reactions, and one of its derivatives, maduropeptin.
Genes underlying PGP and bio-control traits/pathways
The Amycolatopsis sp. BCA-696 genome assembly was analyzed to identify the genes involved in PGP and biocontrol activities. The strain under this study was experimentally validated to produce metabolites such as siderophores and hydrocyanic acid and shows enzymatic activity for cellulase, chitinase, lipase, indole acetic acid, and 1,3-beta-glucanase 16.
Among the siderophores, evidence for the presence of complete biosynthetic pathways for the production of catecholate or mixed types siderophores namely, bacillibactin, enterochelin, mycobactin, etc., were examined in the genome annotation of Amycolatopsis sp. BCA-696. In the RAST annotation, while the enzymes for biosynthesis of siderophore precursors and transporters involved in the export/import of siderophore or Fe-siderophore complex were found (Supplementary Table S4), the genes for biosynthesis of these siderophores from their precursors were largely missing in the (RAST) annotation. Orthology-based search only showed the presence of partial pathways (Supplementary Table S5, Supplementary Fig. S4).
Coming to the biocontrol traits, presence of three key cellulolytic enzymes was observed in Amycolatopsis sp. BCA-696 assembly: Endoglucanase (EC 3.2.1.4), Beta-glucosidase (EC 3.2.1.21), and Cellulose-1,4-beta-cellobiosidase (EC 3.2.1.91; Exoglucanase), wherein the former-two were present in many copies, ranging up to nine (Supplementary Fig. S5, Supplementary Table S6). Regarding the Chitinase activity, all enzymes related to the Chitin degradation pathway were also identified in Amycolatopsis sp. BCA-696 assembly which are: chitinase (EC 3.2.1.14), beta-N-acetylglucosaminidase, chitin deacetylase, chitosanase, exo-1,4-beta-d-glucosaminidase (Supplementary Fig. S6, Supplementary Table S7). Like other hydrolytic enzymes, bacterial lipases also play an important role in biocontrol against many phytopathogens, and also in inducing plant defence response 27. Two enzymes namely, lipase (EC 3.1.1.3) and Diacylglycerol O-acyltransferase (EC 2.3.1.20), were identified in the Amycolatopsis sp. BCA-696 assembly, indicating the presence/absence of underlying genes (Supplementary Fig. S7, Supplementary Table S8).
Examination of biosynthetic genes/enzymes for auxin (IAA) showed the presence of the pathway involving the Tryptamine intermediate. For alternate pathways (for auxin biosynthesis), although a few genes were also present, ultimately the pathways were incomplete (Supplementary Table S9, Supplementary Fig. S8).
Discussion
First genome assembly of an agriculturally important species from the Amycolatopsis genus:
Amycolatopsis genus is well documented for its antibiotic-producing traits28,29 and thus is an obvious focal point for drug discovery programs. To understand the genomic basis behind this(i.e., secondary metabolite production), 150 Amycolatopsis genomes have so far been sequenced (based on data accessed in mid-2023) and about one-fifth of them are either complete or at the chromosome level assembly. The importance of Amycolatopsis in the agricultural sector is, however, not well known. Perhaps, we were the first to report its usefulness as PGP in sorghum and chickpea15, and its antagonistic potential against M. phaseolina mediated charcoal rot disease in sorghum16. Along a similar line, to uncover the genomic components underlying the PGP/biocontrol traits, we reported chromosome-level genome assembly of BCA-696 strain: ~ 9.06 Mb in size with 8,716 protein-coding genes (Fig. 1, Table 1).
Phylogenomic analysis indicated that Amycolatopsis sp. BCA-696 is a species on its own
The taxonomic classification of the BCA-696 strain has been available only till the genus level. The analysis using the whole genome sequence showed that this strain is closer to A. lurida (Fig. 4). However, the poor bootstrap value for the branch of BCA-696 implied that the species-level classification of BCA-696 is still elusive with the existing genomic information.
Like several Amycolatopsis genomes, the Amycolatopsis sp. BCA-696 too had many unique genes
The Amycolatopsis genomes, in particular the BGCs, have been compared in multiple studies22,29,30. A comparison involving 41 genomes showed a core set of size 1212 genes29, which was almost one-third of the size observed in this study (n = 4733), and the difference can be attributed mainly to the different number of genomes involved (41 versus 15). Given the diversity in the biosynthetic potential of secondary metabolites among Amycolatopsis species, the accessory and unique (gene) sets attain a higher importance than the core set. While comparing the Gene Cluster Families (GCFs) in 41 genomes, the conserved features accounted for only a small proportion, and a vast number of GCFs (67%) were represented by a single genome29. In the BCA-696 strain, a prediction of unique features gave 466 genes (Fig. 3), which corroborates earlier reports of a significant fraction of unique genes present in Amycolatopsis genomes.
Unique genes of Amycolatopsis sp. BCA-696 were enriched with antibiotic biosynthesis and resistance functions
While the functional annotation of the majority of the unique genes was unavailable, among the annotated genes antibiotic-related functions were the most prominent. It contained the gene for the biosynthesis of Bialaphos antibiotic (Supplementary Table S2), which has not been reported yet among Amycolatopsis species, and just one Streptomyces species has been known to have capability31,32. This antibiotic has been reported to have fungicidal and herbicidal roles, thus a potential biocontrol agent. In addition, several antibiotic transporters were also found which may play a role in secretion or defense (Supplementary Table S2). Another peculiar feature was the presence of Chloramphenicol and Streptomycin phosphotransferase which are involved in providing resistance against those antibiotics (Supplementary Table S2).
The genes identified for PGP traits indicated inhibition of M. phaseolina potentially due to the hydrolytic enzymes and antibiotics
Plant-growth-promoting bacteria isolated from the rhizosphere are known to produce growth hormones such as auxins and siderophores and hydrolytic enzymes such as chitinase, cellulase, and β-1,3-glucanase and help plants to inhibit pathogens either directly or indirectly33,34,35. Amycolatopsis BCA-696 has been reported to produce biocontrol and PGP traits including siderophore, HCN, chitinase, protease, cellulase, β-1,3-glucanase, lipase, and IAA under in vitro conditions15. The genome sequence analysis indicated that this strain can potentially biosynthesize auxin only through the Tryptamine pathway (Supplementary Table S9) and may need to cooperate with other rhizosphere bacteria for alternate biosynthesis pathways. Moreover, for Cellulase, Lipase, and Chitinase, complete biosynthesis pathways were identified (Supplementary Table S6–S8). Besides, complete or partial biosynthesis pathways for the diverse siderophore molecules were identified along with a number of transporters (Supplementary Tables S3–S5). The presence of biosynthetic genes for the Enediyne family of antibiotics (Supplementary Table S3), which typically act by DNA cleavage36, has been an interesting finding in BCA-696, as some of these secondary metabolites have earlier been shown for antifungal activity among diverse plant and animal pathogens 37,38. Hence, it is concluded that Amycolatopsis BCA-696 potentially produces hydrolytic enzymes or antibiotics that have the potential to inhibit the pathogen M. phaseolina that causes charcoal rot in sorghum.
Future Directions
The usefulness of Amycolatopsis BCA-696, for biocontrol of charcoal rot disease in sorghum, has been demonstrated at both greenhouse and field conditions in our previous study16. We also reported the tolerance of BCA-696 on a wide range of pH (5–11), temperatures (20–40 °C), NaCl concentrations (0–6%), and fungicides (including Bavistin up to 2500 ppm, Thiram up to 3000 ppm, Benlate up to 4000 ppm, Captan up to 3000 ppm and Ridomil up to 3000 ppm15. These traits could help BCA-696 to survive in harsh environments under natural conditions and thus this bioagent can be used in integrated disease management programs. Further, Amycolatopsis BCA-696 needs to be formulated as a bio-inoculant and used for the biocontrol of charcoal rot in other crops. The secondary metabolite(s) responsible for the inhibition of M. phaseolina need to be experimentally characterized. In the absence of a high level of genetic resistance in high-yielding varieties, Amycolatopsis BCA-696 could be effective in controlling charcoal rot disease and related loss in grain and stover quality of sorghum.
Material and methods
Source of actinobacterial strain
A strain of Amycolatopsis sp. BCA-696 (GenBank accession number of 16S ribosomal RNA gene: KM191337), previously reported by us to have the capacity for PGP in sorghum and chickpea15 and for its antagonistic potential against Macrophomina phaseolina that causes charcoal rot disease in sorghum16 was selected for the present study.
Isolation of DNA
DNA of Amycolatopsis sp. BCA-696 was isolated as per the protocols mentioned in Gopalakrishnan et al., 202014. In brief, BCA-696 was inoculated in starch casein broth and incubated for 120 h at 28° C. At the end of incubation, the culture was centrifuged at 10,000g for 10 min at 4 °C and the cells were washed twice with STE buffer (sucrose 0.3 M, Tris/ HCl 25 mM and Na2EDTA 25 mM, pH 8.0). The supernatant was discarded but the pellet (1 g) was re-suspended in 8.55 ml STE buffer and 950 µl lysozyme (20 mg/ml STE buffer) and incubated for 30 min at 30 °C. This was followed by the addition of 500 µl of SDS (10%; w/v) and 50 µl of protease (20 mg/ml) and the mixture was held at 37 °C for one h. At the end of incubation, 1.8 ml of NaCl (5 M) was added with gentle mixing to avoid shearing the DNA, and 1.5 ml of CTAB (10%; w/v) in 0.7 M NaCl (CTAB/NaCl solution) and incubated for 20 min at 65 °C. Once CTAB was added, all the remaining steps were carried out at room temperature. The lysate was extracted twice with an equal volume of phenol/chloroform/isoamyl alcohol (25:24:1; by vol) and centrifuged at 13,000g for 10 min. Finally, the aqueous phase was extracted with chloroform/isoamyl alcohol (24:1, by vol) and transferred to a tube followed by the addition of 600 µl of propan-2-ol and DNA spooled out after 10 min. Alternatively, it was recovered by centrifugation at 12,000g for 10 min. The pellet was washed twice with ethanol (70%; v/v), vacuum dried, and dissolved in 2 ml of TE buffer (10 mM Tris/HCl and 1 mM EDTA, pH 8.0). RNase A (50 mg/ml) was added with incubation at 37 °C for 2 h. The sample was again extracted with phenol as described above. DNA was re-precipitated from the aqueous phase with the addition of 100 µl of 3 M sodium acetate (pH 5.3) and 600 µl of propan-2-ol. The DNA pellet was washed with ethanol (70%; v/v), dried, and dissolved in TE buffer. The purity of the BCA-696 DNA was checked in the agarose gel electrophoresis and quantified using NanoDrop.
Whole genome sequencing
About ~ 5 μg high-quality genomic DNA (free from any contaminant and having A260/280 ratio in the range of ~ 1.8 to 2.0 with DNA concentration ≥ 100 ng/μl) was sent to AgriGenome Labs (Kochi, India) for library preparation and next-generation sequencing using the Illumina platform. The Genomic DNA was fragmented, and a paired-end library with insert size 300 bp, and a mate-pair library with insert size 5 Kbp, were prepared. The DNA fragment libraries were validated by tapestation, and sequenced ~ 9.905 million paired-end (100 bp × 2) and ~ 6.242 million mate pair (250 bp × 2) reads on the Illumina HiSeq 2500.
Genome assembly and its assessment
The Whole-genome sequenced paired and mate-pair reads of the bacterial genome were cleaned (by removing adapters, primer sequences, etc.) using Trimmomatic V.0.3939. The cleaned reads were de novo assembled using SOAPdenovo V2.04 and SPADES V3.10.1 assemblers40,41. Two assemblies were assessed and compared by using QUAST V5.0.2 42 followed by discarding the contigs having < 500 bp length and coverage < 5. The GapCloser V1.0.1 43 was used to close the gaps that emerged during the scaffolding process by the de novo assembler, using the abundant pair relationships of short reads. Further contigs were subjected to Pathosystems Resource Integration Center (PATRIC V3.6.9)44 and KmerFinder V3.2 45 search to find the closely related genomes. In order to do reference genome-based reordering of contigs Amycolatopsis albispora, Amycolatopsis keratiniphila, Amycolatopsis japonica, and Amycolatopsis orientalis genome sequences were obtained from NCBI microbial genomes database followed by scaffolding using Multidraft-Based Scaffolder (MEDUSA V1.6)46. The resulting scaffolds were subjected to an NCBI BLAST database search to check the contamination and scaffold hits other than the Amycolatopsis genus were discarded. The completeness of the genome assembly was examined using the tool ‘Benchmarking Universal Single-Copy Orthologs (BUSCOs) V5.1.3 47. The Circos plot showing the features on the genome (Fig. 1) was generated using Circos V.0.69.8 48.
Genome annotation
The annotation of the assembled genome was carried out by Rapid Annotation using the Subsystem Technology (RAST) server (http://rast.nmpdr.org/rast.cgi), through the RASTtk pipeline25 (Accessed on: April 2020). The genome-based phylogenetic relationships between the BCA-696 strain and its closely related species used to reorganize scaffolds were determined using the reference sequence ALignment-based PHYlogeny (REALPHY) builder method49 (accessed on: April 2020). Figure 2 showing the subsystem classes in BCA-696 was plotted using ggplot2 package50 from Rstudio.
Comparative analysis of Amycolatopsis sp. BCA-696 genome
The pangenome analysis was performed using all Amycolatopsis genomes at the NCBI database with at least scaffold level completion (n = 76, Supplementary Table S10) using Roary pipeline v3.13.051. Having observed an unusually small core set size (< 1%) and a whoppingly large cloud gene set size (> 95%), the pan-genome analysis was repeated on a subset of genomes that were closely related to BCA-696. Species/strains belonging to the clade, identified from the pan-genome-based tree, in which BCA-696 was also present were selected as close relatives. This set (of closely related species/strains) was augmented by including a few more species/strains having contig-level completion. The fourteen close relatives of BCA-696 were: A. orientalis strain B-37 (GCF_000943515.2), A. keratiniphila strain HCCB10007 (GCF_000400635.2), A. regifaucium strain GY080T (GCF_001558125.2), A. coloradensis strain DSM 44225T (GCF_001953865.1), A. roodepoortensis strain ZEL-1 (GCF_024628825.1), A. alba strain DSM 44262T (GCF_000384215.1), A. azurea strain DSM 43854T (GCF_001995215.1), A. decaplanina strain DSM 44594T (GCF_000342005.1), A. lurida strain DSM 43134T (GCF_000749465.2), A. japonica strain DSM 44213T (GCF_000732925.1), A. thailandensis strain JCM 16380T (GCF_002234405.1), A. umgeniensis strain DSM 45272T (GCF_014205155.1), A. oliviviridis strain CGMCC 4.7683 (GCF_014654365.1), and A. pittospori strain PIP199T (GCF_013870525.1) (The type strains are indicated with a “T” in superscript). The genome sequences were annotated with the rapid prokaryotic genome annotation tool Prokka V1.14.652 and implemented Roary pipeline on input annotation files (in GFF format) with a minimum 95% percentage identity for BLASTp. Core gene sequences were aligned with Multiple Alignments using Fast Fourier Transform (MAFFT V7.520)53, and the CD-HIT V4.754 algorithm used to perform clustering was embedded in the Roary pipeline. Further analysis was performed on the gene presence/absence file generated by the Roary analysis.
Orthology-based re-analysis of BCA-696 specific genes
Orthofinder (version 2.5.4)24 was used with default parameters to find common ortholog groups among BCA-696 and 14 other genomes of Amycolatopsis species taken from the NCBI database (Acessed on: October 2022). Genes unique to BCA-696 were selected and mined for functional details using the KEGG PATHWAY Database (Accessed on: October 2022)55 (https://www.genome.jp/kegg/pathway.html).
Phylogenetic relationships of BCA-696
To obtain taxonomic positioning of Amycolatopsis BCA-696 strain in the Amycolatopsis genus based on the genome sequences, the BCA-696 genome was submitted to the Type (Strain) Genome Server (TYGS) (accessed on: Jan 2024)31, and the ten closest genomes were selected by the TYGS to construct the phylogenetic tree with default setting. The TYGS uses overall genome relatedness index (OGRI) methods (GBDP & dDDH).
Prediction of biosynthetic pathways (BGCs) for secondary metabolites and PGP traits
PRISM4 (accessed on: July 2020)56 was used to mine BGCs in the BCA-696 genome. It allows the prediction of a BGC's location and provides insights into the putative structure of the encoded secondary metabolite. BGCs were additionally predicted using another tool namely antiSMASH v7.0.0 57 using 'relaxed' strictness level. Since none of the two tools gave precise details of the secondary metabolites actually being synthesized by the core and/or additional biosynthetic genes, protein sequences of these genes were searched in the KEGG ortholog/pathway database using ‘Automatic KO assignment and KEGG mapping service’ (BlastKOALA). Representative pathways for biosynthesis of Vancomycin and Enediyne from Amycolatopsis keratiniphila, for instance, were accessed at URLs https://www.genome.jp/pathway/aoi01055 and https://www.kegg.jp/pathway/aoi01059, respectively.
Further, pathway information of the PGP traits was obtained from the KEGG PATHWAY Database (Accessed on: July 2020)54, which provided details of the enzymes involved. In order to find orthologs of these KEGG enzymes in the genome of Amycolatopsis sp. BCA-696, the Reciprocal Best BLAST (RBB) search was performed between the proteomes of Amycolatopsis sp. B-696 and a few reference bacterial species (such as E. coli), already have orthologs (KO) of KEGG enzymes.
Data availability
The sequencing data generated in this study has been submitted to the National Centre for Biotechnology Information (NCBI) under the Bioproject ID: PRJNA765508. The genome assembly and annotation can be accessed from NCBI (Genbank accession number CP150484). The genome annotation by the RAST server can be accessed from the author's website (http://sls.uohyd.ac.in/new/fac_details.php?fac_id=34#DataandSoftwares).
References
Leontidou, K. et al. Plant growth promoting rhizobacteria isolated from halophytes and drought-tolerant plants: Genomic characterisation and exploration of phyto-beneficial traits. Sci. Rep. 10, 14857 (2020).
Gu, Y., Ma, Y., Wang, J., Xia, Z. & Wei, H. Genomic insights into a plant growth-promoting Pseudomonas koreensis strain with cyclic lipopeptide-mediated antifungal activity. Microbiologyopen 9, 133 (2020).
Chen, X. H. et al. Comparative analysis of the complete genome sequence of the plant growth-promoting bacterium Bacillus amyloliquefaciens FZB42. Nat. Biotechnol. 25, 1007–1014 (2007).
Olanrewaju, O. S., Ayilara, M. S., Ayangbenro, A. S. & Babalola, O. O. Genome mining of three plant growth-promoting bacillus species from maize rhizosphere. Appl. Biochem. Biotechnol. 193, 3949–3969 (2021).
Chandra, A., Chandra, P. & Tripathi, P. Whole genome sequence insight of two plant growth-promoting bacteria (B. subtilis BS87 and B. megaterium BM89) isolated and characterized from sugarcane rhizosphere depicting better crop yield potentiality. Microbiol. Res. 247, 126733 (2021).
Babalola, O. O., Adeleke, B. S. & Ayangbenro, A. S. Whole genome sequencing of sunflower root-associated Bacillus cereus. Evol. Bioinform. 17, 117693432110389 (2021).
Kwak, Y., Park, G.-S. & Shin, J.-H. High quality draft genome sequence of the type strain of Pseudomonas lutea OK2T, a phosphate-solubilizing rhizospheric bacterium. Stand. Genomic Sci. 11, 51 (2016).
Devi, U. et al. Genomic and functional characterization of a novel Burkholderia sp. strainAU4i from pea rhizosphere conferring plant growth promoting activities. Adv. Genet. Eng. 2015, 1–8 (2015).
Matteoli, F. P. et al. Genome sequencing and assessment of plant growth-promoting properties of a Serratia marcescens strain isolated from vermicompost. BMC Genomics 19, 750 (2018).
Suarez, C. et al. Complete genome sequence of the plant growth-promoting bacterium Hartmannibacter diazotrophicus strain E19T. Int. J. Genomics 2019, 1–12 (2019).
Gupta, A. et al. Whole genome sequencing and analysis of plant growth promoting bacteria isolated from the rhizosphere of plantation crops coconut, cocoa and arecanut. PLoS One 9, e104259 (2014).
Singh, R. P., Nalwaya, S. & Jha, P. N. The draft genome sequence of the plant growth promoting rhizospheric bacterium Enterobacter cloacae SBP-8. Genomics Data 12, 81–83 (2017).
Kang, S.-M. et al. Complete genome sequence of Pseudomonas psychrotolerans CS51, a plant growth-promoting bacterium, under heavy metal stress conditions. Microorganisms 8, 382 (2020).
Subramaniam, G. et al. Complete genome sequence of sixteen plant growth promoting Streptomyces strains. Sci. Rep. 10, 10294 (2020).
Alekhya, G. & Gopalakrishnan, S. Exploiting plant growth-promoting Amycolatopsis sp. in chickpea and sorghum for improving growth and yield. J. Food Legum. 29, 225–231 (2016).
Gopalakrishnan, S., Srinivas, V., Naresh, N., Alekhya, G. & Sharma, R. Exploiting plant growth-promoting Amycolatopsis sp. for bio-control of charcoal rot of sorghum (Sorghum bicolor L.) caused by Macrophomina phaseolina (Tassi) Goid. Arch. Phytopathol. Plant Prot. 52, 543–559 (2019).
Hazen, W., de Bruyn, J. C. & van Dijken, J. P. Nocardia sp. 239, a facultative methanol utilizer with the ribulose monophosphate pathway of formaldehyde fixation. Arch. Microbiol. 135, 205–210 (1983).
Lechevalier, M. P., Prauser, H., Labeda, D. P. & Ruan, J.-S. Two new genera of nocardioform actinomycetes: Amycolata gen. nov. and Amycolatopsis gen. nov. Int. J. Syst. Bacteriol. 36, 29–37 (1986).
Frasch, H.-J. et al. Alternative pathway to a glycopeptide-resistant cell wall in the Balhimycin producer Amycolatopsis balhimycina. ACS Infect. Dis. 1, 243–252 (2015).
Nigam, A. et al. Modification of rifamycin polyketide backbone leads to improved drug activity against rifampicin-resistant Mycobacterium tuberculosis. J. Biol. Chem. 289, 21142–21152 (2014).
Xu, L. et al. Complete genome sequence and comparative genomic analyses of the vancomycin-producing Amycolatopsis orientalis. BMC Genomics 15, 363 (2014).
Kumari, R., Singh, P. & Lal, R. Genetics and genomics of the genus Amycolatopsis. Indian J. Microbiol. 56, 233–246 (2016).
Sánchez-Hidalgo, M., González, I., Díaz-Muñoz, C., Martínez, G. & Genilloud, O. Comparative genomics and biosynthetic potential analysis of two lichen-isolated Amycolatopsis strains. Front. Microbiol. 9, 11 (2018).
Emms, D. M. & Kelly, S. OrthoFinder: Phylogenetic orthology inference for comparative genomics. Genome Biol. 20, 238 (2019).
Aziz, R. K. et al. The RAST server: Rapid annotations using subsystems technology. BMC Genomics 9, 75 (2008).
Schwartz, D. et al. Biosynthetic gene cluster of the herbicide phosphinothricin tripeptide from Streptomyces viridochromogenes Tü494. Appl. Environ. Microbiol. 70, 7093–7102 (2004).
Riseh, R. S., Vatankhah, M., Hassanisaadi, M. & Barka, E. A. Unveiling the role of hydrolytic enzymes from soil biocontrol bacteria in sustainable phytopathogen management. Front. Biosci. 29, 13 (2024).
Bala, S. et al. Reclassification of Amycolatopsis mediterranei DSM 46095 as Amycolatopsis rifamycinica sp. nov.. Int. J. Syst. Evol. Microbiol. 54, 1145–1149 (2004).
Adamek, M. et al. Comparative genomics reveals phylogenetic distribution patterns of secondary metabolites in Amycolatopsis species. BMC Genomics 19, 426 (2018).
Tang, B. et al. A systematic study of the whole genome sequence of Amycolatopsis methanolica strain 239 T provides an insight into its physiological and taxonomic properties which correlate with its position in the genus. Synth. Syst. Biotechnol. 1, 169–186 (2016).
Meier-Kolthoff, J. P. & Göker, M. TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat. Commun. 10, 2182 (2019).
Murakami, T. et al. The bialaphos biosynthetic genes of Streptomyces hygroscopicus: Molecular cloning and characterization of the gene cluster. Mol. Gen. Genet. MGG 205, 42–53 (1986).
Pal, K. K., Tilak, K. V. B. R., Saxcna, A. K., Dey, R. & Singh, C. S. Suppression of maize root diseases caused by Macrophomina phaseolina, Fusarium moniliforme and Fusarium graminearum by plant growth promoting rhizobacteria. Microbiol. Res. 156, 209–223 (2001).
Tokala, R. K. et al. Novel plant-microbe rhizosphere interaction involving Streptomyces lydicus WYEC108 and the pea plant (Pisum sativum ). Appl. Environ. Microbiol. 68, 2161–2171 (2002).
Donate-Correa, J., León-Barrios, M. & Pérez-Galdona, R. Screening for plant growth-promoting rhizobacteria in Chamaecytisus proliferus (tagasaste), a forage tree-shrub legume endemic to the Canary Islands. Plant Soil 266, 261–272 (2005).
Ando, T. et al. A new non-protein enediyne antibiotic N11999A2: Unique enediyne chromophore similar to neocarzinostatin and DNA cleavage feature. Tetrahedron Lett. 39, 6495–6498 (1998).
Cui, B. et al. Antifungal Enediynes, United States Patent US09/679672.2000-10-05.42.
Su, Z.-Z. et al. Evidence for biotrophic lifestyle and biocontrol potential of dark septate endophyte Harpophora oryzae to rice blast disease. PLoS One 8, e61332 (2013).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Luo, R. et al. SOAPdenovo2: An empirically improved memory-efficient short-read de novo assembler. Gigascience 1, 18 (2012).
Bankevich, A. et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477 (2012).
Gurevich, A., Saveliev, V., Vyahhi, N. & Tesler, G. QUAST: quality assessment tool for genome assemblies. Bioinformatics 29, 1072–1075 (2013).
Xu, M. et al. TGS-GapCloser: A fast and accurate gap closer for large genomes with low coverage of error-prone long reads. Gigascience 9, giaa094 (2020).
Snyder, E. E. et al. PATRIC: The VBI PathoSystems resource integration center. Nucleic Acids Res. 35, D401–D406 (2007).
Larsen, M. V. et al. Benchmarking of methods for genomic taxonomy. J. Clin. Microbiol. 52, 1529–1539 (2014).
Bosi, E. et al. MeDuSa: A multi-draft based scaffolder. Bioinformatics 31, 2443–2451 (2015).
Simão, F. A., Waterhouse, R. M., Ioannidis, P., Kriventseva, E. V. & Zdobnov, E. M. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31, 3210–3212 (2015).
Krzywinski, M. I. et al. Circos: An information aesthetic for comparative genomics. Genome Res. https://doi.org/10.1101/gr.092759.109 (2009).
Bertels, F., Silander, O. K., Pachkov, M., Rainey, P. B. & van Nimwegen, E. Automated reconstruction of whole-genome phylogenies from short-sequence reads. Mol. Biol. Evol. 31, 1077–1088 (2014).
Wickham, H. ggplot2: Elegant Graphics for Data Analysis (Springer, 2016).
Page, A. J. et al. Roary: Rapid large-scale prokaryote pan genome analysis. Bioinformatics 31, 3691–3693 (2015).
Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069 (2014).
Katoh, K. & Standley, D. M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 (2013).
Li, W. & Godzik, A. Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22, 1658–1659 (2006).
Kanehisa, M. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30 (2000).
Skinnider, M. A. et al. Comprehensive prediction of secondary metabolite structure and biological activity from microbial genome sequences. Nat. Commun. 11, 6058 (2020).
Blin, K. et al. antiSMASH 70: New and improved predictions for detection, regulation, chemical structures and visualisation. Nucleic Acids Res. 51(W1), W46–W50 (2023).
Acknowledgements
This work was undertaken as part of the CGIAR Research Program on Grain Legumes and Dry Land Cereals (GLDC). ICRISAT is a member of the CGIAR Consortium. This study was also supported by DBT’s Ramalingaswami grant, and author Vivek Thakur is grateful for this. Author Angeo Saji is financially supported by Junior Research Fellowship from the Council of Scientific and Industrial Research (CSIR), Govt. of India. We also thank Mr. PVS Prasad for his technical assistance. We would also like to thank the anonymous reviewers whose critical comments helped in making it comprehensive and scientifically insightful.
Author information
Authors and Affiliations
Contributions
S.G., V.T., A.R., and R.K.S. conceived, designed, supervised the study. V.S. and S.G. maintained the culture of the isolate, and extracted genomic DNA for sequencing for genome sequencing. V.T. generated the sequencing data. P.G., N., P.R., S.S., and A.S. generated the results. P.G., V.T., R.K.S., A.R. along with S.G. analyzed the results. P.G., V.T. and S.G. wrote the first draft; V.T., A.S., R.K.S, A.R., and G.S. reviewed and finalized it. All authors read and approved the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Gandham, P., Vadla, N., Saji, A. et al. Genome assembly, comparative genomics, and identification of genes/pathways underlying plant growth-promoting traits of an actinobacterial strain, Amycolatopsis sp. (BCA-696). Sci Rep 14, 15934 (2024). https://doi.org/10.1038/s41598-024-66835-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-66835-y
- Springer Nature Limited