Abstract
With their long lives and extreme reproductive output, social insect queens have escaped the classic trade-off between fecundity and lifespan, but evidence for a trade-off between fecundity and immunity has been inconclusive. This is in part because pathogenic effects are seldom decoupled from effects of immune induction. We conducted parallel, blind virus infection experiments in the laboratory and in the field to interrogate the idea of a reproductive immunity trade-off in honey bee (Apis mellifera) queens and to better understand how these ubiquitous stressors affect honey bee queen health. We found that queens injected with infectious virus had smaller ovaries and were less likely to recommence egg-laying than controls, while queens injected with UV-inactivated virus displayed an intermediate phenotype. In the field, heavily infected queens had smaller ovaries and infection was a meaningful predictor of whether supersedure cells were observed in the colony. Immune responses in queens receiving live virus were similar to queens receiving inactivated virus, and several of the same immune proteins were negatively associated with ovary mass in the field. This work supports the hypothesized relationship between virus infection and symptoms associated with queen failure and suggests that a reproductive-immunity trade-off is partially, but not wholly responsible for these effects.
Similar content being viewed by others
Introduction
As the sole reproductive female in a colony of thousands of individuals, honey bee (Apis mellifera) queens are the cornerstones of the colonies that they head. Understanding what factors impact their health and reproductive potential is therefore of great interest to the beekeeping industry, which provides pollination services valued annually in the billions of dollars globally1,2. While honey bee queens can live for 5 years or more3, beekeepers regularly replace queens at much younger ages, often after less than 1 year4. It is still unclear, though, exactly what factors and in what combinations contribute to the symptoms of “queen problems” or poor reproductive output, which are regularly reported as one of the main perceived causes of colony loss in North America5,6,7.
Honey bee queens mate over a short period of time with multiple drones at a few weeks of age and store the sperm they acquire for the rest of their lives3. The remaining quality and quantity of this stored sperm is one determining factor of queen quality—if a queen lacks viable sperm with which to fertilize eggs, either because she did not obtain enough through mating, or because the remaining sperm is no longer viable, her productivity will decline8,9. Sperm quality can be affected by abiotic factors, such as temperature stress8,10 and pesticide exposure11. Additionally, the age of larvae that beekeepers graft when rearing new queens can impact the physiology and size of queens12, with older larvae developing into worker-like queens, which impacts their future reproductive potential13,14,15,16. Queens perceived by beekeepers as failing (exhibiting drone laying in worker cells, or producing poor brood patterns) have been shown to have reduced sperm viability8 and we previously reported that they also have smaller ovaries compared to their healthy counterparts of similar age and in similar environments at the same time of year17. In the same study, we also linked natural viral infection to queen problems in field operations, finding that beekeeper-identified failing queens had higher viral RNA copy numbers and that queens with higher viral RNA copy numbers, regardless of their performance status (healthy or failing), also had smaller ovaries17. We later established the causal rather than correlative nature of this relationship wherein injection with Israeli acute paralysis virus (IAPV) caused a reduction in ovary mass—the same phenotype observed in queens identified as “failing” from the field18. However, experiments investigating changes in actual reproductive output in response to viral infection, particularly with the largely asymptomatic infections commonly encountered in the field, have not been conducted.
Many of the honey bee viruses commonly found in colonies can infect queens—including deformed wing virus (DWV), sacbrood virus (SBV), black queen cell virus (BQCV), acute bee paralysis virus (ABPV), and others in addition to IAPV—but their pathological effects are not yet fully defined. Deformed wing virus (DWV) has two common genotypes, A and B, the latter of which was detected more recently, and is potentially replacing genotype A19. Some data derived from worker infection experiments suggest that DWV-B is also more virulent than DWV-A20,21, but Tehel et al.22 found no difference, and Norton et al.23 found that DWV-A was more virulent than DWV-B. DWV is commonly vectored by the parasitic mite Varroa destructor, but is also transmitted horizontally, vertically, and sexually24,25,26. Queens rarely show any overt symptoms of infection, and the deformed wing phenotype seen in workers has only been reported a single time in a queen27. However, the virus has been detected in several queen tissues, including the head, thorax, gut, fat body, ovaries, and spermatheca28,29. High levels of DWV infection have been associated with ovarian degeneration and egg-laying deficiencies in queens30, but few other studies have examined the pathological effects of DWV infection on queens, with most focusing solely on detection of the virus in various organs. Other viruses, including sacbrood virus (SBV), black queen cell virus (BQCV), acute bee paralysis virus (ABPV), and IAPV have also been detected in various queen tissues including the ovaries, but generally only in low titers, and the effects on queen health have been even less well-studied4,29,31,32. A recent study on the prevalence of a newly identified honey bee virus, Solinvivirus-1, found that apiaries testing positive for the virus were nearly twice as likely to contain queenless colonies33, and our previous findings linking viral infection to queen failure suggests that, although infections are often hidden in queens, they may still be compromising queen health and reproductive output.
Determining the mechanism of this effect is also biologically intriguing because while the pathogenic effects of a viral infection could be directly compromising a queen’s health and reproductive output, a reproductive-immunity trade-off may also be at play. A compromise between reproduction and immunity has been well studied in many insects34, but not previously in honey bees. This compromise, which is generally considered to be driven by allocation of limited resources (since both reproduction and immune activation are energetically costly) could be particularly relevant for honey bee queens, who can sustain an enormous reproductive output and can lay up to their own bodyweight in eggs every day35. It is reasonable to assume that sustaining such an energetically intensive process would come with costs, not only to immune function, but to longevity and somatic maintenance in general, as is the case in many insects, many of which are not as specialized for reproduction as the honey bee queen.
However, honey bees and other highly social insects (ants and termites) have escaped the trade-off with lifespan, exhibiting both enormous reproductive output and increased longevity compared to the non-reproductive castes35,36,37,38. This uncoupling of fecundity and longevity appears to be due in part to a rewiring of the endocrine network that signals resources to be directed towards one process or the other in solitary insects37,39,40 and involves insulin/insulin-like growth factor (IIS), juvenile hormone (JH), and vitellogenin (Vg). Given that social insects have escaped at least one compromise, it therefore calls into question whether they may have also evaded the trade-off of reproduction with immunocompetence. However, there is conflicting evidence for other social insects. Black garden ant (Lasius niger) queens appear to have removed the main mediator of a trade-off by uncoupling JH from its role as stimulator of reproduction (as is the case in solitary insects) while maintaining its role as a suppressor of immune activation41. Conversely, a different study in ant queens found evidence for a trade-off after wounding, in which the queens exhibited reduced egg laying with an increase in immune expression and a decrease in germ cell development genes42. In social bee species, the limited evidence so far is similarly conflicting. We have recently suggested that in bumble bees, what appears as evidence for a trade-off between immunity and reproduction in the protein expression patterns of queens may actually be due instead to differing selection pressures at different stages of a queen’s life cycle, which cannot be de-coupled from her reproductive activation43. In honey bees, it has been shown that reproductive potential does not reduce the heat-shock response (a response which includes antiviral immune effectors44), which is unique among the invertebrates that have been studied to date45. In contrast, we have previously found evidence for a trade-off in honey bee queens: sperm viability, another important reproductive metric, is negatively correlated with lysozyme, a conserved immune effector17, and injection with IAPV caused a decrease in the transcriptional expression of vitellogenin and an increase of heat shock protein expression18. We are interested in determining if indeed a trade-off is driving the effects of viral infection we observe because, if so, this means that even the ubiquitous levels of virus infection present in colonies—which may not be “making queens sick” or causing overt disease—could still be reducing the reproductive success of a queen.
Our previous experimental infections with IAPV were conducted over a short time frame (3 days) and measured only ovary mass to quantify the impact to reproduction. However, poor queen performance is a chronic symptom displayed over time, not an acute effect. In this study, we therefore aimed to determine if the effects of experimental virus infection extend to an observable effect on actual reproductive output, beyond just smaller ovaries, by employing more realistic experimental settings and a longer time frame. We conducted two separate experiments in parallel in which we observed queens experimentally infected with a combination of BQCV and DWV-B either in laboratory cages (queen monitoring cages, or QMCs)46, which allowed queens to continue to lay eggs for 1 week following infection, or in field colonies for 6 weeks following infection. To address the potential presence of a reproductive-immunity trade-off, we also injected a group of queens in the cage experiment with a UV-inactivated version of the same viral inoculum to decouple the role of immune activation caused by wounding from viral pathogenesis.
Results
Cage experiment: virus RNA copy numbers
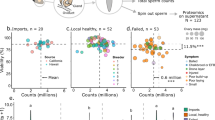
Young, mated honey bee queens were injected with (a) a combination of infectious BQCV and DWV-B, (b) the same mixture of viruses that had been inactivated with ultraviolet light, or (c) saline (injection control) and subsequently kept in incubated cages which enabled them to lay eggs for 7 days following injection. Because queens are likely to have background infections, we assessed the RNA copy numbers of common viruses (BQCV, DWV-A, DWV-B, and SBV) in all groups using RT-qPCR. As expected, there were no differences in viral copy numbers in the inactivated group compared to the saline-injected control, and in the infected group DWV-B was increased and BQCV was marginally increased (Fig. 1a). Correspondingly, the infected group had a significantly higher level of “total virus”, a metric representing the sum of natural logarithm-transformed copy numbers of the individual viruses (Fig. 1b), which we have previously determined to be a more important predictor of ovary mass in queens collected from the field than the levels of any singular virus18. Levels of DWV-A and SBV (which were not present in the inoculum) were similar between groups, as expected, but were difficult to robustly evaluate as few samples fell above the limit of detection. See Table 1 for complete statistical details.
Virus RNA copy numbers of cage experiment queens. Bars show mean values and error bars represent standard deviation; p values are shown relative to the control group. Control queens were injected with saline, infected queens were injected with a combination of BQCV and DWV-B, and queens in the inactivated group were injected with the same combination of viruses that were UV-inactivated (n = 9 for all groups). Queens were kept in queen monitoring cages in an incubator with attendants and sacrificed 7 days after injection. (a) RNA copy numbers of BQCV and DWV-B were higher in the infected group (though for BQCV, the difference was marginally non-significant), while there were no differences in the copy numbers of DWV-A and SBV between groups. The dashed horizontal lines represent the RT-qPCR quantification threshold. (b) The summed total virus copy number was higher in the infected group relative to the control group.
Cage experiment: fertility analysis
The ovary masses of queens injected with live virus were significantly smaller 7 days post infection relative to the saline-injected queens (linear mixed model with batch as a random effect, t = − 3.31, p = 0.0032) (Fig. 2a), which is consistent with what we previously observed after injection with IAPV18. The ovary masses of queens injected with the inactivated virus also tended to be non-significantly reduced relative to the controls, though the effect size was about half that of the live virus group (linear mixed model with batch as random effect, t = − 1.77, p = 0.090). Regardless of the group, ovary mass was negatively associated with levels of BQCV, DWV-B, and total virus load (Fig. 2b, see Table 1 for statistical reporting).
Viral infections reduced ovary mass and egg laying in cage experiment queens. (a) The ovary masses of queens injected with live virus were significantly decreased relative to the control group, with a non-significant effect observed in queens injected with inactivated virus. (b) Ovary mass was negatively associated with levels of BQCV, DWV-B, and total virus load. The dashed vertical lines represent the RT-qPCR quantification threshold. (c) Queens injected with live virus were significantly less likely to recommence egg laying 7 days after injection compared to control queens. (d) The number of eggs laid per day, averaged over days 5–7 after injection, was negatively associated with the total virus RNA copies across all queens. See Table 1 for detailed statistical reporting.
Queens were kept in specialized queen monitoring cages (QMCs) which enabled us to record their egg laying. All queens were confirmed to be laying consistently before being injected, but queens injected with live virus were 75% less likely to recommence laying in the 7 days following injection relative to the control queens (binomial generalized linear model, estimate = − 3.33, z = − 2.51, p = 0.012), with only 2/9 (22.2%) queens in the infected group recommencing laying compared to 8/9 (88.9%) control queens (Fig. 2c). Before injections, queens laid an average of 165 eggs per day (± 46), whereas after injection, queens that did recommence laying produced an average of 65 eggs per day (± 51) (both within the range of what has been observed previously in QMCs)46. Mirroring the results of ovary mass, regardless of group, for every additional unit of total virus infection queens laid approximately 2.5 fewer eggs per day over the final 3 days before being sacrificed (Fig. 2d, linear model, t = − 2.64, p = 0.014).
Cage experiment: hemolymph proteome analysis
To investigate how the immune response of queens injected with UV-inactivated virus differed from those infected with live virus, we performed quantitative proteomics on the hemolymph of queens. Of the 1071 identified protein groups (1% FDR) and 933 protein groups quantified in at least 75% of samples, 23 proteins were differentially expressed in the live infected group, and 15 were differentially expressed in the UV-inactivated group relative to the control (Fig. 3, 5% FDR, Benjamini–Hochberg correction). Between these two groups, 12 proteins are shared, including many which are known to be associated with immunity. IRP30, for example, is a secreted immune protein found in a wide variety of hymenopteran species and is co-expressed with carboxylesterase47,48, the expression of which was also significantly elevated in both groups compared to the controls. Transferrin mediates iron-sequestration as an innate immune strategy to limit the availability of this element for acquisition by pathogens49. Beta-1,3-glucan binding protein is a pattern-recognition receptor in invertebrates for the glucans found on fungal cell walls, which triggers the phenoloxidase cascade, possibly by interacting with a serine protease50,51. Additionally, phenoloxidase activating factor, a modular serine protease, and several of the antimicrobial peptides including defensin, hymenoptaecin, and apidaecin, were all expressed more strongly in queens receiving UV-inactivated virus and live virus groups compared to controls. The only immune effector not shared between groups, lysozyme, was significantly upregulated in only the group receiving the inactivated inoculum. A channel protein from Vairimorpha apis (formerly Nosema apis52 was also differentially expressed, with higher levels in both the inactivated and infected groups.
Hemolymph immune proteins were differentially expressed between groups in cage experiment queens. Each row represents a protein, and each column represents a sample, showing the Uniprot accession number. Proteins that were not quantified in a sample (“NA”) are shown as grey tiles. Proteins in bold were significant in both control-inactivated and control-infected contrasts (5% FDR, Benjamini–Hochberg correction).
Cage experiment: perturbation caused by the cage environment
For experiments in which bees are kept in the laboratory, it is often unknown exactly how disruptive the laboratory system is to their natural functioning. To address this, we sacrificed three “baseline” queens from each of the three batches we received for the cage experiment immediately upon arrival for comparison to the control queens that were subsequently kept in the QMCs. The ovaries of the baseline queens were significantly larger than the ovaries of control queens that were sacrificed 2 weeks later (Supplementary Fig. 1a). This is unsurprising, given that the baseline queens had been actively laying eggs in colonies only ~ 24 h before being sacrificed, while the control queens had been kept in cages which, while allowing them to lay, did restrict their laying compared to a colony environment. To assess the impact of the QMCs more thoroughly, we also performed proteomics on the hemolymph of the baseline queens and found that even though 39 proteins in the hemolymph were differentially expressed in the baseline queens compared to the controls (5% FDR, Benjamini–Hochberg correction), only 3 proteins were overlapping with the proteins differentially expressed in either of the experimental inactivated or infected groups (Supplementary Fig. 1b). This indicates that, even though the cage environment clearly had an impact on the physiology of queens over this 2-week period, those effects did not appear to overlap substantially with the effects we observed as a result of our experimental injections.
Field experiment: virus RNA copies and mite levels
For the field experiment, queens were injected with either saline or a combination of BQCV and DWV-B in the same manner as the cage experiment, placed in colonies, and assessed weekly for 7 weeks following injection. We again assessed the virus loads of the queens at the end of the experiment using RT-qPCR. As shown in Fig. 4a, BQCV was higher in the infected group while DWV-B was actually lower in the infected group (see Table 2 for statistical details). DWV-A was marginally higher in the infected group and SBV was only detected in two queens. Correspondingly, there was no significant difference in the total virus level between the two groups. We also randomly sampled workers from brood frames at both the beginning and the end of the experiment to assess the levels of viruses present in the colony. The viral load in workers at the beginning of the experiment was a significant predictor of the total weight gained by the colony over the course of the experiment (Fig. 4b) while the queen viral load was not. Interestingly, we found that the level of DWV-B in queens was significantly linked to the RNA copies of DWV-B present in the workers at the end of the experiment, though there was no correlation between queen virus RNA copies and worker virus RNA copies for the other viruses after controlling for the effect of site (Fig. 4c). This trend with DWV-B appears to be mainly driven by colonies in the control group. We also examined the correlations of worker viral RNA copies, queen viral RNA copies, and the levels of mites in the colonies at the end of the experiment to gain a better understanding of how these metrics covaried. While mite load is positively correlated with levels of both variants of DWV in both queens and workers, the correlation is not significant after adjusting for multiple hypothesis testing (Bonferroni) (Fig. 4d). There were no differences in mite loads between the experimental groups and colonies generally had low mite loads (see Supplementary Data 1 for mite levels).
Virus RNA copies in field experiment queens and workers. Control queens (n = 15) were injected with saline and infected queens (n = 14) were injected with a combination of BQCV and DWV-B. Queens were kept in nucleus colonies at two field sites in the lower mainland of British Columbia and sacrificed 7 weeks after injection. See Table 2 for detailed statistical reporting. (a) Bars show mean values and error bars represent standard deviation. In the infected group, BQCV and DWV-A RNA copies increased while RNA copies of DWV-B decreased. There were no differences in the RNA copies of SBV between the two groups. The dashed horizontal lines represent the RT-qPCR quantification threshold. (b) The total weight gained by a colony over the 7 weeks was associated with the levels of virus present in the workers at the start of the experiment. (c) The level of DWV-B infection in the queens was strongly correlated with the level of DWV-B present in the workers of the colony at the end of the experiment, and the trend appears to be driven by control group colonies. (d) Correlation matrix showing the Spearman correlation between worker virus RNA copies, queen virus RNA copies, and colony mite counts. Asterisks represent relationships that have p < ɑ/n (Bonferroni correction for multiple hypothesis testing).
Field experiment: supersedure cells, ovary mass, and brood area
There was no difference in the ovary masses of the queens between the two groups, which was unsurprising given the similar levels of infection, but as we observed in the cage trial, there was a negative correlation between ovary mass and the levels of DWV-A and BQCV after controlling for the effect of site (Fig. 5a, see Table 2 for statistical details). To investigate whether ovary mass, as a proxy-metric for queen fertility, could be directly linked to colony population, we assessed the total brood and adult bee area present in the colonies over the course of the experiment. We found that at the termination of the experiment (the time point closest to when queens were sampled), the ratio of brood to adult bees was positively associated with ovary mass (Fig. 5b). This relationship appears to be driven by the correlation between ovary mass and increasing brood area, as there was no significant relationship between ovary mass and adult bee population after controlling for the number of adult bees present at the start of the experiment (see Supplementary Fig. 2). Premature supersedure is a common symptom associated with beekeeper-identified poor or failing queens, and we found that colonies with more heavily infected queens were indeed more likely to build at least one supersedure cell during the experiment, though the statistical significance of this effect was marginal (Fig. 5c; binomial logistic regression/GLM, est. = 0.070, z = 1.93, p = 0.053).
Relationships between ovary mass, virus RNA copies, brood, and supersedure cells in field experiment queens. See Table 2 for complete statistical details. (a) Queen ovary mass is negatively associated with levels of DWV-B, DWV-A, and total virus load. The dashed vertical lines represent the RT-qPCR quantification threshold. (b) The ratio of brood to adult bees, measured as number of frame equivalents, present in the colony is positively associated with the ovary mass of the queens after controlling for the effect of the site and the number of adult bees present at the start of the experiment. (c) The probability of supersedure cells being observed at some point during the experiment increases with increasing levels of virus in the queens. The green points along the curve represent the fitted values of the binomial generalized linear model.
Field experiment: hemolymph proteome changes
While the experimental infection in the field queens did not yield significantly different viral RNA copies between the two groups, we were nonetheless interested in investigating what proteomic changes corresponded to actual virus infection level and ovary size. Quantitative proteomics analysis of hemolymph showed that, of the 1408 protein groups quantified (1% FDR) in at least 75% of samples, 28 proteins were significantly associated with ovary mass (Fig. 6, 5% FDR, Benjamini–Hochberg correction) several of which overlap with proteins differentially expressed among the groups in the cage experiment. These overlapping proteins clustered together (Euclidean distance) and are immune-related. Only 4 proteins were significantly associated with total viral load: an uncharacterized protein (A0A7M7TFJ8), DWV genome polyprotein (G3F401), and the V. apis channel protein (T0LDL0) were elevated with total viral load and phospholipase A1 (A0A7M7IQ52) was downregulated. However, when both ovary mass and viral load were included in the same model, only one protein (beta glucuronidase, A0A7M7MM50, upregulated) was significantly associated with ovary mass and none with virus, reflecting the high level of collinearity between the two variables.
Proteins associated with ovary mass in the hemolymph of the field experiment queens. Each row represents a protein significantly associated with ovary mass (5% FDR, Benjamini–Hochberg method), showing the Uniprot accession number, and each column represents a sample. Total virus RNA copies were not included in this limma model but are shown here to illustrate the correlation between the two variables. Proteins that were not quantified in a sample are shown as grey tiles. Proteins in bold were also differentially expressed between groups in the hemolymph of the cage experiment queens (5% FDR, Benjamini–Hochberg correction). Proteins are clustered via Euclidean distance.
Discussion
In this study, we have further substantiated the relationship between viral infection in honey bees queens and the symptoms associated with beekeeper-identified poor queen quality, while also investigating a potential reproductive-immunity compromise using two parallel infection experiments. In the cage experiment, we found that queens injected with infectious viruses had significantly smaller ovaries and were significantly less likely to begin laying eggs again after infection, while queens that were injected with inactivated viruses had marginally smaller ovaries and had no difference in their likelihood to recommence egg laying. However, both experimental groups exhibited a similar immune response to infection, as determined by the elevated expression of immune proteins in the hemolymph. And, across all three groups, higher levels of viruses were associated with smaller ovaries and fewer eggs laid. The field experiment became a correlational rather than manipulative study, as our experimental infection surprisingly did not alter the overall virus load of the queens (as measured at the end of the experiment). However, the total virus load of the queens, regardless of the group, was indeed negatively associated with ovary mass and positively associated with the presence of supersedure cells, suggesting that there are real impacts of viral infections on queen productivity and possibly worker perception of the queen’s quality. This work connects the proxy-metric for fertility of ovary mass to effects observable at the level of reproductive output (i.e., egg laying) and at the colony level (i.e., relative amount of brood). This validates the use of ovary mass as a metric for current reproductive health, if compared among similar queens in the same environment. Importantly, though, the ovary masses of queens of different ages, sampled at different times, from different locations, and especially those that have been caged and/or transported for different lengths of time, should not be directly compared. In a previous study, we found that the small ovary mass of queens which had been caged for shipping rebounded after being placed back into colonies17.
We were surprised to find that the queens we attempted to infect in the field trial did not actually have higher viral titers than the control group. This contrasts with the cage experiment, where infected queens did have elevated viral loads using the same inoculum; therefore, we know our technique is not fully to blame. This could be due to the lengthened time frame, which could have allowed queens to either recover from the acute effects of infection or acquire new infections from contact with workers. However, it is likely that queens in the cage trials were also nutritionally stressed to some degree compared to queens in the field trial. This could have exacerbated the effects of the experimental infection given that the egg laying output of queens has been shown to be directly related to the nutrition available to her attendants46. Moreover, oogenesis is more severely disrupted in queens kept in cages outside of a colony compared to queens caged inside of a colony, likely due to improved nutrition provided to queens in the colony setting53. Cages had fewer (~ 100) workers present to act as attendants and were supplied with pollen patty supplement, which is less nutritious than the bee bread present in colonies54. Moreover, the attendants that were present in the cages aged beyond the optimal age of nurse bees during the experiment55. While the nutritional stress that queens experience in cages is somewhat artificial, queens could in theory also become nutritionally stressed in colonies during periods of dearth, if worker populations have dwindled due to disease or other stressors, or if mismanagement leads to suboptimal age structure for brood and queen care. We therefore argue that our findings from the cage trial still have the practical utility of highlighting the range of effects that are theoretically possible to occur in colonies.
We endeavored to further investigate whether the impact of virus exposure on reproductive output was due entirely to the pathogenic effects of viral infection or if it was the result of a reproduction immunity trade-off, which assumes nutritional resources are limited to some extent. Such a trade-off is therefore expected to be exacerbated by the reduced nutrition present in the cage environment. Simply comparing the reproductive output of queens challenged with live pathogens fails to fully address this question, given that it is impossible to decouple the effects of the pathogen itself from the effects of the immune response that it has induced. However, by injecting queens with inactivated viruses, we were able to isolate immune effects from pathogenicity. The moderated reduction in ovary size in the queens receiving the inactivated inoculum suggests that honey bees have partially, but not totally, escaped the reproduction-immunocompetence trade-off. This partial escape is likely due in part to social insect queens’ access to far more, but not infinitely more, nutritional resources than solitary reproductive individuals. Additionally, the presence of multi-functional proteins that are involved in both reproduction and immunity (e.g., IRP30; see below) are a tantalizing potential mechanism for reducing the overall cost to sustaining both tasks simultaneously, since resources become effectively shared via common pathways.
Honey bees exhibit a surprising scarcity of immune related genes, possessing only about one-third of those found in Drosophila56. However, Drosophila is also far better studied and there could be unrecognized immune proteins which are either uniquely multi-functional or are yet uncharacterized in social insects. One such protein is IRP30 (immune response protein 30), which was the top significant protein expressed abundantly in both the live virus and inactivated groups in the cage experiment and was negatively associated with ovary mass in hemolymph of the field experiment queens. IRP30 is characterized as an immune effector unique to Hymenoptera and is upregulated in response to a variety of immune challenges including bacteria57, viruses58, and double-stranded (ds)RNA (Nunes et al.59). More recently, it has robustly been shown to also be tied to reproduction: in both workers and queens in bumble bees and honey bees, the expression of IRP30 increases between 20- and 90-fold in egg-laying individuals48. Vitellogenin is another hallmark multi-functional protein, which is not only the major egg yolk protein precursor responsible for provisioning nutrients to developing oocytes, but is also a well-established player in innate immunity in a wide variety of organisms. It functions as a pathogen-associated molecular pattern (PAMP) recognition protein60,61,62, acts as a carrier of pathogen fragments in transgenerational priming63, and possesses antioxidant functions39,64. Vitellogenin expression levels have also been shown to rise in response to Varimorpha ceranae infection, perhaps contributing to a compensatory increase in antioxidant capacity65. The dual role of these proteins in both reproduction and immunity means that a resource allocation conflict should be mitigated, since resources invested in producing these proteins can support both processes.
If the intermediate phenotype of queens receiving inactivated virus was caused by an immune response to the viral particles alone, we would expect queens receiving the live virus to have a stronger response since the number of live viral particles proliferated far beyond the number contained in the single dose of inactivated virus. This is the response we observed for ovary masses, but not immune proteins. The patterns of immune protein expression indicate that both treatments appear to stimulate a very similar immune response, both in the identity of the effectors and the magnitude of expression. Even though this was a viral infection experiment, all the immune-related proteins we identified as significant are canonically associated with the immune response to bacterial and fungal infection. This is unsurprising, given that it has been repeatedly shown there is significant cross-talk among the various immune response pathways, and so-called “antimicrobial” effectors are robustly upregulated in response to viral infection in honey bees44,66. Interestingly, while proteins were differentially expressed between groups in the cage experiment, there were no proteins significantly associated with the total viral load. While this is likely due at least in part to easier statistical ability to detect differences between discrete groups rather than among variable, continuous data with somewhat limited sample numbers, it could also indicate that a perturbation from an individual’s background virus infection in an acute time frame is more impactful to the immune system and ovary mass than absolute virus load (which includes some level of pre-existing infections). Several of these proteins were associated with ovary mass in the field experiment, highlighting how interconnected ovary mass and responses to viral infections are. It is not clear why the immune response in the queens receiving live virus did not have a larger magnitude than the inactivated group, but it is possible that there is an upper limit to immune inducibility that we surpassed with our treatments.
The V. apis protein that was identified in both experiments is intriguing. Vairimorpha apis has been known to frequently co-infect both colonies and individuals with BQCV, and has been shown to act synergistically to increase mortality of adults bees beyond infection with either pathogen individually67, although the way in which these pathogens interact is still unknown. This protein was the only one identified in its protein group, and several other proteins from this pathogen were quantified (although not differentially abundant), which suggests that this is a legitimate finding. The V. ceranae proteome was included in the search database, indicating these are not mis-identified microsporidia proteins either. It is unlikely that the viral inoculum was contaminated with V. apis spores after ultra-centrifugation, given the large size difference between virus particles and spores. Because we also saw this protein in the inactivated group, this suggests that perhaps there is something about stimulating the immune system, either through natural BQCV infection or another stimulant like inactivated virus, which may actually help V. apis invade and proliferate. More research about the relationship between these pathogens and immune stimulation is needed to address this possibility.
Queen “failure” in the field has previously been associated with higher levels of viruses, and we were able to recapitulate symptoms of failure with experimental infections; however, the results of our study also suggest that queen health is intrinsically tied to the circumstances of her colony environment. For instance, levels of virus infection in the colony and in the queen likely have a two-way interaction. Here, we show that in the colony context, it appears that the levels of DWV-B in the workers have a stronger influence on the queen’s DWV-B RNA copies than actual injection with the virus, despite the layers of social immunity that should conceivably shield her from exposure. The presence of Varroa in the colony adds an additional influencing factor, given that DWV is transmitted by mites25 and we found that levels of DWV and mite load were positively correlated, although not significantly. Given that the colonies with the highest levels of DWV-B in the workers and queens were control colonies, it seems unlikely that the relationship between queen and worker infections is driven by infected queens increasing the virus RNA copies in their colonies (though it has previously been shown that virus infection can be vertically transmitted from queen to progeny26,30,68. Many of the common queen failure symptoms can actually be attributed to factors external to the queen, as illustrated by Lee et al.69, who found that switching queens with poor or “failing” brood patterns into colonies with “good” brood patterns significantly improved the brood pattern of the “bad” queen.
Therefore, while viral infection is clearly tied to queen reproductive output, it seems likely that adequate care from her colony can at least partially ameliorate these effects. This is consistent with our hypothesis that honey bee queens may have an increased threshold beyond which a trade-off manifests, rather than a simple presence or absence of a reproduction-immunity trade-off. This could be mediated by multiple factors which are not mutually exclusive: (1) a decreased cost to activating both processes due to the presence of Hymenopteran-specific, multi-functional proteins that function in both reproduction and immunity; (2) the rewiring of specific hormones, which act as a discrete switch between the two processes; or (3) access to an unusually high amount of energetic resources among invertebrates due to social provisioning. Given that the latter element is highly dependent on the health status of the colony, and we found that a compromise between reproduction and immune activation may be partially responsible for some of these effects, this represents a further way in which what we observe as symptoms associated with a queen could be tied to the conditions of the colony. Aside from several issues with queen reproductive health that can be directly attributed to poor mating, aging, or reduced sperm quality, laying blame solely on the queen in cases of queen “failure” belies a significant part of the story involving the health, infection, and nutrition status of a honey bee colony.
Conclusion
Our results indicate that the effects of viral infections in queens translate to the real-world symptoms about which beekeepers are concerned, and these effects are partially, but not entirely, governed by a reproduction-immunity trade-off. Attempted or premature supersedure is an often-cited indicator of queen failure by beekeepers, which we have now shown to be directly correlated to queen virus infections. The acute effects of experimental virus infection were magnified in our laboratory trials, where nutritional stress was likely an exacerbating factor, compared to the field trial where colonies with excess food availability could provide queens with more adequate nutrition. Forage dearth and therefore nutritional stress can, however, also occur in the field, and our data suggest that the effects of virus infection on queens may be worsened under such conditions. These data collectively underscore the likelihood that a reproductive-immunity trade-off may be at play, though possibly moderated by the vast amount of nutritional resources available to social insect queens. This work provides further evidence that virus infections are directly related to symptoms that beekeepers identify in poor queens in the field, highlights the ways in which queen health and her colony environment are inextricably related, and underscores the importance of managing virus levels in beekeeping operations to improve colony health.
Methods
Cage experiment
Young, naturally-mated queens (reared from closely related mothers) that had commenced laying within the previous week were couriered overnight in JZs BZs battery boxes filled with nurses from a British Columbia queen producer (Wild Antho). Queens were received in three batches, each 2 weeks apart, starting in the middle of July. Queens were placed in queen monitoring cages (QMCs)46 along with approximately 11 g of worker bees (~ 100 individuals) collected from a brood frame of a single colony maintained at the University of British Columbia (UBC). The cages were kept in an incubator at 34 °C, with a large water pan to increase humidity, and were supplied ad libitum with fondant (Ambrosia), protein patty (Global Patties), and tap drinking water. A total of n = 9 “baseline” queens, three from each of the three batches received, were anesthetized and frozen immediately upon receipt. Each QMC contained two “egg laying plates” that are made up of 264 hexagonal cells in the dimensions of natural brood comb46. These plates were cleaned and/or replaced every 48–72 h to provide queens with continuous space to lay. Queens were allowed to acclimate to the cages and confirmed to be consistently laying for 6 days before injection, which ensured their ovaries had time to reactivate after caging during shipment. Queens were then injected on Day 6 and observed for 7 more days before sampling. For each batch, queens were randomly assigned to each experimental group (saline injected control, live virus infected, and UV-inactivated virus injected) such that the experimenter (Chapman) was blind to group identifiers. To ensure equal inoculum integrity, the master inoculum was first aliquoted and stored at − 70 °C, with a fresh aliquot used for each batch of queens. In total, n = 9 queens were tested in each group, spread equally across the three experimental batches (3 in each batch). Blinding was maintained until all data had been collected.
Field experiment
Young, age-matched, naturally mated queens (n = 32) were obtained as a single batch of sister queens (grafted from the same donor colony) from the same producer as for the cage experiment and introduced into small, five-frame colonies (referred to by beekeepers as nucleus colonies or “nucs”). The nucs were established on June 7, 2023 almost exclusively from a commercial source of bees (packages obtained from Kintail and Arataki suppliers in New Zealand) that were previously hived in March and allowed to grow until June. Stock imported from New Zealand into Canada is required to have less than 1% Varroa mites (mites per 100 bees), but an oxalic acid mite treatment (3.5% in 50% syrup) was still applied at hiving as a precautionary measure. Some additional stock was pulled from overwintered colonies maintained at the University of British Columbia that had sufficiently low (< 1%) mite loads and an absence of brood disease symptoms. Nucs were produced such that each colony had one frame of honey, two frames of brood, one frame with space for laying (drawn, irradiated comb), and one frame of plastic foundation. Adult bee populations were equalized as much as possible during nuc production but they were further equalized 3 days after queen introduction by adding young adult bees to smaller colonies. The experiment was conducted at two sites 2 km apart (16 nucs at each site) such that no nuc remained at the same site as its parent colony. Nucs were provided with protein supplement (Global Patty) and fondant (Hive Alive) and replenished weekly throughout the experiment to minimize potential nutritional differences among colonies. On July 7, European foulbrood disease was detected in a subset of colonies, therefore all colonies at both sites were treated with Oxytet-25 (oxytetracycline antibiotic) according to the label rate to minimize impact of the disease on the experiment while still managing all colonies equally.
Two weeks after the nucs were produced, baseline colony weights were recorded (supplemental feed was removed prior to weighing), a sample of 15 randomly selected workers from a brood frame was taken for initial worker viral analysis, and both sides of each frame were photographed for subsequent brood area and adult bee coverage calculations. The same day, queens were retrieved then randomly and blindly assigned (with the help of a colleague not otherwise involved in the experiment) to either the saline injected control or live virus infected group (using an aliquot of the same virus mix used for the cage experiments), with balanced groups within each site. Once injected, the queens were returned to their respective colonies within 1–2 h. From that point on, colony weights were recorded weekly and frames were photographed weekly or biweekly (June 22, June 29, July 6, July 19, and Aug 2). During photographing, incidence of brood disease (number of affected cells) was noted and the presence or absence of supersedure cells was recorded before the cells were destroyed to prevent queen loss. During the course of the experiment, 11 colonies grew sufficiently to require an additional box of frames (consisting of a mixture of foundation and irradiated drawn comb), which were weighed and subtracted from the subsequently measured colony weights. At the end of the experiment (Aug 10), we sampled the experimental queens, 15 randomly selected workers from a brood frame (for final worker viral analysis), and ½ cup of bees (~ 300 adults) for Varroa mite counts using alcohol wash method (Jong et al., 1982). A total of 29 queens persisted to the end of the experiment, as in the final 2 weeks one queen was lost by accident and two were superseded. The operators (Chapman and McAfee) remained blind to experimental groups until all data were collected, including image analysis.
Photographs of frames were analyzed using ImageJ to calculate the total area of adult bee coverage and brood coverage. In cases where adult bee coverage was too dense to adequately assess the brood, each side of the frame was photographed, then the bees were gently agitated to expose the brood for a second set of photographs. Top bars were labelled with the colony ID and frame number to associate photos with colonies. In ImageJ, bee area and brood area were traced and calculated, then expressed as a fraction of total frame area, which were summed for each colony.
Virus isolation and propagation
Samples of homogenized honey bees from individual colonies that had been stored at − 80 °C from previous projects were tested by RT-qPCR to identify samples with the highest concentration of the target virus. Those with the highest virus purity relative to other commonly detected viruses in our samples (DWV-B, DWV-A, IAPV, BQCV and SBV) were used for the primary extracts as described by de Miranda et al.70. These extracts were passed through a Corning syringe filter (0.45 µm) to remove bacteria. Primary extracts were used to make 100 µL serial dilutions; DWV primary extracts were diluted with 90 µL of 0.5 M potassium phosphate buffer solution (pH 7.5), BQCV primary extracts were diluted with 0.01 M potassium phosphate buffer (pH 7.0), and three concentrations (105, 104, and 103 gene copies per µL) were used to inoculate pupae.
Pupae that contained low background virus levels were obtained from honey bees that had been initiated from Australian package bees (Aussie bees® sourced from Tasmania) hived on new plastic foundation and held in isolated apiaries in Canada (Star lake Manitoba 49° 45′ 08″ N 95° 15′ 24″ W and Red Deer Lake Ontario 50° 00′ 47″ N 94° 11′ 26″ W) in areas where no other managed honey bees occur. Colonies were sampled using sticky boards and alcohol washes to confirm the absence of Varroa mites. Honey bee pupae collected at the white-eyed to pink-eyed stage were then used to propagate the viruses. A Hamilton® syringe with a 30GxK gauge sterile needle was mounted on a Schley Compact MDL II® insemination stand and then the target virus solution was inoculated into the pupae through the lateral side of the abdomen between the 2nd and 3rd tergites. One µL of virus solution was injected per bee. Inoculated pupae were then placed in a covered Petri dish with paper towel dampened with distilled water, with 20 pupae per dish and incubated at 31.5 °C for 5 days. Pupae were checked regularly, and distilled water was added as required with any dead pupae removed from the dishes. Any pupae that reached the adult stage prior to the end of 5 days were stored at − 80 °C for later processing. After incubation, all healthy pupae (or adults) from each dose (105, 104, or 103 virus particles) were collected in three 50 mL VWR centrifuge tubes. The pupae were stored at − 80 °C until further processing or homogenized immediately after collection. Pupae were placed in 50 mL Spex SamplePrep polycarbonate cryovials with two 9 mm steel beads and homogenized at 1750 rpm for 3 min in a Spex SamplePrep Geno/Grinder® 2010. The homogenate was then used for virus purification.
Virus purification
Extraction buffer, which varied with type of virus, was first added to the propagated virus samples. DWV extraction buffer consisted of 0.5 M potassium phosphate buffer (pH 7.5) and 0.2% ascorbic acid. BQCV buffer consisted of 0.01 M potassium phosphate buffer (pH 7.0) and 0.02% ascorbic acid. To make primary extracts, 2 mL of extraction buffer and 0.5 mL chloroform were added per 1 g of homogenate and samples were kept on ice in a fume hood. Each sample was vortexed for 30 s, then centrifuged at 8000g (8,700 rpm, Eppendorf Centrifuge 5430, placed in the fridge) for 10 min at 15 °C. From each tube, the supernatant was collected into a 5 mL centrifuge tube based on sample origin. Samples were stored at − 80 °C until further purification.
High speed centrifugation was done to further purify the virus samples. The primary extracts were thawed and added to 35 mL Thermo Scientific™ PA Ultracrimp Tubes, with the rest of the volume filled with more extraction buffer. The tubes were centrifuged at 15 °C for 3 h, at 75,000g (27,200 rpm, Sorval Discovery 100SE, Sorval T-865 Rotor). The supernatants were discarded, and the pellets were placed in new 5 mL centrifuge tubes. DWV was resuspended in 5 mL of 0.5 M potassium phosphate buffer (pH 7.5), and BQCV in 0.01 M potassium phosphate (pH 7.0). The solutions were mixed using a spatula and via vortex. The samples were left overnight at 4 °C and then mixed gently in the morning.
The samples were split into 2 mL microcentrifuge tubes to be centrifuged for 15 min at 4 °C, at 8000g (8700 rpm, Eppendorf Centrifuge 5430, placed in the fridge). The supernatants were collected into a new 5 mL centrifuge tube based on sample. The samples were then passed through a 0.45 µm syringe filter into new 5 mL centrifuge tubes, and 20 µL of each sample was taken for viral analysis to determine the final concentrations and purities.
The final extracts had the following purities: DWV-B was 99.99% pure (with other components consisting of 0.000006% DWV-A, 0.00% IAPV, 0.00% BQCV, and 0.000006% SBV), and BQCV was 99.88% pure (with other components consisting of 0.02% DWV-B, 0.02% DWV-A, 0.05% IAPV and 0.02% SBV).
Injection of queens
Queens were anesthetized with carbon dioxide for 3 min (starting when the queen ceased all movement) before being injected on the dorsal side between the 2nd and 3rd abdominal tergites with ~ 100 nL containing either sterile phosphate-buffered saline, a combination of infectious BQCV (~ 1 × 108 copies) and DWV-B (~ 2 × 106 copies), or the same combination and quantity of virus which had been UV-inactivated (cage experiment only). These doses are in line with other experimental infections in honey bees in the literature71,72,73. Injections were performed using the FemoJet Microinjector (Eppendorf) with pulled-capillary glass needles. Injection volumes were calibrated by estimating the droplet volume using a hemocytometer grid under a compound microscope. Everything associated with the injection apparatus was wiped down or soaked in 70% ethanol before and after injections. The saline injected queens were injected first and the live virus group was injected last. To inactivate the virus mixture, a 50 µL aliquot of virus on parafilm (kept on ice) was exposed to 4 consecutive doses of 120,000 mJ of ultraviolet light using a Stratalinker UV Crosslinker (Stratagene) in a manner similar to previously published protocols74,75.
Queen dissection
Queens were sacrificed by anesthetization with carbon dioxide for ~ 5 min. To collect hemolymph for proteomics while avoiding potential contamination from the gut, a leg was removed from the thorax of each queen and a glass capillary was used to collect the exuded hemolymph. After hemolymph was collected, queens were decapitated and the ovaries were removed and weighed on an analytical balance. The thoraxes were stored for viral qPCR analysis. All samples were stored at − 70 °C until subsequent preparation.
Proteomics sample preparation
Hemolymph samples (n = 36 for the cage experiment and n = 29 for the field experiment) were prepared for mass spectrometry essentially as previously described18. Between ~ 5 and ~ 15 µL of hemolymph which had been collected from the queens was directly precipitated by the addition of 500 µL of ice-cold 80% acetone and incubation overnight at − 20 °C. The precipitated protein was pelleted by centrifugation (6000g, 15 min, 4 °C) and the supernatant was discarded. The protein pellet was washed twice with 500 μL of 80% acetone, then the pellet was allowed to air dry (~ 5 min) before solubilization in 15 μL of digestion buffer (6 M urea, 2 M thiourea, 100 mM Tris, pH 8).
The resuspended protein was quantified using a Bradford assay (PIERCE) and then 20 μg of protein was reduced (0.4 μg dithiothreitol, 30 min) and alkylated (2 μg iodoacetamide, 20 min, in the dark). The samples were then digested overnight with 0.8 μg trypsin/LysC mixture (Promega) and diluted with 5 volumes (150 µL) of digestion buffer (50 mM ammonium bicarbonate) after the first 3 h. Digested peptides were acidified with 30 µL of 20% formic acid (to pH < 2.0) and desalted with high-capacity STAGE tips as previously described (Rappsilber et al.76). Eluted samples were dried (SpeedVac, Eppendorf), resuspended in Buffer A (0.1% formic acid, 2% acetonitrile), and then the peptide concentrations were determined using a NanoDrop (Thermo, 205 nm).
Mass spectrometry data acquisition
LC–MS/MS data acquisition was performed exactly as previously described43. A total of 50 ng of digested peptides were injected onto the LC system in randomized order. The digest was separated using NanoElute UHPLC system (Bruker Daltonics) with Aurora Series Gen2 (CSI) analytical column (Ion Opticks, Parkville, Victoria, Australia) heated to 50 °C and coupled to timsTOF Pro (Bruker Daltonics) operated in DIA-PASEF mode. A standard 30 min gradient was run from 2% B to 12% B over 15 min, then to 33% B from 15 to 30 min, then to 95% B over 0.5 min, and held at 95% B for 7.72 min, where buffer B consisted of 0.1% formic acid in 99.4% acetonitrile and buffer A consisted of 0.1% aqueous formic acid and 0.5% acetonitrile in water. Before each run, the analytical column was conditioned with 4 column volumes of buffer A. The NanoElute thermostat temperature was set at 7 °C. The analysis was performed at 0.3 μL/min flow rate.
The trapped ion mobility time-of-flight mass spectrometer (TimsTOF Pro; Bruker Daltonics, Germany) was set to parallel accumulation-serial fragmentation (PASEF) scan mode for data-independent acquisition scanning (100–1700 m/z). The capillary voltage was set to 1800 V, drying gas to 3 L/min, and drying temperature to 180 °C. The MS1 scan was followed by 17 consecutive PASEF ramps containing 22 non-overlapping 35 m/z isolation windows (Table 1), covering the 319.5–1089.5 m/z range. As for TIMS setting, ion mobility range (1/k0) was set to 0.70–1.35 V·s/cm2, 100 ms ramp time and accumulation time (100% duty cycle), and ramp rate of 9.42 Hz; this resulted in 1.91 s of total cycle time. The collision energy was ramped linearly as a function of mobility from 27 eV at 1/k0 = 0.7 V·s/cm2 to 55 eV at 1/k0 = 1.35 V·s/cm2. Mass accuracy was typically within 3 ppm and was not allowed to exceed 7 ppm. For calibration of ion mobility dimension, the ions of Agilent ESI-Low Tuning Mix ions were selected.
Mass spectrometry data processing and analysis
MS data was searched using DIA-NN (v1.8.1)77 using “FASTA digest for library-free search” with “Deep-learning based spectra, RTs and IMs prediction”, allowing for 2 missed cleavages, selecting “Unrelated runs”, setting protein inference to “Protein names from FASTA”, and setting the neural-network classifier to “Double-pass mode”. “N-term M excision” and “C carbamidomethylation” were selected as modifications, and “Use isotopologues”, “MBR”, and “No shared spectra” were all left checked for the algorithm. All other parameters were left set as default. The protein database was the UniProt proteome for Apis mellifera (based on the Amel_HAv3.1 genome assembly), plus common honey bee virus and pathogen proteins from UniProt downloaded on February 6, 2023. To this, a comprehensive list of potential protein contaminants was added78.
All subsequent analyses were performed in R (v 4.3.0). The identified and quantified protein groups from the DIA-NN search were filtered to remove contaminants and proteins identified in fewer than 75% of samples. Normalized LFQ intensity was then log2 transformed. Differential expression analysis was performed using the limma package in R79. For the cage experiment, experimental batch was included as a blocking factor and group (all 4 groups including the baseline queens) was included as a fixed effect. Contrasts were defined with the control queens as reference. For the field experiment, three models were used: just ovary mass as a fixed effect, just total virus RNA copies as a fixed effect, or both. We checked if site should be included as a blocking factor, but the consensus correlation was negative, so it was not included. Empirical Bayes moderation of the standard errors was performed on the resulting outputs, and the false discovery rate was controlled to 5% using the Benjamini–Hochberg correction.
Sample preparation for viral quantification
The thoraces from each queen were placed in separate 2 mL screw-cap homogenizer tubes containing 500 µL of Trizol reagent and three ceramic beads. The samples were homogenized (Precellys 24 homogenizer, Bertin Instruments) 2 times for 30 s at 6500 rpm and placed on ice for 5 min between runs. The homogenate was then centrifuged for 30 s at 14,000g and all 500 µL of supernatant was transferred to a new tube containing 500 µL of 100% ethanol for nucleic acid precipitation. For the worker samples, 5 thoraces from workers were pooled with 1 mL of Trizol reagent and 4 ceramic beads in three tubes for a total of 15 workers sampled from each colony. 250 µL of centrifuged homogenate from each of the 3 tubes was pooled with 500 µL of 100% ethanol for precipitation.
The rest of the RNA extraction was performed using the Direct-zol RNA Miniprep spin-column kit (Zymo Research), including the DNase I treatment, according to manufacturer instructions. The RNA was eluted in 50 µL of RNase-free water and quantified using a Qubit Fluorometer with the Qubit RNA High Sensitivity assay kit (see Supplementary Data 1 for total RNA extract from each thorax). A subset of samples was assessed for contaminants using a NanoDrop One spectrophotometer. The extracted RNA was stored at − 70 °C and thawed only once. RNA was diluted further in RNase-free water and 200 ng was used to produce cDNA. Reverse transcription was carried out using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems), which utilizes random primers, according to the manufacturer’s instructions. cDNA was kept after synthesis at 4 °C for a maximum of 2 weeks.
RT-qPCR for viral quantification
Quantitative real-time PCR was performed on a C1000 Touch Thermal Cycler with a CFX96 Touch Real-Time PCR Detection System (Bio-Rad) using SsoAdvanced™ Universal SYBR® Green Supermix (Bio-Rad). Reactions (10 µL total volume) were set in 96-well plates with 2.5 µL of cDNA at 2 ng/µL (for a total of 5 ng per reaction). Primers were included at a final concentration of 250 nM for both the forward and reverse primers, except for the SBV amplicon for which both primers were at a final concentration of 500 nM. A standard curve in triplicate was included on every plate using a single plasmid containing all the amplicons of interest. See Supplementary Data 2 for the sequences of the plasmid used for the standard curve and the primers. The standard curve spanned 6 orders of magnitude, from 960 copies/µL to 9.6 × 108 copies/µL. The plasmid was dissolved in RNase-free water and the concentration was determined spectrophotometrically using a NanoDrop One. For every plate, the efficiency of the standard curve was between 90 and 110% and the R2 was ≥ 0.99. The qPCR thermocycling conditions were as suggested by the Supermix manufacturer: polymerase activation and initial denaturation for 30 s at 95 °C, followed by denaturation for 10 s at 95 °C, then extension for 20 s at 60 °C for 40 cycles. A melt-curve analysis was performed after every assay following a 65–95 °C gradient at 0.5 °C increments every 2 s, and no unintended amplicons were ever observed. A “no transcript control” was included in triplicate on every plate and was never amplified. During the development of this method, we assessed 3 housekeeping genes (actin, S18, S5) on a subset of 25 samples to verify our sample processing and handling consistency and the standard deviation of the Cqs were under 0.5 for all three genes. Cqs were determined by the regression mode in the CFX Manager Software (v 2.1) rather than by a single threshold. Starting quantity (SQ) in copies was calculated within the software based on the standard curve. These values were then further processed in R (v 4.3.0) for statistical analysis. Samples with a Cq greater than the average Cq of the lowest point on the standard curve for that plate were removed. The SQ’s of the remaining samples were then averaged across replicates for each sample. Viral quantity is reported as copies/ng of starting total RNA.
Statistical analysis
All data analysis was performed in R (v 4.3.0). For analyzing the differences in virus RNA copies and ovary mass between groups, analyzing the relationship between ovary mass and virus RNA copies, and assessing the relationship between the number of eggs laid and the total virus RNA copies of queens in the cage experiment, we used a linear mixed model (lmer in the lme4 package80) in addition to lmerTest81 to obtain p values using Satterthwaite's degrees of freedom method, with experimental batch as a random effect. To assess the differences in the likelihood of queens beginning to lay eggs again after infection in the cage experiment, we used a generalized linear model with a binomial family function (glm in the base R “stats” package) and group as a factor (3 levels). For the field experiment, all models (see Table 2 for detailed model structures), except for the model examining the presence of supersedure cells, were fit using a simple linear model including site as a fixed effect (2 levels). The supersedure cell model was fit with a generalized linear model with a binomial family function (glm in base R “stats” package) and site as a fixed effect. All models were assessed for goodness-of-fit using the DHARMa package82, which utilizes simulation-based residuals to check for typical model misspecification problems such as over/underdispersion and residual autocorrelation.
Data availability
All proteomics raw data, search results, and search parameters are available on MassIVE (www.massive.ucsd.edu, accession MSV000094113 for the field experiment and MSV000094114 for the cage experiment) which are associated with Figs. 3 and 6 and Supplementary Fig. 1. Sample metadata, including viral abundances, are available in Supplementary Data 1. The sequence of the plasmid used for the standard curve and primers used for the viral RT-qPCR assay are available in Supplementary Data 2. Source code underlying figures and data analysis are available freely upon request.
References
Gallai, N., Salles, J.-M., Settele, J. & Vaissière, B. E. Economic valuation of the vulnerability of world agriculture confronted with pollinator decline. Ecol. Econ. 68, 810–821 (2009).
Khalifa, S. A. M. et al. Overview of bee pollination and its economic value for crop production. Insects 12, 688 (2021).
Winston, M. L. The Biology of the Honey Bee (Harvard University Press, 1987).
Amiri, E., Strand, M., Rueppell, O. & Tarpy, D. Queen quality and the impact of honey bee diseases on queen health: Potential for interactions between two major threats to colony health. Insects 8, 48 (2017).
Canadian Association of Professional Apiculturists (CAPA). Statement on Honey Bee Wintering Losses in Canada for 2023 (2024).
Seitz, N. et al. A national survey of managed honey bee 2014–2015 annual colony losses in the USA. J. Apic. Res. 54, 292–304 (2022).
vanEngelsdorp, D., Hayes, J. Jr., Underwood, R. M. & Pettis, J. A survey of honey bee colony losses in the U.S., fall 2007 to spring 2008. PLoS ONE 3, e4071 (2008).
Pettis, J. S., Rice, N., Joselow, K., vanEngelsdorp, D. & Chaimanee, V. Colony failure linked to low sperm viability in honey bee (Apis mellifera) queens and an exploration of potential causative factors. PLoS ONE 11, e0147220 (2016).
Tarpy, D. R. & Olivarez, R. Jr. Measuring sperm viability over time in honey bee queens to determine patterns in stored-sperm and queen longevity. J. Apic. Res. 53, 493–495 (2014).
McAfee, A. et al. Vulnerability of honey bee queens to heat-induced loss of fertility. Nat. Sustain. 3, 367–376 (2020).
Chaimanee, V., Evans, J. D., Chen, Y., Jackson, C. & Pettis, J. S. Sperm viability and gene expression in honey bee queens (Apis mellifera) following exposure to the neonicotinoid insecticide imidacloprid and the organophosphate acaricide coumaphos. J. Insect Physiol. 89, 1–8 (2016).
Woyke, J. Correlations between the age at which honeybee brood was grafted, characteristics of the resultant queens, and results of insemination. J. Apic. Res. 10, 45–55 (1971).
De Souza, D. A., Hartfelder, K. H. & Tarpy, D. R. Effects of larval age at grafting and juvenile hormone on morphometry and reproductive quality parameters of in vitro reared honey bees (Hymenoptera: Apidae). J. Econ. Entomol. 112, 2030–2039 (2019).
Dedej, S., Hartfelder, K., Aumeier, P., Rosenkranz, P. & Engels, W. Caste determination is a sequential process: Effect of larval age at grafting on ovariole number, hind leg size and cephalic volatiles in the honey bee (Apis mellifera carnica). J. Apic. Res. 37, 183–190 (1998).
Rangel, J., Keller, J. J. & Tarpy, D. R. The effects of honey bee (Apis mellifera L.) queen reproductive potential on colony growth. Insectes Sociaux 60, 65–73 (2013).
Tarpy, D. R. & Mayer, M. K. The effects of size and reproductive quality on the outcomes of duels between honey bee queens (Apis mellifera L.). Ethol. Ecol. Evol. 21, 147–153 (2009).
McAfee, A., Chapman, A., Pettis, J. S., Foster, L. J. & Tarpy, D. R. Trade-offs between sperm viability and immune protein expression in honey bee queens (Apis mellifera). Commun. Biol. 4, 48 (2021).
Chapman, A. et al. Fertility costs of cryptic viral infections in a model social insect. Sci. Rep. 12, 15857 (2022).
Paxton, R. J. et al. Epidemiology of a major honey bee pathogen, deformed wing virus: Potential worldwide replacement of genotype A by genotype B. Int. J. Parasitol. Parasites Wildl. 18, 157–171 (2022).
McMahon, D. P. et al. Elevated virulence of an emerging viral genotype as a driver of honeybee loss. Proc. R. Soc. B Biol. Sci. 283, 20160811 (2016).
Natsopoulou, M. E. et al. The virulent, emerging genotype B of deformed wing virus is closely linked to overwinter honeybee worker loss. Sci. Rep. 7, 5242 (2017).
Tehel, A. et al. The two prevalent genotypes of an emerging infectious disease, deformed wing virus, cause equally low pupal mortality and equally high wing deformities in host honey bees. Viruses 11, 114 (2019).
Norton, A. M., Remnant, E. J., Buchmann, G. & Beekman, M. Accumulation and competition amongst deformed wing virus genotypes in naïve Australian honeybees provides insight into the increasing global prevalence of genotype B. Front. Microbiol. 11, 620 (2020).
de Miranda, J. R. & Fries, I. Venereal and vertical transmission of deformed wing virus in honeybees (Apis mellifera L.). J. Invertebr. Pathol. 98, 184–189 (2008).
de Miranda, J. R. & Genersch, E. Deformed wing virus. J. Invertebr. Pathol. 103, S48–S61 (2010).
Ravoet, J., De Smet, L., Wenseleers, T. & de Graaf, D. C. Vertical transmission of honey bee viruses in a Belgian queen breeding program. BMC Vet. Res. 11, 61 (2015).
Williams, G. R. et al. Deformed wing virus in western honey bees (Apis mellifera) from Atlantic Canada and the first description of an overtly-infected emerging queen. J. Invertebr. Pathol. 101, 77–79 (2009).
Amiri, E., Meixner, M. D. & Kryger, P. Deformed wing virus can be transmitted during natural mating in honey bees and infect the queens. Sci. Rep. 6, 33065 (2016).
Francis, R. M., Nielsen, S. L. & Kryger, P. Patterns of viral infection in honey bee queens. J. Gen. Virol. 94, 668–676 (2013).
Gauthier, L. et al. Viruses associated with ovarian degeneration in Apis mellifera L. queens. PLoS ONE 6, e16217 (2011).
Amiri, E., Herman, J. J., Strand, M. K., Tarpy, D. R. & Rueppell, O. Egg transcriptome profile responds to maternal virus infection in honey bees, Apis mellifera. Infect. Genet. Evol. 85, 104558 (2020).
Chen, Y., Pettis, J. S. & Feldlaufer, M. F. Detection of multiple viruses in queens of the honey bee Apis mellifera L. J. Invertebr. Pathol. 90, 118–121 (2005).
Ryabov, E. V. et al. Apis mellifera solinvivirus-1, a novel honey bee virus that remained undetected for over a decade, is widespread in the USA. Viruses 15, 1597 (2023).
Schwenke, R. A., Lazzaro, B. P. & Wolfner, M. F. Reproduction-immunity trade-offs in insects. Annu. Rev. Entomol. 61, 239–256 (2016).
Rueppell, O., Aumer, D. & Moritz, R. F. Ties between ageing plasticity and reproductive physiology in honey bees (Apis mellifera) reveal a positive relation between fecundity and longevity as consequence of advanced social evolution. Curr. Opin. Insect Sci. 16, 64–68 (2016).
Blacher, P., Huggins, T. J. & Bourke, A. F. G. Evolution of ageing, costs of reproduction and the fecundity–longevity trade-off in eusocial insects. Proc. R. Soc. B Biol. Sci. 284, 20170380 (2017).
Remolina, S. C. & Hughes, K. A. Evolution and mechanisms of long life and high fertility in queen honey bees. AGE 30, 177–185 (2008).
von Wyschetzki, K., Rueppell, O., Oettler, J. & Heinze, J. Transcriptomic signatures mirror the lack of the fecundity/longevity trade-off in ant queens. Mol. Biol. Evol. 32, 3173–3185 (2015).
Corona, M. et al. Vitellogenin, juvenile hormone, insulin signaling, and queen honey bee longevity. Proc. Natl. Acad. Sci. U.S.A. 104, 7128–7133 (2007).
Rodrigues, M. A. & Flatt, T. Endocrine uncoupling of the trade-off between reproduction and somatic maintenance in eusocial insects. Curr. Opin. Insect Sci. 16, 1–8 (2016).
Pamminger, T., Treanor, D. & Hughes, W. O. H. Pleiotropic effects of juvenile hormone in ant queens and the escape from the reproduction–immunocompetence trade-off. Proc. R. Soc. B Biol. Sci. 283, 20152409 (2016).
von Wyschetzki, K., Lowack, H. & Heinze, J. Transcriptomic response to injury sheds light on the physiological costs of reproduction in ant queens. Mol. Ecol. 25, 1972–1985 (2016).
McAfee, A., Chapman, A., Bao, G., Tarpy, D. R. & Foster, L. J. Investigating trade-offs between ovary activation and immune protein expression in bumble bee (Bombus impatiens) workers and queens. Proc. Biol. Sci. 291, 20232463 (2024).
McMenamin, A. J., Daughenbaugh, K. F. & Flenniken, M. L. The heat shock response in the western honey bee (Apis mellifera) is antiviral. Viruses 12, 245 (2020).
Shih, S. R., Huntsman, E. M., Flores, M. E. & Snow, J. W. Reproductive potential does not cause loss of heat shock response performance in honey bees. Sci. Rep. 10, 19610 (2020).
Fine, J. D. et al. Quantifying the effects of pollen nutrition on honey bee queen egg laying with a new laboratory system. PLoS ONE 13, e0203444 (2018).
Albert, Š et al. Evidence of a novel immune responsive protein in the Hymenoptera. Insect Biochem. Mol. Biol. 41, 968–981 (2011).
Dong, J., Wu, J., Han, L., Huang, J. & Wang, D. Novel characteristics of immune responsive protein IRP30 in the bumble bee Bombus lantschouensis (Hymenoptera: Apidae). J. Insect Sci. 20, 11 (2020).
Iatsenko, I., Marra, A., Boquete, J.-P., Peña, J. & Lemaitre, B. Iron sequestration by transferrin 1 mediates nutritional immunity in Drosophila melanogaster. Proc. Natl. Acad. Sci. U.S.A. 117, 7317–7325 (2020).
Ma, C. & Kanost, M. R. A β1,3-glucan recognition protein from an insect, Manduca sexta, agglutinates microorganisms and activates the phenoloxidase cascade*. J. Biol. Chem. 275, 7505–7514 (2000).
Vargas-Albores, F. & Yepiz-Plascencia, G. Beta glucan binding protein and its role in shrimp immune response. Aquaculture 191, 13–21 (2000).
Tokarev, Y. S. et al. A formal redefinition of the genera Nosema and Vairimorpha (Microsporidia: Nosematidae) and reassignment of species based on molecular phylogenetics. J. Invertebr. Pathol. 169, 107279 (2020).
Aamidor, S. E. et al. Reproductive plasticity and oogenesis in the queen honey bee (Apis mellifera). J. Insect Physiol. 136, 104347 (2022).
Watkins de Jong, E., DeGrandi-Hoffman, G., Chen, Y., Graham, H. & Ziolkowski, N. Effects of diets containing different concentrations of pollen and pollen substitutes on physiology, Nosema burden, and virus titers in the honey bee (Apis mellifera L.). Apidologie 50, 845–858 (2019).
Lass, A. & Crailsheim, K. Influence of age and caging upon protein metabolism, hypopharyngeal glands and trophallactic behavior in the honey bee (Apis mellifera L.). Insectes Sociaux 43, 347–358 (1996).
Evans, J. D., Aronstein, K., Chen, Y. P. & Hetru, C. Immune pathways and defence mechanisms in honey bees Apis mellifera. Insect Mol. Biol. 12, 645–656 (2006).
Randolt, K. et al. Immune-related proteins induced in the hemolymph after aseptic and septic injury differ in honey bee worker larvae and adults. Arch. Insect Biochem. Physiol. 69, 155–167 (2008).
Fujiyuki, T. et al. Distribution of Kakugo virus and its effects on the gene expression profile in the brain of the worker honeybee Apis mellifera L. J. Virol. 83, 11560–11568 (2009).
Nunes, F. M. F. et al. Non-target Effects of Green Fluorescent Protein (GFP)-derived Double-Stranded RNA (dsRNA-GFP) Used in Honey Bee RNA Interference (RNAi) Assays. Insects 4, 90–103 (2013).
Leipart, V. et al. Structure prediction of honey bee vitellogenin: A multi-domain protein important for insect immunity. FEBS Open Bio 12, 51–70 (2022).
Sun, C. & Zhang, S. Immune-relevant and antioxidant activities of vitellogenin and yolk proteins in fish. Nutrients 7, 8818–8829 (2015).
Sun, W. et al. Vitellogenin receptor expression in ovaries controls innate immunity in the Chinese mitten crab (Eriocheir sinensis) by regulating vitellogenin accumulation in the hemolymph. Fish Shellfish Immunol. 107, 480–489 (2020).
Harwood, G., Amdam, G. & Freitak, D. The role of Vitellogenin in the transfer of immune elicitors from gut to hypopharyngeal glands in honey bees (Apis mellifera). J. Insect Physiol. 112, 90–100 (2019).
Seehuus, S.-C., Norberg, K., Gimsa, U., Krekling, T. & Amdam, G. V. Reproductive protein protects functionally sterile honey bee workers from oxidative stress. Proc. Natl. Acad. Sci. 103, 962–967 (2006).
Alaux, C. et al. Pathological effects of the microsporidium Nosema ceranae on honey bee queen physiology (Apis mellifera). J. Invertebr. Pathol. 106, 380–385 (2011).
Durand, T., Bonjour-Dalmon, A. & Dubois, E. Viral co-infections and antiviral immunity in honey bees. Viruses 15, 1217 (2023).
Bailey, L., Ball, B. V. & Perry, J. N. Association of viruses with two protozoal pathogens of the honey bee. Ann. Appl. Biol. 103, 13–20 (1983).
Chen, Y., Evans, J. & Feldlaufer, M. Horizontal and vertical transmission of viruses in the honey bee, Apis mellifera. J. Invertebr. Pathol. 92, 152–159 (2006).
Lee, K. V., Goblirsch, M., McDermott, E., Tarpy, D. R. & Spivak, M. Is the brood pattern within a honey bee colony a reliable indicator of queen quality?. Insects 10, 12 (2019).
de Miranda, J. R. et al. Standard methods for virus research in Apis mellifera. J. Apic. Res. 52, 1–56 (2013).
Al Naggar, Y. & Paxton, R. J. Mode of transmission determines the virulence of black queen cell virus in adult honey bees, posing a future threat to bees and apiculture. Viruses 12, 535 (2020).
Dubois, E. et al. Outcomes of honeybee pupae inoculated with deformed wing virus genotypes A and B. Apidologie 51, 18–34 (2020).
Tehel, A., Streicher, T., Tragust, S. & Paxton, R. J. Experimental infection of bumblebees with honeybee-associated viruses: No direct fitness costs but potential future threats to novel wild bee hosts. R. Soc. Open Sci. 7, 200480 (2020).
Mathew, A. M., Mun, A. B. & Balakrishnan, A. Ultraviolet inactivation of chikungunya virus. Intervirology 61, 36–41 (2018).
Soh, T. K. et al. A validated protocol to UV-inactivate SARS-CoV-2 and herpesvirus-infected cells. PLoS ONE 18, e0274065 (2023).
Rappsilber, J., Ishihama, Y. & Mann, M. Stop and Go Extraction Tips for Matrix-Assisted Laser Desorption/Ionization, Nanoelectrospray, and LC/MS Sample Pretreatment in Proteomics. Anal. Chem. 75, 663–670 (2003).
Demichev, V., Messner, C. B., Vernardis, S. I., Lilley, K. S. & Ralser, M. DIA-NN: Neural networks and interference correction enable deep proteome coverage in high throughput. Nat. Methods 17, 41–44 (2020).
Frankenfield, A. M., Ni, J., Ahmed, M. & Hao, L. Protein contaminants matter: Building universal protein contaminant libraries for DDA and DIA proteomics. J. Proteome Res. 21, 2104–2113 (2022).
Ritchie, M. E. et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015).
Bates, D., Mächler, M., Bolker, B. & Walker, S. Fitting linear mixed-effects models using lme4. J. Stat. Softw. 67, 1–48 (2015).
Kuznetsova, A., Brockhoff, P. B. & Christensen, R. H. B. lmerTest package: Tests in linear mixed effects models. J. Stat. Softw. 82, 1–26 (2017).
Hartig, F. DHARMa: Residual Diagnostics for Hierarchical (Multi-Level/Mixed) Regression Models. R package (2022).
Acknowledgements
Honey bee research in LJF’s group is supported by an NSERC Discovery Grant. Mass spectrometry infrastructure is supported by the Canada Foundation for Innovation, Genome Canada and Genome BC (264PRO, 374PRO) and computational infrastructure is supported by a Digital Research Alliance of Canada Resource Allocation to LJF. A Project Apis m. grant to AM, AC, LJF and DRT funded this research. We would like to acknowledge Emily Huxter (Wild Antho) for providing the queens, Scott Covey for help developing the RT-qPCR methods, and the UBC proteomics core facility team—Jason Rogalski, Renata Moravcova, and Jeanne Yuan—for running the mass spectrometry samples, instrument maintenance, and technical expertise. Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture. USDA is an equal opportunity provider and employer.
Author information
Authors and Affiliations
Contributions
A.M. and A.C. conceptualized the experiments and analyses with input from L.J.F. and D.R.T. A.C. wrote the first draft of the manuscript, made the figures, and interpreted the data with assistance from A.M. L.J.F., D.R.T., R.W.C., J.F., A.M., Z.R. and K.P. provided editing assistance. A.C. conducted the cage experiment and A.M. conducted the field experiment with assistance from A.C. J.F. provided the queen monitoring cages and provided guidance on their use. R.W.C., K.P., and Z.R. produced the purified virus for use in experimental infections. A.C. conducted the proteomics and RT-qPCR virus analysis. Grants supplied to A.C., A.M., L.J.F., and D.R.T. funded the work.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Chapman, A., McAfee, A., Tarpy, D.R. et al. Common viral infections inhibit egg laying in honey bee queens and are linked to premature supersedure. Sci Rep 14, 17285 (2024). https://doi.org/10.1038/s41598-024-66286-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-66286-5
- Springer Nature Limited