Abstract
The incidence of vulvar carcinoma varies by race; however, it is a rare disease, and its genomic profiles remain largely unknown. This study examined the characteristics of vulvar squamous cell carcinoma (VSCC) in Japanese patients, focusing on genomic profiles and potential racial disparities. The study included two Japanese groups: the National Cancer Center Hospital (NCCH) group comprised 19 patients diagnosed between 2015 and 2023, and the Center for Cancer Genomics and Advanced Therapeutics group comprised 29 patients diagnosed between 2019 and 2022. Somatic mutations were identified by targeted or panel sequencing, and TP53 was identified as the most common mutation (52–81%), followed by HRAS (7–26%), CDKN2A (21–24%), and PIK3CA (5–10%). The mutation frequencies, except for TP53, were similar to those of Caucasian cohorts. In the NCCH group, 16 patients of HPV-independent tumors were identified by immunohistochemistry and genotyping. Univariate analysis revealed that TP53-mutated patients were associated with a poor prognosis (log-rank test, P = 0.089). Japanese VSCC mutations resembled those of Caucasian vulvar carcinomas, and TP53 mutations predicted prognosis regardless of ethnicity. The present findings suggest potential molecular-targeted therapies for select VSCC patients.
Similar content being viewed by others
Introduction
The incidence of vulvar carcinoma in Japan is 0.2–0.4 per 100,000 women per year1. By contrast, the incidence of vulvar carcinoma is as high as 2.5 per 100,000 women in the USA2. A higher prevalence has been reported in South Africa and certain European countries such as Germany, whereas the incidence is low in Western Asia and the Middle East3,4. Vulvar squamous cell carcinoma (VSCC) was traditionally considered as a disease affecting postmenopausal women. However, its incidence is increasing among younger people5, which is attributed to an increase in the rate of human papillomavirus (HPV) infection5,6. HPV is responsible for 22–40% of vulvar carcinomas worldwide7. When limited to squamous cell carcinoma, the prevalence of HPV varies from 0 to 86% in different regions, with a pooled prevalence of 39.7%8. A study in Japan reported 76% non-HPV VSCC, which may suggest a low prevalence of HPV9. A disparity in the prevalence of squamous cell carcinoma between East Asian and Western countries has been identified. Studies in Japan1,9 and Korea10 reported that approximately 50–60% of vulvar carcinomas are squamous cell carcinomas, which is a lower proportion than that reported by studies in Western countries (75–90%)7. These findings suggest that the characteristics of VSCC vary by race and region. However, because of the rarity of vulvar carcinoma and the limited number of Asian population-based studies, the clinicopathological and genomic profiles of VSCC in Asian populations remain unexplored.
The prognosis of VSCC is relatively good for localized disease; however, patients with advanced or recurrent disease have a dismal prognosis. Shin et al. reported a 5-year survival rate of 65.1% for patients with squamous cell carcinoma in Korea10, which is comparable with the rates reported in Europe (69.9% in the Netherlands11 and 69.9% in the UK12). Advances in surgical techniques and chemoradiotherapy have improved the chance of survival for patients with localized disease. However, the prognosis for patients with groin recurrence, distant recurrence, or advanced VSCC has not improved in the past decades, with a reported 5-year survival rate of 25–50%13. Clear guidelines for the treatment of recurrent VSCC are lacking14, and elucidating the genomic mutational profiles of VSCC is essential to identify potential therapeutic targets.
Recent advances in next-generation sequencing (NGS) have improved our understanding of the characteristics of this rare cancer. Genes that are frequently mutated in VSCC include TP53 (35–62%), PIK3CA (9–35%), HRAS (6–14%), and CDKN2A (11–38%), although the prevalence of these alterations varies among studies15,16,17,18,19,20,21,22,23,24. Most studies are based on analysis of Western VSCC patients; therefore, genomic analyses focusing on Asian VSCC patients are needed.
This study investigated the genomic mutational profiles of Japanese patients with VSCC to determine the differences in potential somatic mutations among races and identify somatic mutations related to therapeutic targets or prognosis.
Results
Patient characteristics
Nineteen VSCC patients who met the inclusion criteria were enrolled in the NCCH group. The patient characteristics are summarized in Table 1. The age indicates the age at diagnosis. Most of the samples were HPV-independent tumors (84.2%). In this group, all patients with HPV negative tumors had early stage disease. Half of the patients had early-stage disease at the time of diagnosis, whereas the other half had advanced-stage disease. Among the 19 patients, eight experienced recurrence (42.1%) and five died (26.3%).
The clinical characteristics of the 29 patients in the C-CAT group are also summarized in Table 1. The age also indicates the age at diagnosis. Because C-CAT collects data from patients with advanced or progressive disease, most cases were relapsed (96%).
Of the 19 cases, p53 IHC was performed in 16. All three HPV-associated tumors exhibited wild-type staining patterns. In comparison, in 13 cases considered HPV-independent tumors, diffuse strong positive staining was observed in eight cases, and a wild-type pattern was observed in five cases (Supplementary Table 1).
Among 19 cases, three were classified as HPV-associated tumors based on p16 block-type positivity, and 13 were classified as HPV-independent. Regarding preinvasive lesions, classic vulvar intraepithelial neoplasia (VIN) 3 was observed in three cases of HPV-associated tumors. In comparison, differentiated VIN was observed in nine cases of HPV-independent tumors, and differentiated exophytic vulvar intraepithelial lesions (DE-VIL) were observed in two cases. Lymphovascular invasion was observed in 12 of 18 cases. Lymph node dissection was performed in 16 of the 19 cases, with a median number of excised lymph nodes of 14 (range, 1–56). Lymph node metastasis was observed in 10 of 17 evaluable cases, with three cases showing extracapsular invasion of the lymph nodes. Surgical margins were evaluated in 18 primary tumor resection cases, excluding one lymph node metastasis resection, with 13 cases showing negative margins, five positive horizontal margins, and one positive vertical margin (Supplementary Table 1).
Genomic alterations in VSCC detected in the two groups
In the NCCH group, mutation patterns were observed in all HPV-independent tumors, whereas no mutations were found in HPV-dependent tumors (Fig. 1). The gene with the highest frequency of somatic mutations was TP53 (81%), followed by HRAS (26%), CDKN2A (21%), and PIK3CA (5%) (Fig. 1 and Supplementary Table 2). The rate of TP53 mutations in recurrent cases was remarkably high (89%). The candidate somatic mutations in the two groups are listed in Supplementary Tables 2 and 3. In both groups, most TP53 mutations occurred at the DNA-binding domain (DBD), a 191-amino-acid protease-resistant fragment (residues 102–292) containing hotspot mutations at R175, R248, R273, and R282. Most TP53 mutations are located in the DBD25, and the mutation profile was similar to those previously reported for VSCC.
In the C-CAT group, TP53 was also the most frequently mutated gene (52%), followed by CDKN2A (24%), PIK3CA (10%), and HRAS (7%) (Fig. 2 and Supplementary Table 3). In the C-CAT and NCCH groups, TP53 mutations were detected in > 50% of patients, showing a trend toward a higher mutation proportion than previously reported (Table 2). The frequency and type of mutations in other genes were similar to those observed in Western populations. Regarding copy number alterations, PIK3CA, SOX2, BLC6, and MAP3K13 amplification was detected in three patients (10%) and CCND1 alterations were observed in two patients (7%). CDKN2A loss was detected in two different cases (7%).
The gene mutation frequencies from previously reported genomic analyses of VSCC are summarized in Table 2. The most commonly observed genomic alterations in these studies were in TP53 (35–62%), PIK3CA (9–35%), CDKN2A (11–38%), and HRAS (6–14%).
TP53 mutation as a prognostic factor
In the NCCH group, the frequency of TP53 mutations was 8/9 in patients with recurrent disease and 5/9 in patients without recurrent disease. TP53 mutation correlated with a worse prognosis regarding relapse-free survival (log-rank test, P = 0.089) (Fig. 3A), and this association was also observed in patients without HPV infection (log-rank test, P = 0.090) (Fig. 3B). However, did not correlate with worse overall survival in the NCCH and C-CAT groups (log-rank test, P = 0.45, P = 0.13) (Fig. S3).
Correlation between TP53 gene mutational status and recurrence-free survival in the NCCH group. (A) Kaplan–Meier survival curves showing the clinical outcomes of 19 patients. (B) Kaplan–Meier survival curves show the clinical outcomes of 16 HPV-independent patients. TP53 wt = TP 53 wild-type; TP53 mt = TP53 mutated; HPV neg/TP53 wt = HPV-independent/TP53 wild-type; HPV neg/TP53 mt = HPV-independent/TP53 mutated.
Discussion
In this study, we investigated the genomic mutational profiles of 48 Japanese VSCC patients and compared them with those of Caucasian patients with VSCC data obtained from previous genomic analyses. The mutational pattern was comparable between Japanese and Caucasian VSCC patients. TP53 was the most frequently mutated gene, followed by HRAS, CDKN2A, and PIK3CA. TP53 mutation correlated with clinical outcome, which supports previous studies identifying TP53 mutation as a prognostic factor.
The frequency of common gene mutations in the present groups did not deviate significantly from that reported previously, except for TP53 mutations, which were observed more frequently. TP53, a crucial tumor suppressor gene, is the most frequently mutated gene in human cancers26. Corey et al. detected TP53 mutations in approximately 80% of VSCCs without HPV infection24, which is consistent with the present findings. Racial disparity regarding the frequency of TP53 mutations in VSCC may be due to differences in the prevalence of HPV infection. Kang et al.7 suggested that there is geographical variation in HPV prevalence in vulvar carcinoma. The HPV infection rate in the present group was 16%, whereas that reported previously is approximately 40%8. Therefore, the higher frequency of TP53 mutations in this group may be due to the low proportion of HPV-associated VSCC, which is characterized by a low frequency of TP53 mutation. The present findings suggest that genetic factors alone do not account for the regional variation in the epidemiology of vulvar carcinoma.
TP53 mutation is significantly associated with advanced disease and a poor prognosis for various cancer types27,28. In this group, the recurrence rate was notably higher in cases with TP53 mutations. Although the number of VSCC patients analyzed was not sufficient to establish TP53 alteration as a poor prognostic factor, the correlation between TP53 alterations and clinical outcomes in VSCC was examined in previous studies (Table 3)18,19,20,21,22. In previous studies, TP53 alterations were detected used Sanger sequencing, targeted panel sequencing, and IHC. Nooij et al.18 reported worse recurrence-free survival in an HPV negative/p53 mutated group (P = 0.044) when comparing three groups based on TP53 mutational status and HPV status (i.e., HPV positive, HPV negative/p53 wildtype, and HPV negative/p53 mutated groups). Kortekaas et al.19 and Lérias et al.20 also showed significant differences in recurrence-free survival (P < 0.001) and rate of recurrence (P = 0.0005) according to TP53 mutation status. By contrast, Tessier-Cloutier et al.22 showed that TP53 mutation alone was not an independent prognostic factor (P = 0.23 for OS and P = 0.35 for PFS), whereas TP53 and PIK3CA double mutation was associated with a poor prognosis (P = 0.0016 for OS and P = 0.034 for PFS). Choschzick et al.21 did not identify TP53 as an independent prognostic factor (P > 0.05 for cumulative survival) when comparing three groups with different TP53 mutation and HPV status. In brief, although TP53 mutation correlates with worse prognosis in several studies19,20,21, other studies report opposite findings22,23. TP53 mutation may be a poor prognostic factor for VSCC, although further studies are needed to confirm the role of TP53 mutation as a critical predictor of worse clinical outcomes.
In VSCC, TP53 was the most frequently mutated gene, followed by HRAS, CDKN2A, and PIK3CA, regardless of ethnicity. TP53 mutations, which are present in many cancer types, are considered targets for novel cancer therapies28,29,30. A recent study by Guiley31 reported compounds that directly target mutant p53; however, further studies will be required to determine whether these compounds are therapeutically effective. HRAS, a member of the RAS family of genes, is also known as the mouse sarcoma virus and is associated with human cancer. Tipifarnib is a farnesyltransferase inhibitor that suppresses HRAS function and is used as a targeted therapy for HRAS-mutated tumors26,32, 33. Ho et al. investigated the efficacy of tipifarnib in head and neck squamous cell carcinoma patients with a mutation in HRAS34, reporting an objective response rate of 55%. Loss of the tumor suppressor gene CDKN2A, which encodes p16 and p14, is a frequent occurrence in VSCC. Hsu et al. reported that CDK4/6 inhibitors are a breakthrough therapy for a subset of sarcomas35. Therapeutic strategies targeting CDKN2A loss hold great potential for the treatment of various cancer types including VSCC36. Pharmacological inhibition of the p16 targets CDK4/6 is a prime example of such a strategy37, although additional studies are needed prior to its clinical application. BYL-719 is a PIK3CA inhibitor with low drug-drug interactions, suggesting its potential for combinatorial therapy of recurrent/advanced cervical cancer38. Alpelisib (BYL-719) plus fulvestrant was recently approved for the treatment of ER-positive breast cancer patients who develop resistance to ER-targeted therapy39. An expansion in the indications for molecular-targeted drugs for VSCC is expected.
This study had several limitations. The number of patients was limited because of the rarity of this cancer. Some clinical data in the C-CAT group were not available, and further validation studies are needed. Additionally, because genomic testing requires a certain amount of tissue, early-stage disease cases and small tumors were not included in the analysis, which may have led to selection bias in the NCCH group.
In conclusion, we found similarities in the somatic mutation profiles of VSCC between Japanese and Caucasian populations. This suggests that the genetic etiology of VSCC does not differ significantly according to ethnic background. Analysis of the correlation between TP53 mutation status and clinical outcomes confirmed previous findings identifying TP53 mutation as a prognostic factor. Molecular-targeted therapies may be indicated for selected VSCC patients based on somatic mutational profiles. This study, which included a specific number of Asian patients, will contribute to our understanding of this less investigated yet clinically crucial disease.
Materials and methods
Characteristics of 48 patients with VSCC (divided into two groups)
The clinical data used in this study were obtained retrospectively from patient records. The study included 48 Japanese patients with primary VSCC who were divided into two independent groups. The decision trees for patient selection are provided in Fig. S1. Forty-two vulvar carcinoma patients were treated at the National Cancer Center Hospital (NCCH) between January 2015 and January 2023. The tumors were diagnosed pathologically according to the 2021 International Federation of Gynecology and Obstetrics criteria, and classified according to the World Health Organization classification of tumors. Of the 42 patients, 23 were excluded due to the histological type or insufficient material; therefore, 19 patients were eligible for inclusion in the NCCH group. Among the 91 patients who were diagnosed between 2019 and 2022 and registered at the Center for Cancer Genomics and Advanced Therapeutics (C-CAT) databank (ver. 20221218), 30 patients with vaginal cancer and 32 patients with other histological types were excluded. Ultimately, 29 patients with VSCC were included in the C-CAT group. In this study, there was no overlap between patients in the NCCH group and those in the C-CAT group. C-CAT is a national data center for cancer genomic medicine in Japan that was established in 201940. Comprehensive genomic profiling tests used were FoundationOne® CDx and OncoGuide™ NCC Oncopanel. As of June 30, 2022, C-CAT has aggregated 36,340 data points of clinical/genomic information, and is open for sharing with academic institutions and industries.
This study was performed in accordance with the Declaration of Helsinki and approved by the institutional review board of the National Cancer Center Research Institute (approval numbers 2017-136, 2020-067) and the C-CAT Data Utilization Review Board (approval number CDU2021-001N). All participants provided written informed consent.
Immunohistochemistry (IHC) of p16
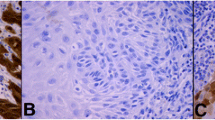
IHC was performed using p16 as a surrogate marker to identify HPV-associated tumors. H&E (Fig. S2A and C) and IHC staining were performed on representative whole sections of tumor from formalin-fixed, paraffin-embedded (FFPE) specimens, and the paraffin blocks were sectioned at a thickness of 4 μm. IHC was performed using a Ventana Benchmark Ultra automated immunostainer (Tucson, AZ, USA) and a mouse monoclonal antibody specific for p16 (E6H4, prediluted, Roche diagnostics) according to standard protocols and with appropriate positive and negative controls. A case was classified as an HPV-associated tumor only if it showed block-positive p16 staining, with diffuse nuclear or nuclear and cytoplasmic positivity (Fig. S2B). Negative results (Fig. S2D) were defined as total absence of staining or weak, focal, and discontinuous staining, which indicated HPV-independent tumors.
The results of p53 immunohistochemical staining analyses were available for 16 out of 19 cases. Staining was performed using a p53 monoclonal antibody (DO7, prediluted, Agilent/Dako, Glostrup, Denmark) in all cases.
DNA preparation and targeted sequencing
FFPE tissue blocks from a VSCC sample were retrieved and serially sectioned at 10 μm thickness on slides. The slides were deparaffinized, and the H&E-stained sections were used for macrodissection. Genomic DNA was extracted from 19 VSCC tissue samples using the QIAamp DNA FFPE tissue kit according to the manufacturer's instructions (Qiagen, Hilden, Germany). Genomic DNA (50 ng) purified from tumor tissues was used for library construction using the Ion AmpliSeq™ Cancer Hotspot Panel v2 (Thermo Fisher Scientific, Waltham, MA, USA). Sequencing was performed on the Ion Proton platform (Thermo Fisher Scientific). For quality control, samples with a mean read depth of coverage > 1000 and a base quality score of 20 (with < 1% probability of being incorrect), which accounted for 90% of total reads, were selected41,42,43,44.
Data were analyzed using Torrent Suite Software v5.0.4 (Thermo Fisher Scientific). First, somatic mutations were selected using the following criteria: (i) variant allele frequency of somatic mutations in tumor tissues > 4%; (ii) removal of single nucleotide polymorphisms if they showed a threshold allele frequency of ≥ 0.01 in either the NHLBI GO Exome Sequencing Project (ESP6500) (http://evs.gs.washington.edu/EVS/) or the Japanese Multi Omics Reference Panel (jMorp, 20210907) (https://jmorp.megabank.tohoku.ac.jp/ijgvd/)45; and (iii) registration of mutations as “pathogenic/likely pathogenic variants” in ClinVar46 or “oncogenic/likely oncogenic mutations” in OncoKB9. Finally, the selected variants were analyzed by manual inspection using the Integrative Genomics Viewer (IGV; http://www.broadinstitute.org/igv/)47.
HPV genotyping by Sanger sequencing
The 19 samples were subjected to HPV genotyping. Genomic DNA (10 ng) was amplified by PCR targeting two distinct HPV genomic regions using TaKaRa Taq DNA polymerase (Takara Bio Inc., Shiga, Japan). The PCR reaction was performed as described previously41. Sanger sequencing was performed using an ABI 3130xl DNA Sequencer (Applied Biosystems). The similarity between the obtained sequences and various HPV genotypes in the GenBank database was determined using the Basic Local Alignment Search Tool (BLAST) (https://blast.ncbi.nlm.nih.gov/Blast.cgi).
Statistical analysis
Statistical analysis was performed using EZR version 1.61 (Saitama Medical Center, Jichi Medical University, Saitama, Japan), a graphical user interface for R (The R Foundation for Statistical Computing, Vienna, Austria)48. The level of statistical significance was set at P < 0.05. Cumulative survival was estimated using the Kaplan–Meier method, and differences in survival between groups were analyzed using the log-rank test.
Data availability
The C-CAT dataset used in this study was obtained from the C-CAT database (https://www.ncc.go.jp/jp/c_cat/use/index.html). The datasets used and analyzed during this study are available from the corresponding author upon reasonable request.
References
Tanaka, Y. et al. Trends in incidence and long-term survival of Japanese women with vulvar cancer: A population-based analysis. Int. J. Clin. Oncol. 24, 1137–1142 (2019).
Miller, B. A., Chu, K. C., Hankey, B. F. & Ries, L. A. Cancer incidence and mortality patterns among specific Asian and Pacific Islander populations in the U.S. Cancer Causes Control. 19, 227–256 (2008).
Bucchi, L., Pizzato, M., Rosso, S. & Ferretti, S. New insights into the epidemiology of vulvar cancer: Systematic literature review for an update of incidence and risk factors. Cancers 14, 389 (2022).
Bray, F., Laversanne, M., Weiderpass, E. & Arbyn, M. Geographic and temporal variations in the incidence of vulvar and vaginal cancers. Int. J. Cancer. 147, 2764–2771 (2020).
Olawaiye, A. B., Cuello, M. A. & Rogers, L. J. Cancer of the vulva: 2021 update. Int. J. Gynaecol. Obstet. 155(Suppl 1), 7–18 (2021).
Xiao, X. et al. Vulvar cancer in China: Epidemiological features and risk analysis. J. Cancer. 8, 2950–2958 (2017).
Kang, Y. J. et al. Vulvar cancer in high-income countries: Increasing burden of disease. Int. J. Cancer. 141, 2174–2186 (2017).
Faber, M. T. et al. Prevalence and type distribution of human papillomavirus in squamous cell carcinoma and intraepithelial neoplasia of the vulva. Int. J. Cancer. 141, 1161–1169 (2017).
Osakabe, M. et al. Characteristics of vulvar squamous cell carcinoma in Japanese women. Pathol. Int. 57, 322–327 (2007).
Shin, D. W., Bae, J., Ha, J., Lee, W. M. & Jung, K. W. Trends in incidence and survival of patients with vulvar cancer in an Asian country: Analysis of the Korean Central Cancer Registry 1999–2018. Gynecol. Oncol. 164, 386–392 (2022).
Schuurman, M. S. et al. Trends in incidence and survival of Dutch women with vulvar squamous cell carcinoma. Eur. J. Cancer. 49, 3872–3880 (2013).
Lai, J. et al. Vulval cancer incidence, mortality and survival in England: Age-related trends. BJOG. 121, 728–738 (2014) (discussion 739).
Nooij, L. S. et al. Risk factors and treatment for recurrent vulvar squamous cell carcinoma. Crit. Rev. Oncol. Hematol. 106, 1–13 (2016).
Koh, W. J. et al. Vulvar cancer, Version 1.2017, NCCN clinical practice guidelines in oncology. J. Natl. Compr. Cancer Netw. 15, 92–120 (2017).
Zieba, S. et al. Somatic mutation profiling of vulvar cancer: Exploring therapeutic targets. Gynecol. Oncol. 150, 552–561 (2018).
Salama, A. M., Momeni-Boroujeni, A., Vanderbilt, C., Ladanyi, M. & Soslow, R. Molecular landscape of vulvovaginal squamous cell carcinoma: New insights into molecular mechanisms of HPV-associated and HPV-independent squamous cell carcinoma. Mod. Pathol. 35, 274–282 (2022).
Weberpals, J. I. et al. Vulvar squamous cell carcinoma (VSCC) as two diseases: HPV status identifies distinct mutational profiles including oncogenic fibroblast growth factor receptor 3. Clin. Cancer Res. 23, 4501–4510 (2017).
Carreras-Dieguez, N. et al. Molecular landscape of vulvar squamous cell carcinoma. Int. J. Mol. Sci. 22, 7069 (2021).
Nooij, L. S. et al. Genomic characterization of vulvar (pre)cancers identifies distinct molecular subtypes with prognostic significance. Clin. Cancer Res. 23, 6781–6789 (2017).
Kortekaas, K. E. et al. Vulvar cancer subclassification by HPV and p53 status results in three clinically distinct subtypes. Gynecol. Oncol. 159, 649–656 (2020).
Lerias, S. et al. CD274 (PD-L1), CDKN2A (p16), TP53, and EGFR immunohistochemical profile in primary, recurrent and metastatic vulvar cancer. Mod. Pathol. 33, 893–904 (2020).
Choschzick, M. et al. Role of TP53 mutations in vulvar carcinomas. Int. J. Gynecol. Pathol. 30, 497–504 (2011).
Tessier-Cloutier, B. et al. Molecular characterization of invasive and in situ squamous neoplasia of the vulva and implications for morphologic diagnosis and outcome. Mod. Pathol. 34, 508–518 (2021).
Corey, L. et al. The genomic landscape of vulvar squamous cell carcinoma. Int. J. Gynecol. Pathol. 42, 515–522 (2023).
Kandoth, C. et al. Mutational landscape and significance across 12 major cancer types. Nature. 502, 333–339 (2013).
Gilardi, M. et al. Tipifarnib as a precision therapy for HRAS-mutant head and neck squamous cell carcinomas. Mol. Cancer Ther. 19, 1784–1796 (2020).
Wang, X. & Sun, Q. TP53 mutations, expression and interaction networks in human cancers. Oncotarget. 8, 624–643 (2017).
Chen, X. et al. Mutant p53 in cancer: From molecular mechanism to therapeutic modulation. Cell Death Dis. 13, 974 (2022).
Brosh, R. & Rotter, V. When mutants gain new powers: News from the mutant p53 field. Nat. Rev. Cancer. 9, 701–713 (2009).
Silwal-Pandit, L. et al. TP53 mutation spectrum in breast cancer is subtype specific and has distinct prognostic relevance. Clin. Cancer Res. 20, 3569–3580 (2014).
Guiley, K. Z. & Shokat, K. M. A small molecule reacts with the p53 somatic mutant Y220C to rescue wild-type thermal stability. Cancer Discov. 9, 56–69 (2023).
Untch, B. R. et al. Tipifarnib inhibits HRAS-driven dedifferentiated thyroid cancers. Cancer Res. 78, 4642–4657 (2018).
Odeniyide, P. et al. Targeting farnesylation as a novel therapeutic approach in HRAS-mutant rhabdomyosarcoma. Oncogene. 41, 2973–2983 (2022).
Ho, A. L. et al. Tipifarnib in head and neck squamous cell carcinoma with HRAS mutations. J. Clin. Oncol. 39, 1856–1864 (2021).
Hsu, J. Y., Seligson, N. D., Hays, J. L., Miles, W. O. & Chen, J. L. Clinical utility of CDK4/6 inhibitors in sarcoma: Successes and future challenges. JCO Precis. Oncol. 6, e2100211 (2022).
Peguero, J. et al. Tissue/site-agnostic study of ribociclib for tumors with cyclin D-CDK4/6 pathway genomic alterations: A phase II, open-label, single-arm basket study. JCO Precis. Oncol. 3, 1–10 (2019).
Goel, S., Bergholz, J. S. & Zhao, J. J. Targeting CDK4 and CDK6 in cancer. Nat. Rev. Cancer. 22, 356–372 (2022).
Bogani, G. et al. BYL719 (alpelisib) for the treatment of PIK3CA-mutated, recurrent/advanced cervical cancer. Tumori. 109, 244–248 (2023).
Boyd, D. C. et al. Discovering synergistic compounds with BYL-719 in PI3K overactivated basal-like PDXs. Cancers 15, 1582 (2023).
Kohno, T. et al. C-CAT: The national datacenter for cancer genomic medicine in Japan. Cancer Discov. 12, 2509–2515 (2022).
Hiranuma, K. et al. Rare FGFR fusion genes in cervical cancer and transcriptome-based subgrouping of patients with a poor prognosis. Cancer Med. 12, 17835–17848 (2023).
Murakami, N. et al. Distribution of genetic alterations in high-risk early-stage cervical cancer patients treated with postoperative radiation therapy. Sci. Rep. 11, 10567 (2021).
Kuno, I. et al. TP53 mutants and non-HPV16/18 genotypes are poor prognostic factors for concurrent chemoradiotherapy in locally advanced cervical cancer. Sci. Rep. 11, 19261 (2021).
Hirose, S. et al. Genomic alterations in STK11 can predict clinical outcomes in cervical cancer patients. Gynecol. Oncol. 156, 203–210 (2020).
Tadaka, S. et al. jMorp updates in 2020: Large enhancement of multi-omics data resources on the general Japanese population. Nucleic Acids Res. 49, D536–D544 (2021).
Landrum, M. J. et al. ClinVar: Improving access to variant interpretations and supporting evidence. Nucleic Acids Res. 46, D1062–D1067 (2018).
Robinson, J. T., Thorvaldsdottir, H., Wenger, A. M., Zehir, A. & Mesirov, J. P. Variant review with the integrative genomics viewer. Cancer Res. 77, e31–e34 (2017).
Kanda, Y. Investigation of the freely available easy-to-use software “EZR” for medical statistics. Bone Marrow Transplant. 48, 452–458 (2013).
Acknowledgements
The authors thank Hitoshi Ichikawa, Maiko Matsuda, Yoko Shimada, Sachiyo Mitani, and other physicians and staff members at the National Cancer Center Hospital for their assistance and support. We thank Bioedit Ltd. for assistance with English language editing.
Funding
This work was supported by a Grant-in-Aid for Young Scientists (B) Number 20K18207, a Grant-in-Aid for Scientific Research (C) Number 20K09636, 19K16572 and 22K09563, a Grant-in-Aid for Scientific Research (B) Number 20H03695, BRIDGE (programs for bridging the gap between R&D and the ideal society (Society 5.0), and the National Cancer Center Research and Development Fund (2023-J-2, NCC Biobank, and NCC Core Facility).
Author information
Authors and Affiliations
Contributions
EF and SK interpreted the data, performed the experiments, and wrote the manuscript. EF, MKK, MI, KT, HY, and TK diagnosed and treated the patients, and provided clinical information. SK and HY contributed to concept development and the study design, and edited the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Fujii, E., Kato, M.K., Yamaguchi, M. et al. Genomic profiles of Japanese patients with vulvar squamous cell carcinoma. Sci Rep 14, 13058 (2024). https://doi.org/10.1038/s41598-024-63913-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-63913-z
- Springer Nature Limited