Abstract
Analyze the relationship between genetic variations in the MTHFR gene at SNPs (rs1801131 and rs1801133) and the therapy outcomes for Iraqi patients with rheumatoid arthritis (RA). The study was conducted on a cohort of 95 RA Iraqi patients. Based on their treatment response, the cohort was divided into two groups: the responder (47 patients) and the nonresponder (48 patients), identified after at least three months of methotrexate (MTX) treatment. A polymerase chain reaction-restriction fragment length polymorphism (PCR–RFLP) technique was employed to analyze the MTHFR variations, specifically at rs1801133 and rs1801131. Overall, rs1801131 followed both codominant and dominate models, in which in the codominant model, GG [OR (95% CI) 0.11 (0.022–0.553)] and TG [OR (95% CI) 0.106 (0.021–0.528)] predict responders compared to the TT genotype; meanwhile, for the dominate model, the presence of both GG and TG genotypes [OR (95% CI) 0.108 (0.023–0.507)] together predict responders compared to the TT genotype. The Ars1801133Grs1801131 haplotype was significantly associated with responders [OR (95% CI): 0.388 (0.208–0.723)], while the Grs1801133Trs1801131 haplotype was associated marginally with nonresponders [OR (95% CI) 1.980 (0.965–4.064)]. In the final multivariate analysis, GG/TGrs1801131 genotypes were independently related to responders after adjustment for patients, disease, and treatment characteristics, while TTrs1801131 genotypes were associated with nonresponders. The Iraqi RA patients showed genetic polymorphism in MTHFR gene rs1801131 with T carrier allele associated with nonresponders to MTX therapy. The rs1801131 followed both codominant and dominant models. The G-carried allele for rs1801131 showed an independent association with responder to MTX therapy after adjustment for patients, disease, and treatment characteristics.
Similar content being viewed by others
Introduction
Rheumatoid arthritis (RA) is a persistent and widespread inflammatory condition that causes pain and swelling in joints on both sides of the body1. Insufficient management of chronic inflammation can lead to irreversible damage to the joint tissues2. Methotrexate (MTX) acts as a folic acid inhibitor, exhibiting anti-inflammatory and anti-proliferative activities by closely resembling folic acid concerning its structure and chemical properties. MTX is widely employed in the management of RA due to its substantial effectiveness, excellent safety record, and cost efficiency3,4. Although MTX is commonly used to treat RA, a substantial percentage of patients, ranging from 30 to 50%, do not respond well to MTX medication or experience a recurrence with inadequate response to re-treatment. This event results in the emergence of drug resistance, requiring either the cessation of MTX or the implementation of alternate pharmacological treatments5. The changes in gene expression or activity within the folic acid-MTX metabolic pathway may explain the variability in drug pharmacokinetics and response to MTX observed between patients6.
Folate metabolism is linked to the enzyme methylenetetrahydrofolate reductase (MTHFR). Methionine-methylation of homocysteine (Hcy) is made possible by the MTHFR enzyme, which catalyzes the conversion of 5,10-methylenetetrahydrofolate (5,10-CH2-THF) to 5-methyltetrahydrofolate (5-MTHF)7. This enzyme's regulatory genes have been intensively researched because of their importance in folic acid metabolism7.
Carrying heterozygous or homozygous variants of MTHFR polymorphisms is linked to a genetically set inclination towards higher levels of homocysteine in the blood8. These individuals are frequently diagnosed with mild hyperhomocysteinemia, with fasting plasma levels typically below 30 µmol/l in cases of dietary folate insufficiency8. The reduction in MTHFR activity is particularly noticeable in individuals who have both the rs1801133 variant in one allele of the MTHFR gene and the rs1801131 variant in the other allele9.
Folic acid is essential for preventing chromosomal breakage and hypomethylation of DNA. The activity is hindered when there is a low concentration of Vitamin B12 (B12) because it reduces the activity of methionine synthase, lowering the concentration of S-adenosyl methionine (SAM). This reduction in SAM may lead to a decrease in DNA methylation and prevent folate from being available for the conversion of dUMP to dTMP10.
The presence of a mutation in the MTHFR gene (rs1801133) can have a protective effect against cancer. However, it can also elevate the chance of developmental abnormalities, such as Down syndrome and neural tube anomalies11,12,13. The MTHFR mutation is likely to reduce the inclusion of uracil in DNA, thereby minimizing chromosome breakage and rearrangement. However, its effect on DNA methylation is relatively minor. This suggests that chromosome breakage and rearrangement may be more important than hypomethylation in cancer development. However, the significance of hypomethylation may vary depending on the intake of folate and the extent to which it causes aneuploidy, a potential carcinogenic event14,15. Nevertheless, the MTHFR mutation's influence on DNA methylation status could be significant throughout the intricately regulated development process, where the coordinated and timely expression of genes is crucial and potentially more sensitive to the correct DNA methylation status. The observations regarding the rs1801133 and rs1801131 MTHFR polymorphism demonstrate the potential significance of utilizing folate supplements to address metabolic constraints16,17. It is anticipated that a similar approach will apply to Vitamin B12 for deficiencies in other crucial enzymes like methionine synthase and methionine synthase reductase18. The presence of a mutation in the MTHFR gene (rs1801133) can have a protective effect against cancer. However, it can also elevate the chance of developmental abnormalities, such as Down syndrome and neural tube anomalies. The MTHFR mutation is likely to reduce the inclusion of uracil in DNA, thereby minimizing chromosome breakage and rearrangement. However, its effect on DNA methylation is relatively minor. This suggests that chromosome breakage and rearrangement may be more important than hypomethylation in cancer development. However, the significance of hypomethylation may vary depending on the intake of folate and the extent to which it causes aneuploidy, a potential carcinogenic event. Nevertheless, the MTHFR mutation's influence on DNA methylation status could be significant throughout the intricately regulated development process, where the coordinated and timely expression of genes is crucial and potentially more sensitive to the correct DNA methylation status. The observations regarding the C677T and A1298C MTHFR polymorphism demonstrate the potential significance of utilizing folate supplements to address metabolic constraints. A similar approach is expected to be applicable to Vitamin B12 for deficiencies in other crucial enzymes like methionine synthase and methionine synthase reductase10.
The enzyme expressed by the MTHFR gene on chromosome 1p36.3 plays an important role in cellular metabolism. Several polymorphic locations in the MTHFR gene have been found in previous research, with the SNP sites rs1801133 and rs1801131 being the most often investigated. Mutation in rs1801131 is linked to disruption of MTHFR enzymatic activities, which affect MTX activities in RA patients; however, not all ethnic groups have been tested to confirm this disruption19,20,21. Neither homozygotes nor heterozygotes for rs1801131 were associated with alteration in folic acid levels22. Investigating the link between MTX treatment response and MTHFR rs1801131 and rs1801133 polymorphisms yielded highly varied findings, leaving this pivotal connection doubtful. There is a lack of studies examining the effect of MTHFR genetic polymorphism and its consequence on MTX treatment response in Iraqi RA patients, which was the main drive behind this study.
The current study evaluates the association between genetic mutations in the MTHFR gene in rs1801133 and rs1801131 on the therapeutic outcomes for RA Iraqi patients, which has been reported previously in other nationality, and this is the first study to examine Arabic Iraqi RA patients with these polymorphisms.
Methods
Study design
A study was conducted using a sample of 95 patients from Iraq who had been diagnosed with rheumatoid arthritis (RA) based on the revised 2010 criteria set by the American College of Rheumatology (ACR) and the European League Against Rheumatism (EULAR) for the classification of RA23.
Patients were divided into two groups according to a previous study24, in which the responder group was defined as DAS28 ≤ 3.2, and the nonresponder DAS28 > 3.2 after at least three months of MTX treatment24,25,26. Responder groups included 48 patients and 47 patients as nonresponders.
The study was prepared per STREGA guidelines for reporting genetic association studies27.
Study settings
The participants in this study were selected from the Rheumatology Department of Diwaniya Teaching Hospital. The study was conducted from June 1, 2022, to March 1, 2023.
Inclusion criteria
Adult patients (age ≥ 18 years) with confirmed RA according to revised 2010 ACR/EULAR RA classification criteria23, and all patients should be on MTX for at least three months.
Exclusion criteria
Patients with comorbidities, concurrent connective tissue diseases, patients on concomitant disease-modifying anti-rheumatic drugs (DMARDs) or biological drugs, incomplete data, patients with chronic infectious diseases, cancer, hepatic or renal impairment, endocrinopathy, hematological disorders, and cardiac conditions.
Variables
Disease activity (DAS28-ESR)
In the present study, DAS28-ESR (four variables) was used to examine the disease activity of RA patients28. This score is a validated method for such purpose and has validated many studies and recognized by ACR/EULAR29,30,31,32, and it was calculated based on the following equation28:
Patients were classified as remission (< 2.6), low (≥ 2.6 – ≤ 3.2), moderate (> 3.2 – ≤ 5.1), and high (> 5.1)32.
Laboratory analysis
Measurement of erythrocyte sedimentation rate (ESR) was conducted using the modified Westergren method, which involves the use of whole blood that has been anticoagulated with EDTA (VISION Pro ESR Analyzer, Shenzhen YHLO Biotech Co., Ltd)33,34,35, while rheumatoid factor (RF) was measured using a Chemistry Analyzer, Smart—150, Geno Lab-TEK Corporation, Canada.
Disease response
Patients were divided into two groups according to a previous study24, in which the responder group was defined as DAS28 ≤ 3.2, and nonresponder DAS28 > 3.2 after three months of MTX treatment24,25,26.
Sample size
Power analysis was used to calculate the sample size, G*Power version (3.1.9.7)36,37. The total sample size was 97, with an alpha probability of 0.05, power of 88%, and one-tailed using one sample case variance.
DNA extraction
The genomic DNA was extracted from 5 ml of venous blood obtained from the individuals and subsequently stored at a temperature of − 20 °C until further analysis. The isolation of DNA relies on inorganic methods38. Following agarose gel electrophoresis (1% agarose) using the Bio-Rad Experion Automated Electrophoresis System (RRID: SCR_019691), all images were analyzed using the CS analyzer v3 software by ATTO, Japan. The DNA samples were then validated using spectrophotometry with the Shimadzu UV-1800 UV/Visible Scanning Spectrophotometer; 115 VAC (Germany)39,40, see Fig. 1.
Primer selection
The process of designing PCR primers for MTHFR variants (rs1801133 and rs1801131) is outlined in Table 1. Here is a brief summary: The primers were designed using the NCBI-primer BLAST online software [NCBI BLAST (RRID: SCR_004870)]. The specificity of the produced primers for their target sequences was confirmed by performing a BLAST against the human genome. The primers pair was then selected based on criteria such as product length, melting temperature similarity, primers length, and specificity. The primer's capacity to generate a secondary structure was assessed using the Oligo Calc online software [OligoCalc (RRID: SCR_022663)]. The primer would be deemed unsuitable if it included five or more bases capable of forming self-dimerization or if it had four bases capable of creating a hairpin structure. The generation of primer dimers was assessed for each primer pair using the "Multiple Primer Analyzer" online software provided by Thermo Fisher Scientific Inc.©. The software's sensitivity was set to a value of 2, and any primer pair that produced dimers at this level of sensitivity was deemed unsuitable and excluded41,42,43.
Genotyping of polymorphisms
The MTHFR variations (rs1801133 and rs1801131) were analyzed using a polymerase chain reaction-restriction fragment length polymorphism (PCR–RFLP) technique. The Macrogen Company from Korea supplied the forward and reverse primers in lyophilized state. All polymerase chain reaction (PCR) processes need the use of a PCR thermal cycler, which is manufactured in Germany. The reactions were modified to a total volume of 20 µl, including of Master mix probes and 10–20 ng/µl of genomic DNA (refer to Table 2). The amplification technique is outlined in Table 3.
Restriction fragment length polymorphism analysis for MTHFR rs1801133 and rs1801131 genotyping
Restriction fragment length polymorphism (RFLP) analysis was performed; the basic principle of RFLP involves the fragmentation of a DNA sample via a specific restriction enzyme that acts on a specific restriction site, producing different fragment lengths for different variants due to loss or gain of a restriction site by mutations. The fragments with different lengths of digested DNA are separated using gel electrophoresis depending on the size of these fragments44.
Suitable restriction enzymes (Table 4) were selected using SnapGene viewer software (V6.0.5). The reagents and conditions utilized for the restriction reactions are presented in Table 5. The reaction mixtures were incubated overnight at 37 °C in a water bath for complete digestion by the restriction enzyme.
Electrophoresis of the digested products
The digestion yields of rs1801133 were assessed utilizing electrophoresis via running 5 μl of each sample in 2% agarose gel, while these of rs1801131 using 8% polyacrylamide gel and ethidium bromide nucleic acid staining solution. Applying a 100-V current for 1 h separated the digestion products. The electrophoretic bands were later visualized through a U.V. transilluminator for agarose gel. The electrophoretic bands in polyacrylamide gel were detected using white light. The determination of genotypes was based on the band patterns.
The amplification of the MTHFR gene exhibited an amplicon of 90 bp that contains the target SNP rs1801133. The product digestion by a restriction enzyme revealed three bands’ patterns. The product digestion by a restriction enzyme explored three patterns. One uncut fragment (90 bp) band represents the wild genotype (GG). In contrast, two bands (30 bp) and (60 bp), represent the homozygous genotype (TT), and three bands (30 bp), (60 bp), and (90 bp) represent the heterozygous genotype (TG), as seen in Fig. 2.
Genotyping of rs1801131 polymorphism by PCR–RFLP technique, the PCR product (90 bp) was digested with MboI restriction enzyme, T allele would be recognized by restriction enzyme and cut the amplicon to produce 30 and 60 bp fragment, on the other hand, the G allele did not recognize by the restriction enzyme and produce the original 90 bp fragment. Genotype pattern: TT genotype (30 and 60 bp fragments) as shown in lanes 2, 8, and 10; GG genotype pattern (90 bp fragment) as shown in lanes 1, 4 and 6; TG genotype (30, 60, and 90 bp fragments) as shown in lanes 3, 5, 7 and 9. lane marked with L: 100 + 50 DNA ladder. The product restriction product was resolved on a polyacrylamide gel. [Bio-Rad Experion Automated Electrophoresis System (RRID: SCR_019691)].
The amplification of the MTHFR gene exhibited an amplicon of 254 bp that contains the target SNP rs1801133. The product digestion by a restriction enzyme revealed three band patterns. One uncut fragment (254 bp) band represents the wild genotype (AA). In contrast, two bands (118 bp) and (136 bp) represent the homozygous genotype (GG), and three bands (254 bp), (118 bp) and (136 bp) represent the heterozygous genotype (AG), as seen in Fig. 3.
Genotyping of rs1801133 polymorphism by PCR–RFLP technique, the PCR product (254 bp) was digested with PspN4I restriction enzyme, G allele would be recognized by restriction enzyme and cut the amplicon to produce 118 and 136 bp fragments, on the other hand, the G allele did not recognize by the restriction enzyme and produce the original 254 bp fragment. Genotype pattern: GG genotype (118 and 136 bp fragments) as shown in lanes 1, 2, 7, 8, 11, 12, and 13. AA genotype pattern (254 bp fragment) as shown in lanes 4, 5, 6, and 9. AG genotype (118, 136, and 254 bp fragments) as shown in lanes 3 and 10. The lane was marked with an L: 100 bp DNA ladder. The restriction product was resolved on 2% agarose gel. [Bio-Rad Experion Automated Electrophoresis System (RRID: SCR_019691)].
Ethics approval
The study was approved by "The University of Baghdad-College of Pharmacy Research Ethics Committee" (Approval number: RECAUBCP 6620226, date: 6th June 2022), and written informed consent was obtained from all participants in the study, in accordance with the Helsinki Declaration and its later amendments.
Statistical analysis
An analysis was conducted on the genotyping results to calculate the frequency of alleles and genotypes. The Hardy–Weinberg Equilibrium (HWE) Calculator for 2 Alleles is an online tool that calculates the difference in distribution between the observed frequency of genotypes and the expected frequency of genotypes45. Both rs1801133 (p-value = 0.018) and rs1801131 (p-value = 0.304) adhere to Hardy–Weinberg Equilibrium (HWE) with a p-value of 0.0057 used for allele frequency less than 100, assuming a type I error (α) of 0.0146.
The analysis of haplotypes was conducted using the SHEsis online software, which utilizes the partition-ligation-combination-subdivision EM algorithm for inferring haplotypes using markers that have multiple alleles47,48.
The Shapiro–Wilk test is used to evaluate the adherence of variables to a normal distribution. The chi-square test is employed to ascertain categorical variables. The Fisher exact test is employed as an alternative to the chi-square test in situations when the sample size is below 20 or when there are two or more categories with anticipated frequencies below 5, as the chi-square test is not suitable in such scenarios. The Fisher–Freeman–Halton exact test of independence is used for n x k tables, which is an expansion of the traditional 2 × 2 Fisher exact test where the expected frequency is less than 5%49. The study utilized independent t-tests to analyze the differences in means between the two groups, assuming that both groups follow a normal distribution and do not include any significant outliers. The Mann–Whitney U test is a non-parametric statistical test utilized to assess whether there is a significant difference between two independent groups. The U test was utilized to assess the differences in the median values between the two groups, especially when the data does not follow a normal distribution50. The study utilized binary logistic regression analysis to calculate the odds ratio (OR) and their related 95% confidence intervals. This statistical method is appropriate for cases when the result variable may be categorized into two binary levels.
In addition, the Wald test, which conforms to a Chi-Square distribution with one degree of freedom, was employed to assess the relative magnitude of the parameters. In the framework of multivariate binary logistic regression analysis, an unconditional model was used to investigate the different factors related to patients (model 1), disease (model 2), and therapy (model 3). In addition, the possible interaction between different single nucleotide polymorphisms (SNPs) was taken into account by using multivariate analysis. The statistical analysis was conducted using GraphPad Prism version 10.0.0 for Windows [GraphPad Prism (RRID: SCR_002798)]. A p-value less than 0.05 was considered significant when acceptable.
Consent to participate
Written informed consent obtained.
Results
The study comprised 95 patients diagnosed with rheumatoid arthritis (RA), with an average age of 43.1 ± 10.6 years. The majority of the patients were female (85.3%), approximately 35.8% were smokers. Among the patients, 45.2% had low disease activity, followed by 41.1% with moderate activity, 9.5% with high activity, and 4.2% in remission. Out of the total, 49.5% of the participants were classified as responders, which means their Disease Activity Score (DAS28) was less than or equal to 3.2 after three months of treatment with MTX. The other participants, whose DAS28 was greater than 3.2, were classified as nonresponders24,25,26.
There was no significant difference in age, sex, and smoking; meanwhile, ESR, RF, DAS28, MTX dose and duration, duration of disease, and side effect (SE) were significantly lower in responders compared to nonresponders, as illustrated by Table 6.
Genotyping was done for all RA patients (95) with no genotyping failure. For both SNPs, the results are reported in electrophoresis see Figs. 2 and 3.
Three genetic models were taken into consideration during the analyses to determine the relationship between genetic polymorphism and RA disease activity:
-
a codominant (three distinct genotypes for rs1801131: TT, TG, and GG. and for rs1801133: GG, AG, and AA),
-
a dominant (For the minor allele, heterozygotes and homozygotes were paired together for rs1801131: GG plus TG vs. TT. And for rs1801133: AA plus AG vs. GG.)
-
recessive model (GG vs. TG plus TT are homozygotes for the minor allele for rs1801131. And for rs1801133: AA vs AG plus GG).
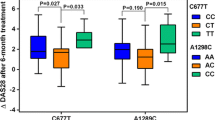
The distribution of genotypes for the rs1801131 SNP was significantly different among the three genotypes. For the single alleles, the TT allele showed the highest frequency in nonresponders, and the T allele showed the highest frequency in nonresponders. Meanwhile, the genotype distribution for the (rs1801133) SNP was not significantly different among the three genotypes and for the single alleles, as illustrated by Table 7.
Overall, rs1801131 SNP followed both codominant and dominate models, in which in the codominant model, GG and TG predict responders compared to the TT genotype; meanwhile, for the dominate model, the presence of both GG and TG genotype together predict responders compared to the TT genotype. There was no significant difference in the distribution of three genotypes according to the three genetics models for rs1801133 SNP, as illustrated by Table 8.
The AG haplotype was significantly associated with responders with OR: 0.388 (p-value = 0.003), while the GT haplotype was associated marginally with nonresponders OR: 1.98 (p-value = 0.060), as illustrated by Table 9.
In the final multivariate analysis, GG/TGrs1801131 genotypes were independently associated with responders after adjustment for patients, disease, and treatment characteristics, while TTrs1801131 genotypes were related to nonresponders, as illustrated by Table 10.
Discussion
The genetic role in predisposition, development, severity, and response for therapeutic intervention of RA is well documented, and MTX is one of the most intriguing subjects for pharmacogenomics research. Pharmacogenetic studies facilitate the discovery of response predictors for optimal efficacy with the least harm43,51,52,53,54,55.
In Iraq, the prevalence survey for rheumatoid arthritis was done during the summer of 1975 in persons aged 16 and over in areas of Iraq; definite Rheumatoid arthritis was observed in 1% of the population56. Additional studies reported prevalence as high as 1.93%57, and 2.34%58.
Regarding the demographic variables, the mean age of the patients was 43.1 ± 10.6 years, with females representing the majority of the patients (85.3%). In Iraq, the published literature showed similarities to observed demographic data in 470 RA patients in Basrah city, revealing a mean age of 49.9 ± 11.9 years, and 81.9% of the patients were female59. In another study involving 1039 RA patients in Babel city, 82.09% were female and had a mean age of 44.87 ± 3.63 years60. In a study in Erbil City, the mean age was 47.4 ± 10.6 years, with females representing 72.30% of the patients61. In a study done in Baghdad city, the mean age was 46.5 years, with 58.1% of the patients being females62. In a large study that involved several Iraqi cities, the mean age was 48.1 ± 12.1 years, and 83% were females63. The consistency of the current study regarding age and sex with the previous Iraqi study indicates the selection of RA patients in the current study is a good representative of all RA patients; thus, the current study finding can be generalized.
Pharmacogenomics studies concerning RA have emphasized the identification of genetic markers that may predict a patient's clinical response or the development of side effects for a given therapy, as well as the assessment of potential interactions between a patient's genetic profile and environmental factors64,65,66,67.
This field holds promise for advancing personalized medicine and enhancing RA treatment approaches. In the present study, the genetic association between MTHFR gene rs1801131 and rs1801133 SNP polymorphism and MTX response was sought; it was observed that the rs1801131 genotype follows both codominant and dominant genetic model, in which GG/TGrs1801131 genotype in codominant/dominate models predict responder to treatment, while TTrs1801131 genotype predict nonresponder. In the final multivariate analysis, the GG/TGrs1801131 genotypes were independently associated with responders after adjustment for patients, disease, and treatment characteristics, while TTrs1801131 genotypes were associated with nonresponders. This phenomenon could be attributed to decreased MTHFR activity, resulting in lower levels of 5-MTHF and other folate cofactors. This reduction may lead to a decrease in adenosine release, partly explaining the lack of response to MTX.
In haplotyping analysis, the AG haplotype for rs1801133–rs1801131 was found to be significantly associated with responders, while the GT haplotype for rs1801133–rs1801131 was associated marginally with nonresponders OR: 1.98 (p-value = 0.060).
This is the first study that examined the Arabic Iraqi population; however, recently, in another study that examined a different ethnic group (namely Kurdish) Iraqi RA patients, the authors concluded that genetic polymorphisms of MTHFR SNP (rs1801133 and rs1801131) are associated with MTX efficacy but not toxicity in RA patients68. The current study only found that rs1801131 was associated with treatment response, regarding the Kurdish-Iraqi study68, and in our study, no association between rs1801133 and rs1801131 with sex was found.
Similar to the current study, in Urano et al., the rs1801131 allele was associated with lower doses of MTX. At the same time, rs1801133 showed no significant difference between GG and TG allele (GG showed more association) and had a significantly higher association with good response (more reduction of ESR and CRP), while RS1801133 showed no association69. In haplotyping analysis, 677C–1298G was associated with lower doses of MTX69.
Another similar study reported that rs1801133 and rs1801131 alleles were associated with response in multivariate analysis; the following diplotypes CC/GG, TA/AG, CC/AG, CA/AG, and CA/GG were significantly associated with a lower probability of response compared to CA/GT (rs1801133–rs1801131)70. In the study conducted by Xiao et al., it was shown that the allele frequency of rs1801131G in the clinical response group was substantially greater compared to the non-response group (21.0% vs. 8.1%, p < 0.05). Furthermore, it was found that patients with the GG or TG genotype had a more pronounced clinical reaction than those with the TT genotype71.
Sharaki et al. examined 50 Arabic Egyptian patients with RA of similar age, sex, and disease duration. The authors read MTHFR SNP (rs1801131), in their multivariate analysis, found that MTHFR rs1801131 (TG + GG genotypes) independently associated with higher odds of non-response to MTX (OR 39.1, 95% CI 2.0–776.6), however they did not examine the effect of genetic polymorphism aside from MTHFR rs180113124, which is to the contrary to the current study. In Algerian RA patients, no association was found between RS1801133 polymorphism and MTX efficiency. In contrast, the T allele of the T1298G polymorphism was associated with good and moderate response (p = 0.02, OR 3.28, 95% CI [1.11–9.42])72. It might be linked to a decline in MTHFR function, which also results in lower quantities of 5-MTHF and other folate cofactors, reducing adenosine releases and partially explaining MTX non-response73,74. Kato et al. studied 150 RA Japanese patients, and the mean age was more than 59 years, with similar DAS. The authors examined several SNPs, including the MTHFR genes rs1801131 and rs1801133. Contrary to the current study, the TT genotype for rs1801131 SNP had lower DAS28 compared to the TG/GG genotype (3.15 vs. 3.92, p-value = 0.04), which indicates a better response conferred by the T allele75, in Wessels et al. study that examined 205 RA Netherlands patients with older age and similar sex distribution. The authors examined the same MTHFR gene rs1801131 and rs1801133 SNP polymorphism; for rs1801133 SNP no the genetic polymorphism did not associate with MTX treatment response, while the TT genotype of rs1801131 was associated with a good response76, their findings are best applied to patients with early RA who have not received DMARDs. Van Ede et al. found no significant difference in the average change in DAS44 in MTHFR 677 T allele carriers versus noncarriers77. Research has demonstrated that the length of a condition and the prior treatment received have a significant impact on the result of methotrexate (MTX) treatment78,79, and their cohort included patients with persistent RA who had previously taken other DMARDs.
In Kumagai et al. study, MTHFR rs1801133 and rs1801131 polymorphisms showed no association with MTX-related efficacy80. A meta-analysis comprising ten studies found that the MTHFR gene RS1801131 polymorphism did not significantly impact the outcome of methotrexate (MTX) treatment in patients with rheumatoid arthritis (RA). However, subgroup analysis revealed a significant correlation between the MTHFR gene rs1801131 polymorphism and reduced efficacy of MTX in the South Asian population. Specifically, individuals with the GG genotype had a lower odds ratio (OR) of 0.45 (95% confidence interval [CI] 0.23–0.89, p = 0.021) compared to those with the GT + TT genotypes. Furthermore, the presence of the MTHFR gene rs1801131 polymorphism in the group receiving partial folate supplementation was found to be associated with a decrease in the effectiveness of MTX (GG vs. GT + TT: odds ratio = 0.43, 95% confidence interval 0.20–0.92, p = 0.029)81.
The reasons behind the higher prevalence of MTX and MTHFR response in female patientscould be explained by various theories, such as the hormonal differences between females and males and genetic factors that could be unique to the Iraqi Arabic female population.
Study limitations
Reproducing findings in genetic association research is intrinsically intricate, resulting in difficulties in making significant comparisons across many studies. The strongest evidence of a correlation is in reproducing this association in a separate population that possesses the same genetic makeup, physical characteristics, and direction of influence76. The majority of research examining genetic connections suffer from insufficient statistical power as a result of the limited number of individuals with the homozygous mutant genotype76.
One of the challenges encountered when assessing the effectiveness of treatment in rheumatoid arthritis (RA) is the requirement for agreement on the measures used to determine efficacy. There exists a divergence of viewpoints regarding issues such as the duration and experimental approach required82,83,84,85,86,87. In therapeutic applications, it is desirable to establish definitive threshold values that can be used to categorize patients as either responders or nonresponders, hence facilitating the development of treatment protocols. Nevertheless, within clinical practice, clinicians are inclined to modify treatment regimens in response to any observed alteration in efficacy measurements88,89.
Conclusion
The Iraqi RA patients showed genetic polymorphism in rs1801131 SNP with T carrier allele associated with nonresponder to MTX therapy; the rs1801131 SNP followed both codominant and dominate models. The G-carried allele for rs1801131 showed an independent association with responder to MTX therapy after adjustment for patients, disease, and treatment characteristics. While rs1801133 SNP did not associate with MTX responsiveness (Supplementary figures).
Data availability
Zenodo: MTHFR gene polymorphism. https://doi.org/10.5281/zenodo.8392701. Data are available under the terms of the Creative Commons Attribution 4.0 International license (CC-BY 4.0).
References
Mohammed, S. I., Zalzala, M. H. & Gorial, F. I. The effect of TNF-alpha gene polymorphisms at -376 G/A, -806 C/T, and -1031 T/C on the likelihood of becoming a non-responder to etanercept in a sample of Iraqi Rheumatoid arthritis patients. Iraqi J. Pharm. Sci. 31, 113–128. https://doi.org/10.31351/vol31iss2pp113-128 (2022).
Mikhael, E. M. & Ibrahim, T. Neutrophil/lymphocyte ratio is not correlated with disease activity in rheumatoid arthritis patients. Iraqi J. Pharm. Sci. 22, 9–14. https://doi.org/10.31351/vol22iss2pp9-14 (2017).
Abdul-Wahab, F. K. & Al-Shawi, N. N. Effects of vitamin D3 on methotrexate-induced jejunum damage in rats. Iraqi J. Pharm. Sci. 29, 260–267. https://doi.org/10.31351/vol29iss1pp260-267 (2020).
Faiq, M., Kadhim, D. & Gorial, F. Assessing quality of life among sample of Iraqi patients with rheumatoid arthritis. Int. J. Res. Pharm. Sci. 10, 2856–2863 (2019).
Inoue, K. & Yuasa, H. Molecular basis for pharmacokinetics and pharmacodynamics of methotrexate in rheumatoid arthritis therapy. Drug Metab. Pharmacokinet. 29, 12–19. https://doi.org/10.2133/dmpk.dmpk-13-rv-119 (2014).
Dervieux, T., Greenstein, N. & Kremer, J. Pharmacogenomic and metabolic biomarkers in the folate pathway and their association with methotrexate effects during dosage escalation in rheumatoid arthritis. Arthritis Rheum. 54, 3095–3103. https://doi.org/10.1002/art.22129 (2006).
Murto, T. et al. Folic acid supplementation and methylenetetrahydrofolate reductase (MTHFR) gene variations in relation to in vitro fertilization pregnancy outcome. Acta Obstet. Gynecol. Scand. 94, 65–71. https://doi.org/10.1111/aogs.12522 (2015).
Li, W.-X., Dai, S.-X., Zheng, J.-J., Liu, J.-Q. & Huang, J.-F. Homocysteine metabolism gene polymorphisms (MTHFR C677T, MTHFR A1298C, MTR A2756G and MTRR A66G) jointly elevate the risk of folate deficiency. Nutrients 7, 6670–6687 (2015).
Baba, S. M. et al. Methylenetetrahydrofolate reductase gene C677T and A1298C polymorphic sequence variations influences the susceptibility to chronic myeloid leukemia in Kashmiri population. Front. Oncol. 9, 612. https://doi.org/10.3389/fonc.2019.00612 (2019).
Fenech, M. The role of folic acid and vitamin B12 in genomic stability of human cells. Mutat. Res. Fund. Mol. Mech. Mutag. 475, 57–67. https://doi.org/10.1016/S0027-5107(01)00079-3 (2001).
Fowler, B. Genetic defects of folate and cobalamin metabolism. Eur. J. Pediatr. 157, S60–S66. https://doi.org/10.1007/PL00014306 (1998).
Stabler, S. P., Lindenbaum, J. & Allen, R. H. Vitamin B-12 deficiency in the elderly: Current dilemmas. Am. J. Clin. Nutr. 66, 741–749. https://doi.org/10.1093/ajcn/66.4.741 (1997).
Rosenberg, I. H. & Rosenberg, L. E. The implications of genetic diversity for nutrient requirements: The case of folate. Nutr. Rev. 56, S47-53. https://doi.org/10.1111/j.1753-4887.1998.tb01687.x (1998).
Chen, J. et al. A methylenetetrahydrofolate reductase polymorphism and the risk of colorectal cancer. Cancer Res. 56, 4862–4864 (1996).
Ma, J. et al. Methylenetetrahydrofolate reductase polymorphism, dietary interactions, and risk of colorectal cancer. Cancer Res. 57, 1098–1102 (1997).
Skibola, C. F. et al. Polymorphisms in the methylenetetrahydrofolate reductase gene are associated with susceptibility to acute leukemia in adults. Proc. Natl. Acad. Sci. USA 96, 12810–12815. https://doi.org/10.1073/pnas.96.22.12810 (1999).
Ames, B. N. Cancer prevention and diet: Help from single nucleotide polymorphisms. Proc. Natl. Acad. Sci. USA 96, 12216–12218. https://doi.org/10.1073/pnas.96.22.12216 (1999).
Wilson, A. et al. A common variant in methionine synthase reductase combined with low cobalamin (vitamin B12) increases risk for spina bifida. Mol. Genet. Metab. 67, 317–323. https://doi.org/10.1006/mgme.1999.2879 (1999).
Ashfield-Watt, P. A. et al. Methylenetetrahydrofolate reductase 677C–>T genotype modulates homocysteine responses to a folate-rich diet or a low-dose folic acid supplement: A randomized controlled trial. Am. J. Clin. Nutr. 76, 180–186. https://doi.org/10.1093/ajcn/76.1.180 (2002).
Yamada, K., Chen, Z., Rozen, R. & Matthews, R. G. Effects of common polymorphisms on the properties of recombinant human methylenetetrahydrofolate reductase. Proc. Natl. Acad. Sci. USA 98, 14853–14858. https://doi.org/10.1073/pnas.261469998 (2001).
Froese, D. S. et al. Mutation update and review of severe methylenetetrahydrofolate reductase deficiency. Hum. Mutat. 37, 427–438. https://doi.org/10.1002/humu.22970 (2016).
Hekmatdoost, A. et al. Methyltetrahydrofolate vs folic acid supplementation in idiopathic recurrent miscarriage with respect to methylenetetrahydrofolate reductase C677T and A1298C polymorphisms: A randomized controlled trial. PLoS ONE 10, e0143569. https://doi.org/10.1371/journal.pone.0143569 (2015).
Kay, J. & Upchurch, K. S. ACR/EULAR 2010 rheumatoid arthritis classification criteria. Rheumatology 51(Suppl 6), 5–9. https://doi.org/10.1093/rheumatology/kes279 (2012).
Sharaki, O. A., Elgerby, A. H., Nassar, E. S. & Khalil, S. S. E. Impact of methylenetetrahydrofolate reductase (MTHFR) A1298C gene polymorphism on the outcome of methotrexate treatment in a sample of Egyptian rheumatoid arthritis patients. Alexandria J. Med. 54, 633–638. https://doi.org/10.1016/j.ajme.2017.11.008 (2018).
Fransen, J. & van Riel, P. L. The disease activity score and the EULAR response criteria. Clin. Exp. Rheumatol. 23, S93-99 (2005).
Fransen, J., Creemers, M. C. & Van Riel, P. L. Remission in rheumatoid arthritis: Agreement of the disease activity score (DAS28) with the ARA preliminary remission criteria. Rheumatology 43, 1252–1255. https://doi.org/10.1093/rheumatology/keh297 (2004).
Little, J. et al. Strengthening the reporting of genetic association studies (STREGA): An extension of the STROBE statement. PLoS Med. 6, e22. https://doi.org/10.1371/journal.pmed.1000022 (2009).
Prevoo, M. L. et al. Modified disease activity scores that include twenty-eight-joint counts: Development and validation in a prospective longitudinal study of patients with rheumatoid arthritis. Arthritis Rheum. 38, 44–48. https://doi.org/10.1002/art.1780380107 (1995).
van Riel, P. L. The development of the disease activity score (DAS) and the disease activity score using 28 joint counts (DAS28). Clin. Exp. Rheumatol. 32, 65–74 (2014).
Saag, K. G. et al. American College of Rheumatology 2008 recommendations for the use of nonbiologic and biologic disease-modifying antirheumatic drugs in rheumatoid arthritis. Arthritis Rheum. 59, 762–784. https://doi.org/10.1002/art.23721 (2008).
Singh, J. A. et al. 2012 update of the 2008 American College of Rheumatology recommendations for the use of disease-modifying antirheumatic drugs and biologic agents in the treatment of rheumatoid arthritis. Arthritis Care Res. 64, 625–639. https://doi.org/10.1002/acr.21641 (2012).
Fraenkel, L. et al. 2021 American college of rheumatology guideline for the treatment of rheumatoid arthritis. Arthritis Care Res. 73, 924–939. https://doi.org/10.1002/acr.24596 (2021).
Horsti, J. & Kovanen, M. Using EDTA as an anticoagulant for ESR to replace citrate. Kliin Lab. 17, 97–100 (2000).
ICSH. ICSH recommendations for measurement of erythrocyte sedimentation rate International Council for Standardization in Haematology (Expert Panel on Blood Rheology). J. Clin. Pathol. 46, 198–203. https://doi.org/10.1136/jcp.46.3.198 (1993).
Koepke, J., Van Assendelft, O., Bull, B. & Richardson-Jones, A. Standardization of EDTA anticoagulation for blood counting procedures. Labmedica 1989, 5 (1988).
Faul, F., Erdfelder, E., Lang, A. G. & Buchner, A. G*Power 3: A flexible statistical power analysis program for the social, behavioral, and biomedical sciences. Behav. Res. Methods 39, 175–191. https://doi.org/10.3758/bf03193146 (2007).
Faul, F., Erdfelder, E., Buchner, A. & Lang, A.-G. Statistical power analyses using G*Power 3.1: Tests for correlation and regression analyses. Behav. Res. Methods 41, 1149–1160. https://doi.org/10.3758/BRM.41.4.1149 (2009).
Chacon-Cortes, D. & Griffiths, L. Methods for extracting genomic DNA from whole blood samples: Current perspectives. J. Bioreposit. Sci. Appl. Med. 2, 1–9. https://doi.org/10.2147/BSAM.S46573 (2014).
Tataurov, A. V., You, Y. & Owczarzy, R. Predicting ultraviolet spectrum of single stranded and double stranded deoxyribonucleic acids. Biophys. Chem. 133, 66–70. https://doi.org/10.1016/j.bpc.2007.12.004 (2008).
Hegazi, M. A. M., Seleem, A., El-Adawy, E. H. & Elhussini, M. E. A. Association of IGF-I gene polymorphism with diabetic nephropathy in Egyptians with type 2 diabetes. Egypt. J. Intern. Med. 30, 191–196. https://doi.org/10.4103/ejim.ejim_48_18 (2018).
Kibbe, W. A. OligoCalc: An online oligonucleotide properties calculator. Nucleic Acids Res. 35, W43-46. https://doi.org/10.1093/nar/gkm234 (2007).
Qu, W. et al. MFEprimer-2.0: A fast thermodynamics-based program for checking PCR primer specificity. Nucleic Acids Res. 40, W205-208. https://doi.org/10.1093/nar/gks552 (2012).
Al-Radeef, M. Y., Fawzi, H. A. & Allawi, A. A. ACE gene polymorphism and its association with serum erythropoietin and hemoglobin in Iraqi hemodialysis patients. Appl. Clin. Genet. 12, 107–112. https://doi.org/10.2147/tacg.s198992 (2019).
Pingoud, A. & Jeltsch, A. Structure and function of type II restriction endonucleases. Nucleic Acids Res. 29, 3705–3727 (2001).
Schaid, D. J., Batzler, A. J., Jenkins, G. D. & Hildebrandt, M. A. Exact tests of Hardy–Weinberg equilibrium and homogeneity of disequilibrium across strata. Am. J. Hum. Genet. 79, 1071–1080. https://doi.org/10.1086/510257 (2006).
Wigginton, J. E., Cutler, D. J. & Abecasis, G. R. A note on exact tests of Hardy–Weinberg equilibrium. Am. J. Hum. Genet. 76, 887–893. https://doi.org/10.1086/429864 (2005).
Li, Z. et al. A partition-ligation-combination-subdivision EM algorithm for haplotype inference with multiallelic markers: Update of the SHEsis. Cell Res. 19, 519–523. https://doi.org/10.1038/cr.2009.33 (2009).
Shi, Y. Y. & He, L. SHEsis, a powerful software platform for analyses of linkage disequilibrium, haplotype construction, and genetic association at polymorphism loci. Cell Res. 15, 97–98. https://doi.org/10.1038/sj.cr.7290272 (2005).
Ghent, A. W. A method for exact testing of 2X2, 2X3, 3X3, and other contingency tables, employing binomial coefficients. Am. Midl. Nat. 88, 15–27. https://doi.org/10.2307/2424485 (1972).
Hintze, J. L. & Nelson, R. D. Violin plots: A box plot-density trace synergism. Am. Stat. 52, 181–184. https://doi.org/10.1080/00031305.1998.10480559 (1998).
Owen, S. A. et al. Genetic polymorphisms in key methotrexate pathway genes are associated with response to treatment in rheumatoid arthritis patients. Pharmacogenom. J. 13, 227–234. https://doi.org/10.1038/tpj.2012.7 (2013).
Muss, T., Al-Faham, M., Gorial, F. & Zghair, A. Galectin-8 gene polymorphism among Iraqi patients with rheumatoid arthritis. Res. J. Pharm. Tech. 12, 1643–1645. https://doi.org/10.5958/0974-360X.2019.00274.9 (2019).
Hussain, M. A. T. D. A. A. Role investigation of interleukin- IL-17 rs763780 T/C gene polymorphism with iraqi rheumatoid arthritis patients. Iraqi J. Biotechnol. 21, 103–114 (2022).
Ad’hiah, A. H., Mahmood, A. S., Al-kazaz, A.-K.A. & Mayouf, K. K. Gene expression and six single nucleotide polymorphisms of interleukin-6 in rheumatoid arthritis: A case-control study in Iraqi patients. Alexandria J. Med. 54, 639–645. https://doi.org/10.1016/j.ajme.2018.08.001 (2018).
Al-Saffar, E. A. & Al-Saadi, B. Q. Study the association of IRAK1 gene polymorphism and some immunological markers with the risk of rheumatoid arthritis incidence in sample of Iraqi patients. Iraqi J. Biotechnol. 21, 46–60 (2022).
Al-Rawi, Z. S., Alazzawi, A. J., Alajili, F. M. & Alwakil, R. Rheumatoid arthritis in population samples in Iraq. Ann. Rheum. Dis. 37, 73–75. https://doi.org/10.1136/ard.37.1.73 (1978).
Ali Mohammed Hussein, A. Incidence of Rheumatoid Arthritis [2001 to 2011]. (2013).
Rashid, M. K. Prevalence rate of rheumatoid arthritis among patients attending rheumatology consultation clinic at Baquba Teaching Hospital. Diyala J. Med. 24, 54–65 (2023).
Mathkhor, A. J., Abdullah, A. H. & Khoudhairy, A. S. Demographic, clinical, and serological features of Iraqi patients with rheumatoid arthritis: Evaluation of 470 patients. Int. J. Clin. Rheumatol. 16, 99–103 (2021).
Alkazzaz, A. M. H. Incidence of rheumatoid arthritis [2001 to 2011]. Iraqi Postgrad. Med. J. 12, 568–572 (2013).
Albarzinji, N., Ismael, S. A. & Albustany, D. Association of rheumatoid arthritis and its severity with human leukocytic antigen-DRB1 alleles in Kurdish region in North of Iraq. BMC Rheumatol. 6, 4. https://doi.org/10.1186/s41927-021-00229-9 (2022).
Isho Gorial, F., Naema, S., Ali, H. & Hussain, S. The influence of rheumatoid arthritis on work productivity among a sample of Iraqi patients. Al Rafidain J. Med. Sci. 1, 110–117. https://doi.org/10.54133/ajms.v1i.47 (2021).
Al-Ani, N. et al. Clinical outcomes in Iraqi patients with rheumatoid arthritis following earlier or later treatment with etanercept. Open Access Rheumatol. 13, 57–62. https://doi.org/10.2147/oarrr.S300838 (2021).
Awni, A. A., Hamed, Z. O., Abdul-Hassan Abbas, A. & Abdulamir, A. S. Effect of NLRP3 inflammasome genes polymorphism on disease susceptibility and response to TNF-α inhibitors in Iraqi patients with rheumatoid arthritis. Heliyon 9, e16814. https://doi.org/10.1016/j.heliyon.2023.e16814 (2023).
Younis, S. S., Issa, N. K. & Sulaiman, D. M. MTHFR gene polymorphisms in Iraqi Kurdish rheumatoid arthritis patients: Relation to methotrexate response and toxicity. Iraqi J. Sci. 63, 5186–5196. https://doi.org/10.24996/ijs.2022.63.12.9 (2022).
Mohammed, S., Zalzala, M. & Gorial, F. Association of tumor necrosis factor-alpha promoter region gene polymorphism at positions -308G/A, -857C/T, and -863C/A with etanercept response in Iraqi rheumatoid arthritis patients. Arch. Rheumatol. 37, 613–625. https://doi.org/10.46497/ArchRheumatol.2022.9272 (2022).
Mohammed, M. M., Hamadi, S. A. & Jasim, G. A. Impact of metabolic factors on the incidence and intensity of knee osteoarthritis in a sample of Iraqi patients. J. Glob. Pharm. Technol. 10, 971–980 (2018).
Relation to Methotrexate Response and Toxicity. MTHFR gene polymorphisms in Iraqi Kurdish rheumatoid arthritis patients. Iraqi J. Sci. 63, 5186–5196. https://doi.org/10.24996/ijs.2022.63.12.9 (2022).
Urano, W. et al. Polymorphisms in the methylenetetrahydrofolate reductase gene were associated with both the efficacy and the toxicity of methotrexate used for the treatment of rheumatoid arthritis, as evidenced by single locus and haplotype analyses. Pharmacogenetics 12, 183–190. https://doi.org/10.1097/00008571-200204000-00002 (2002).
Kurzawski, M., Pawlik, A., Safranow, K., Herczynska, M. & Drozdzik, M. 677C>T and 1298A>C MTHFR polymorphisms affect methotrexate treatment outcome in rheumatoid arthritis. Pharmacogenomics 8, 1551–1559. https://doi.org/10.2217/14622416.8.11.1551 (2007).
Xiao, H. et al. Associations between the genetic polymorphisms of MTHFR and outcomes of methotrexate treatment in rheumatoid arthritis. Clin. Exp. Rheumatol. 28, 728–733 (2010).
Berkani, L. M. et al. Association of MTHFR C677T and A1298C gene polymorphisms with methotrexate efficiency and toxicity in Algerian rheumatoid arthritis patients. Heliyon 3, e00467. https://doi.org/10.1016/j.heliyon.2017.e00467 (2017).
van Ede, A. E., Laan, R. F., Blom, H. J., De Abreu, R. A. & van de Putte, L. B. Methotrexate in rheumatoid arthritis: An update with focus on mechanisms involved in toxicity. Semin. Arthritis Rheum. 27, 277–292. https://doi.org/10.1016/s0049-0172(98)80049-8 (1998).
van Ede, A. E., Laan, R. F., Blom, H. J., De Abreu, R. A. & van de Putte, L. B. Seminars in Arthritis and Rheumatism 277–292 (Elsevier, 2023).
Kato, T., Hamada, A., Mori, S. & Saito, H. Genetic polymorphisms in metabolic and cellular transport pathway of methotrexate impact clinical outcome of methotrexate monotherapy in Japanese patients with rheumatoid arthritis. Drug Metab. Pharm. 27, 192–199. https://doi.org/10.2133/dmpk.dmpk-11-rg-066 (2012).
Wessels, J. A. et al. Efficacy and toxicity of methotrexate in early rheumatoid arthritis are associated with single-nucleotide polymorphisms in genes coding for folate pathway enzymes. Arthritis Rheum. 54, 1087–1095. https://doi.org/10.1002/art.21726 (2006).
van Ede, A. E. et al. The C677T mutation in the methylenetetrahydrofolate reductase gene: A genetic risk factor for methotrexate-related elevation of liver enzymes in rheumatoid arthritis patients. Arthritis Rheum. 44, 2525–2530. https://doi.org/10.1002/1529-0131(200111)44:11%3c2525::aid-art432%3e3.0.co;2-b (2001).
Hoekstra, M. et al. Factors associated with toxicity, final dose, and efficacy of methotrexate in patients with rheumatoid arthritis. Ann. Rheum. Dis. 62, 423–426. https://doi.org/10.1136/ard.62.5.423 (2003).
Anderson, J. J., Wells, G., Verhoeven, A. C. & Felson, D. T. Factors predicting response to treatment in rheumatoid arthritis: The importance of disease duration. Arthritis Rheum. 43, 22–29. https://doi.org/10.1002/1529-0131(200001)43:1%3c22::aid-anr4%3e3.0.co;2-9 (2000).
Kumagai, K., Hiyama, K., Oyama, T., Maeda, H. & Kohno, N. Polymorphisms in the thymidylate synthase and methylenetetrahydrofolate reductase genes and sensitivity to the low-dose methotrexate therapy in patients with rheumatoid arthritis. Int. J. Mol. Med. 11, 593–600 (2003).
Bagheri-Hosseinabadi, Z., Imani, D., Yousefi, H. & Abbasifard, M. MTHFR gene polymorphisms and susceptibility to rheumatoid arthritis: A meta-analysis based on 16 studies. Clin. Rheumatol. 39, 2267–2279. https://doi.org/10.1007/s10067-020-05031-5 (2020).
Pincus, T. Assessment of long-term outcomes of rheumatoid arthritis: How choices of measures and study designs may lead to apparently different conclusions. Rheum. Dis. Clin. N. Am. 21, 619–654 (1995).
Albert, D. A. et al. Criteria for improvement in rheumatoid arthritis: alternatives to the American College of Rheumatology 20. The Journal of rheumatology 31, 856–866 (2004).
Pincus, T. & Stein, C. M. ACR 20: Clinical or statistical significance?. Arthritis Rheum. 42, 1572–1576. https://doi.org/10.1002/1529-0131(199908)42:8%3c1572::aid-anr2%3e3.0.co;2-g (1999).
van Riel, P. L., van Gestel, A. M. & van de Putte, L. B. Development and validation of response criteria in rheumatoid arthritis: Steps towards an international consensus on prognostic markers. Br. J. Rheumatol. 35(Suppl 2), 4–7. https://doi.org/10.1093/rheumatology/35.suppl_2.4 (1996).
Felson, D. T. Whither the ACR20?. J. Rheumatol. 31, 835–837 (2004).
Baqer, M. S. & Mohammed, M. M. Evaluation of the anti-inflammatory effect of curcumin as an additive therapy to meloxicam in management of knee osteoarthritis. Int. J. Drug Deliv. Technol. 12, 310–315. https://doi.org/10.25258/ijddt.12.1.57 (2022).
Hoekstra, M., van de Laar, M. A., Bernelot Moens, H. J., Kruijsen, M. W. & Haagsma, C. J. Longterm observational study of methotrexate use in a Dutch cohort of 1022 patients with rheumatoid arthritis. J. Rheumatol. 30, 2325–2329 (2003).
Maetzel, A. et al. Meta-analysis of treatment termination rates among rheumatoid arthritis patients receiving disease-modifying anti-rheumatic drugs. Rheumatology 39, 975–981. https://doi.org/10.1093/rheumatology/39.9.975 (2000).
Funding
The affiliated authors funded the project.
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. [Qassim Mahdi Mutlak], and [Ali Abdulhussain Kasim] performed material preparation, data collection, and analysis. The first draft of the manuscript was written by [Qassim Mahdi Mutlak], and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Mutlak, Q.M., Kasim, A.A. Impact of MTHFR gene polymorphism on the outcome of methotrexate treatment in a sample of Iraqi rheumatoid arthritis patients. Sci Rep 14, 15119 (2024). https://doi.org/10.1038/s41598-024-65199-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-65199-7
- Springer Nature Limited