Abstract
The FZP gene plays a critical role in the formation of lateral branches and spikelets in rice panicle architecture. This study investigates the qSBN7 allele, a hypomorphic variant of FZP, and its influence on panicle architectures in different genetic backgrounds. We evaluated two backcross inbred lines (BILs), BC5_TCS10sbn and BC3_TCS10sbn, each possessing the homozygous qSBN7 allele but demonstrating differing degrees of spikelet degeneration. Our analysis revealed that BC5_TCS10sbn had markedly low FZP expression, which corresponded with an increase in axillary branches and severe spikelet degeneration. Conversely, BC3_TCS10sbn exhibited significantly elevated FZP expression, leading to fewer secondary and tertiary branches, and consequently decreased spikelet degeneration. Compared to BC5_TCS10sbn, BC3_TCS10sbn carries three additional chromosomal substitution segments from its donor parent, IR65598-112-2. All three segments significantly enhance the expression of FZP and reduce the occurrence of tertiary branch and spikelet degeneration. These findings enhance our understanding of the mechanisms regulating FZP and aid rice breeding efforts.
Similar content being viewed by others
Introduction
The number of spikelets per panicle is a crucial factor in determining rice grain yield. This number varies depending on various components of a rice panicle, such as panicle length, the number of primary, secondary, and tertiary branches, and the number of lateral and terminal spikelets. These variations result from subtle changes in panicle architecture during development. During the reproductive stage, the apical meristem of the rice shoot evolves into the rachis meristem, which is responsible for producing axillary meristems that develop into primary branches. Subsequently, the basal meristem of these primary branches becomes secondary branches, while the upper lateral and terminal meristems directly form spikelet meristems. All meristems, including the lateral and terminal meristems of the secondary branches, develop into spikelets. Some cultivars with high spikelet numbers may develop tertiary branches from the basal meristem of secondary branches. Eventually, meristems of these tertiary branches also transform into spikelets1,2. Researchers have identified several genes associated with panicle architecture, such as APO1, APO2, FZP, Gn1a, IPA1/WFP, LAX1, LAX2, RCN1, RCN2 and TAW13,4,5,6,7,8,9,10,11,12. In addition to panicle architecture, spikelet degeneration is another factor affecting the number of spikelets per panicle. Spikelet degeneration results from genetic defects in specific genes, including ASA, DPS1, FZP, OsALMT7, PAA3, TSD1, TUTOU1 and TUTOU25,13,14,15,16,17,18,19, as well as from resource-limited conditions such as shading, drought, and heat stress20,21,22,23.
Our previous study identified a beneficial, hypomorphic allele of FZP under the qSBN7 quantitative trait loci (QTL), which increases the number of spikelets per panicle and grain yield24. FZP encodes an AP2/ERF transcription factor, regulates the balance between branches and spikelet meristem formation5,25,26,27,28. A nonfunctional FZP variant leads to an exceptionally high number of secondary and higher-order branches, resulting in severe spikelet degeneration5. By contrast, several FZP hypomorphic alleles increase the number of secondary branches and spikelets without apparent degeneration, thus demonstrating their value for the breeding of elite rice varieties24,29,30. Nevertheless, introgression of the same FZP hypomorphic allele (qSBN7) into the indica rice variety TCS10 resulted in severe panicle degeneration and aborted spikelets24. The current study aims to gain further insights into the impact of FZP on panicle architecture under different genetic backgrounds.
Results
Phenotypic characterization of the BC5_TCS10sbn
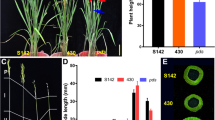
BC5_TCS10sbn, a BC5 backcross inbred line (BIL) which was developed using the IR65598-112-2 rice variety as the donor parent, and TCS10 as the recurrent parent. To elucidate the phenotypic characteristics of BC5_TCS10sbn, we compared its agronomic traits during the vegetative and reproductive stages with TCS10. The plant morphology of BC5_TCS10sbn closely resembled that of TCS10 (Fig. 1a); however, the panicle of BC5_TCS10sbn exhibited severe degeneration (Fig. 1b and Supplementary Fig. S1). We observed no significant difference in the primary branch number per panicle between BC5_TCS10sbn and TCS10 (Fig. 1c). Nevertheless, BC5_TCS10sbn displayed higher number of secondary and tertiary branches per panicle (Fig. 1d,e). Compared with TCS10, the panicles of BC5_TCS10sbn bore only 66 normal spikelets and exhibited severe degeneration in spikelets (Fig. 1f–h).
Phenotypic characterization of TCS10 and BC5_TCS10sbn. (a) Overall plant morphology of TCS10 and BC5_TCS10sbn. Scale bar = 20 cm. (b) Structure of panicles from TCS10 and BC5_TCS10sbn. Scale bar = 4 cm. (c–h) Comparison of number of primary, secondary, and tertiary branches and number of normal and degenerated spikelets per main panicle, including the ratio of degenerated spikelets for TCS10 and BC5_TCS10sbn. Values in (c–h) are means ± SD. Student’s t test was used to examine p values. ** indicates significance at 1% level; ns denotes not significant.
To determine the timing and location of degeneration, we investigated the development of panicles from 1 to 22 cm. In TCS10 panicles, spikelets were observed to form and rapidly elongate from 3 to 22 cm (Fig. 2a,c,e–i). Conversely, in BC5_TCS10sbn panicles, more than half of the spikelets were replaced by numerous continuous bract-like structures (Fig. 2l). The remaining spikelets in BC5_TCS10sbn demonstrated a gradual delay in development from 3 to 15 cm (Fig. 2b,d–h). In BC5_TCS10sbn panicles, only a few spikelets positioned at the basal or the apical of secondary and tertiary branches rapidly elongated from 15 to 22 cm and developed into survival spikelets after heading (Fig. 2j,k and Supplementary Fig. S1). These findings indicated that the panicle architecture of BC5_TCS10sbn closely resembled that of severe fzp mutants (fzp-3, fzp-14 and abp1) identified in previously studies. The rice panicle structure observed in TCS10 and BC5_TCS10sbn is illustrated in Fig. 3.
Panicle morphology of TCS10 and BC5_TCS10sbn at different developmental stages. (a–j) Panicle morphology of TCS10 and BC5_TCS10sbn from 1–22 cm. Scale bar = 1.5 cm. (k) Normal spikelets (green arrows),degenerated spikelets (white arrows) and continuous bract-like structures (blue arrows) were observed in the 22 cm panicle of BC5_TCS10sbn. Scale bar = 1.0 cm. (l) Continuous bract-like structures (blue arrows) were observed in the 22 cm panicle of BC5_TCS10sbn. Scale bar = 1.0 cm.
Development of BC3_TCS10sbn line with slightly degenerated panicle
qSBN7 allele was introgressed from a new plant type variety, IR65598-112-2, which presents high spikelet number without significant degradation. In order to determine the genetic factors responsible for the differences in degeneration between BC5_TCS10sbn and IR65598-112-2, a BC3-derived BIL (BC3_TCS10sbn) with slightly degraded panicles from the same parents as BC5_TCS10sbn were developed (Fig. 4a,b). Unlike BC5_TCS10sbn, BC3_TCS10sbn’s panicles harbored almost three times the number of normal spikelets, ranging between 190 and 220 (Fig. 4a,c). Furthermore, we evaluated five yield components between TCS10 and BC3_TCS10sbn. BC3_TCS10sbn exhibited a significant increase in secondary branches and normal spikelets per panicle, while displaying a decrease in 1000-grain weight compared to TCS10 (Supplementary Figure S2). However, no significant difference in the percentage of filled grains and panicle weight was observed between two lines (Supplementary Figure S2).
Phenotypic characterization and FZP expression in BC3_TCS10sbn and BC5_TCS10sbn. (a) Panicle morphology of two backcross inbred lines (BILs). Scale bar = 4 cm. (b) Plant morphology of two BILs. Scale bar = 20 cm. (c) Number of normal spikelets per main panicle in the two BILs across two crop seasons. (d, e) Graphical genotypes of the two BILs. White squares: homozygous chromosomal segments of TCS10; black squares: homozygous chromosomal segment of IR65598-112–2. (f) Transcript levels of FZP in TCS10, BC5_TCS10sbn and BC3_TCS10sbn at the 1-mm panicle stage.
To determine the genetic differences between BC5_TCS10sbn and BC3_TCS10sbn, we conducted a whole-genome survey of IR65598-112-2, TCS10, BC5_TCS10sbn, and BC3_TCS10sbn using next-generation sequencing. Our results indicated that BC5_TCS10sbn only carried an introgression segment (28.30–29.69 Mb) from IR65598-112-2 on chromosome 7, with approximately 99.7% of the recurrent parent genome recovered (Fig. 4d). Conversely, BC3_TCS10sbn had three substituted segments from IR65598-112-2, specifically IG2 (10.33–29.32 Mb) on chromosome 2, IG4 (0–21.21 Mb) on chromosome 4, and IG7 (9.08–29.69 Mb) on chromosome 7, resulting in an 84.1% recovery of BC3_TCS10sbn (Fig. 4e). This suggests that the genes contributing to the phenotypic differences between the two BILs likely reside within these segments.
Evaluation of the causal factors leading to phenotypic differences between BC5_TCS10sbn and BC3_TCS10sbn
There are two potential explanations for the variation between two BILs. One explanation is that BC3_TCS10sbn contains genes that control FZP and suppress the growth of secondary and tertiary branches. The other explanation is that BC3_TCS10sbn carries genes that enhance photosynthesis or carbohydrate translocation, leading to an increase in the number of survival spikelets. To understand which explanation is more realistic, we conducted quantitative PCR (qPCR) to assess the transcript levels of FZP in TCS10, BC5_TCS10sbn, and BC3_TCS10sbn at the 1-mm panicle stage. Our findings reveal that BC5_TCS10sbn exhibited a 98.2% decrease in FZP expression compared with TCS10 (Fig. 4f), whereas BC3_TCS10sbn exhibited intermediate FZP expression levels, distinct from TCS10 and BC5_TCS10sbn (Fig. 4f). A further comparison of panicle characteristics between BC5_TCS10sbn and BC3_TCS10sbn revealed that BC3_TCS10sbn had more normal spikelets and primary branches than BC5_TCS10sbn, but fewer secondary branches, tertiary branches, and the ratio of degenerated spikelet (Table 1). This indicates that BC3_TCS10sbn might possess genes that enhance FZP expression, thereby limiting axillary differentiation and the occurrence of degenerate spikelets.
We also assessed whether source-related traits varied between BC5_TCS10sbn and BC3_TCS10sbn. A comparative analysis of various traits, including culm diameter, flag leaf length, flag leaf width, and chlorophyll content of the flag leaf, revealed a significant difference only in culm diameter between the two BILs (Table 1). This finding suggests that BC3_TCS10sbn may possess genes influencing culm diameter, which may influence carbohydrate allocation and contribute to the production of more regular spikelets. Thus, increased in FZP expression and changed in culm diameter are likely to jointly contribute to the reduced number of degenerate spikelets in BC3_TCS10sbn.
Detection of genomic regions associated with degenerated spikelets traits between two BILs
To identify genomic regions related to degenerated spikelet traits, we developed three chromosome segment substitution lines (CSSLs). Each line contained homozygous substituted segments from IR65598-112-2. We analyzed these lines for genomic regions related to degenerated spikelet traits and performed qPCR to examine FZP transcript levels in BC5_TCS10sbn, BC3_TCS10sbn, and the three CSSLs. Our results demonstrated that each CSSL exhibited an increased number of normal spikelets, fewer tertiary branches, and a lower ratio of degenerated spikelets compared with BC5_TCS10sbn (Fig. 5a–i). Additionally, FZP expression was higher in all three CSSLs than in BC5_TCS10sbn (Fig. 5k). This expression exhibited a strong positive correlation with the number of normal spikelets per panicle and a negative correlation with the ratio of degenerated spikelets. These results suggest that the substituted segments from IR65598-112-2 in each CSSL could interact with FZP to repress the formation of tertiary branches. Furthermore, CSSL2 and CSSL7 exhibited significantly larger culm diameters in BC3_TCS10sbn (Fig. 5j). This observation indicates that CSSL2 and CSSL7 contain QTLs capable of increasing culm diameter, which in turn may influence the degree of spikelet degeneration.
Panicle characterization and FZP expression levels in BC5_TCS10sbn, BC3_TCS10sbn, and three CSSLs (chromosome segment substitution lines). (a–e) Panicle morphology of BC5_TCS10sbn, CSSL2, CSSL4, CSSL7 and BC3_TCS10sbn. Scale bar = 4 cm. (f–j) Comparative analyses of BC5_TCS10sbn, BC3_TCS10sbn, and the three CSSLs in terms of normal spikelet number, secondary branch number, tertiary branch number per main panicle, ratio of degenerated spikelets, and culm diameter. (k) FZP transcript levels in BC5_TCS10sbn, BC3_TCS10sb, and three CSSLs. ns indicates no significant difference; *, ** indicates significant differences compared with BC5_TCS10sbn at α = 0.05, 0.01, respectively, based on the Dunnett’s multiple range test.
Discussion
FZP is an essential gene that regulates rice panicle architecture. To date, researchers have identified 21 FZP alleles in rice5,24,25,26,27,28,29,30,31,32,33,34,35. In general, null alleles in FZP mutants lead to the formation of higher-order rachis branches rather than spikelets, leading to an absence of normal spikelets5,25,31,32,35. Severe hypomorphic mutants of FZP exhibit an increased number of secondary and tertiary panicle branches and produce partially normal spikelets due to their partial functionality. However, these mutants often have various degenerated or defective spikelets5,26,27,28. The use of such alleles in rice breeding is impractical due to these drawbacks. Conversely, mild hypomorphic alleles of FZP provide a desirable balance between spikelet and branch development, enhancing both without causing degeneration24,29,30. Fujishiro et al.30 used a backcrossing strategy to introduce the qSrn7/FZP (a mild hypomorphic variant of FZP) from Kasalath into the Koshihikari cultivar, resulting in a 40–60% increase in yield. Wang et al.24 improved the yield of rice variety, TN13 by 10.9% through marker-assisted backcrossing strategy to introduce the qSBN7 (a mild hypomorphic variant of FZP) from IR65598-112-2. These findings underscore the value of utilizing these mild hypomorphic alleles of FZP in developing new elite cultivars.
Our previous study highlighted that the qSBN7 allele displays distinct panicle architectures in different genetic backgrounds24. In the japonica genetic backgrounds of NIL_TN13sbn, BSI325, IR76904-7-19, and IR65598-112-2, qSBN7 exhibits a phenotype resembling that of mild hypomorphic mutants. However, in the indica genetic background of BC5_TCS10sbn, it showed a phenotype analogous to severe hypomorphic mutants24. This difference in phenotype is likely attributed to specific gene–gene interactions of qSBN7 in these different genetic backgrounds. Thus, understanding the underlying mechanisms of spikelet degeneration in BC5_TCS10sbn is pivotal for the effective application of the qSBN7 allele in a broad spectrum of rice breeding programs.
The present study successfully identified three genomic regions from IR65598-112-2, namely IG2, IG4, and IG7, that can effectively reduce the occurrence of degenerated spikelets in BC3_TCS10sbn. Individually, IG2, IG4, and IG7 reduced the ratio of degenerated spikelets by 32.3%, 17.1%, and 15.7%, respectively (Fig. 5). When combined, these segments resulted in a remarkable 68.1% reduction in degenerated spikelets (Fig. 5). Our comprehensive analysis indicates that each segment harbored QTLs that regulated FZP expression, consequently diminishing the number of tertiary branches and the ratio of degenerated spikelets. Previous studies have established a strong correlation between FZP expression levels, the number of secondary branches, and the occurrence of degenerated spikelets24. In the genetic background of TN13, an increase in secondary branches and spikelets without significant spikelet degeneration, was observed when FZP expression reached over 40% of the wild-type level. However, pronounced spikelet degeneration occurred when FZP expression fell below 30% of the wild-type level24. In this study, BC5_TCS10sbn exhibited FZP expression levels of approximately 1.8% of the wild-type (TCS10) (Fig. 4f), which was associated with severe spikelet degeneration (Fig. 1). Conversely, BC3_TCS10sbn, which had less spikelet degeneration, exhibited FZP expression levels at approximately 39% of TCS10 (Fig. 4). Additionally, the study observed a significant increased in FZP expression and a decreased in the number of tertiary branches in IG2, IG4, and IG7 compared with BC5_TCS10sbn (Fig. 5). Therefore, these observations suggest that these genomic regions harbored QTLs that influenced FZP expression. Elevated FZP expression in BC3_TCS10sbn appears to reduce the number of tertiary branches, ultimately decreasing the number of degenerated spikelets. In previous studies, two genes, OsBZR1 and OsARF6, have been identified as regulators of FZP expression29,33. Comparing their physical map positions on the rice chromosome, only OsBZR1 is within the IG2 segment. Nonetheless, no synonymous substitutions were detected in the coding region of OsBZR1 between TCS10 and IR65598-112-2, suggesting other mechanisms may be at play in modulating FZP expression through these chromosomal segments.
This study observed a strong correlation between FZP expression levels and reduced spikelet degeneration. However, factors related to the efficiency of carbohydrate biosynthesis, distribution, and transfer might also contribute to the decrease in spikelet degeneration in BC3_TCS10sbn. In BC3_TCS10sbn, CSSL2, and CSSL7, we observed a significantly wider panicle culm compared with BC5_TCS10sbn (Fig. 5j). Studies have indicated that wider culms are often associated with higher carbohydrate transfer efficiency, and plants with wider culms typically have a higher number of grains per panicle36,37,38,39,40. Although we observed a negative correlation between culm diameter and spikelet degeneration, varieties with wider culm diameters exhibited lower levels of spikelet degeneration. Whether there is a causal relationship between culm diameter and spikelet degeneration remains to be uncovered.
The number of grains per panicle is the most crucial factor in determining rice yield. To increase grain yield, breeders have been selecting rice varieties with more secondary branches and grain numbers. In this study, BC3_TCS10sbn was found to have a significant increase in normal spikelets per panicle compared to TCS10. However, there was no significant increase in panicle weight in BC3_TCS10sbn compared to TCS10 (Supplementary Figure S2). The expression level of FZP in rice can affect spikelet number per panicle, but an increase in spikelet number per panicle does not necessarily lead to an increase in final rice yield. When spikelet number per panicle is increased in rice, it often leads to trade-offs among yield components7,8,24,29. Our previous study showed that the NIL_TN13sbn carrying qSBN7 increased spikelet number per panicle but reduced 1000-grain weight and fertility rate24. In the present study, the BC3_TCS10sbn carrying qSBN7 only reduced 1000-grain weight compared to TCS10, while there was no significant difference in fertility rate (Supplementary Figure S2). It is worth noting that varieties with high grain numbers often exhibit lower fertility rates, which may be related to ethylene biosynthesis41,42. The qSBN7 exhibits different trade-off characteristics in different varieties, whether this phenomenon is related to ethylene regulation under different genetic backgrounds needs to be further confirmed.
Our study reveals that the combination of three minor chromosomal substitution segments significantly influenced panicle architecture differences between the two BILs. However, even with the pyramiding of these three segments in BC3_TCS10sbn, we still observed a 10–40% rate of spikelet degeneration or replacement by continuous bract-like structures (Fig. 5i and Table 1). This suggests the presence of additional QTL differences between IR65598-112-2 and BC3_TCS10sbn that may affect spikelet degeneration. In future studies, we aim to conduct fine mapping and RNA-seq analysis to further uncover why qSBN7 causes spikelet degeneration in BC5_TCS10sbn. This will contribute to the widespread application of qSBN7 in rice breeding programs to enhance grain yield.
Materials and methods
Plant materials
Through backcross breeding, we developed a BC5-derived BIL (BC5_TCS10sbn), which have the homozygous qSBN7 allele of IR65598-112-2 cultivars, in the genetic background of indica cultivar TCS1024 (previously named NIL_TCS10sbn). To obtain a BIL with reduced spikelet degeneration, a BC3F2 population derived from the same recurrent and donor parent was developed. Three BC3F2 individuals with minor panicle degeneration were selected and self-pollinated twice to produce three BC3F4 BILs. After visual evaluation, we chose one BIL, BC3_TCS10sbn, which carries the homozygous qSBN7 alleles of IR65598-112-2 and exhibits slight panicle degeneration, for further experiments.
To develop three CSSLs, we crossed the BC3_TCS10sbn with BC5_TCS10sbn. Three CSSLs, CSSL2 (with a homozygous introgression segment on chromosome 2), CSSL4 (with a homozygous introgression segment on chromosome 4), and CSSL7 (with a homozygous introgression segment on chromosome 7), were developed by screening 260 BC4F2 plants using seven Kompetitive Allele Specific PCR (KASP) markers (Supplementary Table S1) located at the introgression segments of IR65598-112-2 in BC3_TCS10sbn. The KASP genotyping master mix was supplied by LGC Genomics (Middlesex, UK). The KASP analysis was carried out according to the manufacturer’s protocol. The breeding process for our plant materials is detailed in Supplementary Fig. S3.
Growing conditions and phenotypic evaluation
In the current study, fifty-two plants from each line were grown in paddy fields in Chia-Yi, Taiwan (23°42’N, 120°28’E), with a spacing of 20 cm between plants and 30 cm between rows. The rice plants were cultivated in a well-irrigated paddy field according to conventional management practices. Each field received 160 kg ha−1 of nitrogen fertilizer.
To assess the yield components of TCS10 and BC3_TCS10sbn, each line was cultivated with three replications. At maturity, several traits were evaluated, such as the number of secondary branches per panicle, the number of normal spikelets per panicle, the percentage of filled grains, 1000-grain weight, and panicle weight. Six individuals from each line were randomly selected for measurement. Differences between the lines were analyzed using Student's t test.
For phenotypic assessment of panicle-related and source-related traits, we selected sixteen individuals from the middle of a row for each line. We measured panicle-related traits on the main panicles, identified as the tallest tillers of each plant. Panicle-related traits were measured by manually counting. In the present study, we categorized all observed degenerated organs, including degenerated spikelets and bract-like structures on branches, as degenerated spikelets. The number of degenerated spikelets and the ratio of degenerated spikelets of the panicle were analyzed using the following formulae.
The number of degenerated spikelets = Number of degenerated spikelets + Number of continuous bract-like structures.
The ratio of degenerated spikelets = 100×(Number of degenerated spikelets + Number of continuous bract-like structures)/(Number of normal spikelets + Number of degenerated spikelets + Number of continuous bract-like structures).
The diameter of the rachis base of these main panicles served as the measure for culm diameter. We evaluated chlorophyll content using a Soil Plant Analysis Development chlorophyll meter (Konica–Minolta, Osaka, Japan), adhering to the instructions in its operation manual.
Whole genome sequencing and analysis
To investigate the genetic backgrounds of BC3_TCS10sbn and BC5_TCS10sbn, the whole genome DNA libraries of TCS10, IR65598-112-2, BC3_TCS10sbn, and BC5_TCS10sbn were constructed and sequenced as 150-bp pair-end reads at Genomics (Taipei, Taiwan). Around 10 Gb of raw reads were obtained each on an Illumina NovaSeq 6000 platform. Paired-end reads were trimmed to remove adapters and low-quality bases (< 20) and filtered for reads < 100-bp using Trimmomatic 0.3643. Filtered short reads were then mapped to the Nipponbare reference genome sequences IRGSP1.044 using BWA-MEM45. Single nucleotide polymorphisms (SNPs) were called for the entire data set of TCS10, IR65598-112–2, BC3_TCS10sbn, and BC5_TCS10sbn using the mpileup command of SAMtools46. SNPs were filtered using the filter command of bcftools46 according to the following criteria: quality score (QUAL) > 30.0 and depth (DP) > 20 across all samples.
RNA isolation and quantitative PCR analysis
For RNA extraction, we harvested inflorescences at the 1 mm stage of development. Three inflorescences from a single plant were collected as one biological replicate. We extracted total RNA using the RNeasy Plant Mini Kit (QIAGEN, Valencia, CA) with RNase-free DNase I (QIAGEN, Valencia, CA). To evaluate the expression level of FZP, we synthesized complementary DNA (cDNA) from 0.6 μg of total RNA using the iScrip cDNA Synthesis Kit (Bio-Rad, Hercules, CA). The transcriptional levels of FZP were quantified through qPCR analysis. Each assay incorporated three biological and three technical replicates. The qPCR was performed on a CFX96 Connect Real-Time PCR detection system (Bio-Rad). We selected the rice ubiquitin gene UBQ5 (LOC_Os01g22490) as the internal control. The primers and probe are described in a previous study24.
Data availability
The raw DNA sequence data is available in the NCBI Sequence Read Archive with accession number PRJNA1086197.
References
Ikeda, K., Sunohara, H. & Nagato, Y. Developmental course of inflorescence and spikelet in rice. Breed. Sci. 54, 147–156 (2004).
Itoh, J. et al. Rice plant development: from zygote to spikelet. Plant Cell Physiol. 46, 23–47 (2005).
Ikeda, K., Ito, M., Nagasawa, N., Kyozuka, J. & Nagato, Y. Rice ABERRANT PANICLE ORGANIZATION 1, encoding an F-box protein, regulates meristem fate. Plant J. 51, 1030–1040 (2007).
Ikeda-Kawakatsu, K., Maekawa, M., Izawa, T., Itoh, J. & Nagato, Y. ABERRANT PANICLE ORGANIZATION 2/RFL, the rice ortholog of Arabidopsis LEAFY, suppresses the transition from inflorescence meristem to floral meristem through interaction with APO1. Plant J. 69, 168–180 (2012).
Komatsu, M., Chujo, A., Nagato, Y., Shimamoto, K. & Kyozuka, J. FRIZZY PANICLE is required to prevent the formation of axillary meristems and to establish floral meristem identity in rice spikelets. Development 130, 3841–3850 (2003).
Ashikari, M. et al. Cytokinin oxidase regulates rice grain production. Science 309, 741–745 (2005).
Jiao, Y. et al. Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat. Genet. 42, 541–544 (2010).
Miura, K. et al. OsSPL14 promotes panicle branching and higher grain productivity in rice. Nat. Genet. 42, 545–549 (2010).
Komatsu, M., Maekawa, M., Shimamoto, K. & Kyozuka, J. The LAX1 and FRIZZY PANICLE 2 genes determine the inflorescence architecture of rice by controlling rachis-branch and spikelet development. Dev. Biol. 231, 364–373 (2001).
Tabuchi, H. et al. LAX PANICLE2 of rice encodes a novel nuclear protein and regulates the formation of axillary meristems. Plant Cell 23, 3276–3287 (2011).
Nakagawa, M., Shimamoto, K. & Kyozuka, J. Overexpression of RCN1 and RCN2, rice TERMINAL FLOWER 1/CENTRORADIALIS homologs, confers delay of phase transition and altered panicle morphology in rice. Plant J. 29, 743–750 (2002).
Yoshida, A. et al. TAWAWA1, a regulator of rice inflorescence architecture, functions through the suppression of meristem phase transition. Proc. Natl. Acad. Sci. 110, 767–772 (2013).
Zhou, D. et al. APICAL SPIKELET ABORTION (ASA) controls apical panicle development in rice by regulating salicylic acid biosynthesis. Front. Plant Sci. 12, 636877. https://doi.org/10.3389/fpls.2021.636877 (2021).
Zafar, S. A. et al. DEGENERATED PANICLE AND PARTIAL STERILITY 1 (DPS1) encodes a cystathionine β-synthase domain containing protein required for anther cuticle and panicle development in rice. New Phytol. 225, 356–375 (2020).
Heng, Y. et al. OsALMT7 maintains panicle size and grain yield in rice by mediating malate transport. Plant Cell 30, 889–906 (2018).
Yang, F. et al. Panicle apical abortion 3 controls panicle development and seed size in rice. Rice 14, 68 (2021).
Cai, Z. et al. Thermo-sensitive spikelet defects 1 acclimatizes rice spikelet initiation and development to high temperature. Plant Physiol. 191, 1684–1701 (2023).
Bai, J. et al. Rice TUTOU1 encodes a suppressor of cAMP receptor-like protein that is important for actin organization and panicle development. Plant Physiol. 169, 1179–1191 (2015).
Zhu, Z. C., Luo, S., Lei, B., Li, X. Y. & Cheng, Z. J. Locus TUTOU2 determines the panicle apical abortion phenotype of rice (Oryza sativa L.) in tutou2 mutant. J. Integr. Agric. 21, 621–630 (2022).
Saini, H. S. & Westgate, M. E. Reproductive development in grain crops during drought. Adv. Agron. 68, 59–96 (1999).
Yao, Y., Yamamoto, Y., Yoshida, T., Nitta, Y. & Miyazaki, A. Response of differentiated and degenerated spikelets to top-dressing, shading and day/night temperature treatments in rice cultivars with large panicles. Soil Sci. Plant Nutr. 46, 631–641 (2000).
Wang, Z. Q., Zhang, W. Y. & Yang, J. C. Physiological mechanism underlying spikelet degeneration in rice. J. of integr. Agric. 17, 1475–1481 (2018).
Ali, A., Xu, P., Riaz, A. & Wu, X. Current advances in molecular mechanisms and physiological basis of panicle degeneration in rice. Int. J. Mol. Sci. 20, 1613. https://doi.org/10.3390/ijms20071613 (2019).
Wang, S. S., Chung, C. L., Chen, K. Y. & Chen, R. K. A novel variation in the FRIZZLE PANICLE (FZP) gene promoter improves grain number and yield in rice. Genetics 215, 243–252 (2020).
Bai, X. et al. Regulatory role of FZP in the determination of panicle branching and spikelet formation in rice. Sci. Rep. 6, 19022. https://doi.org/10.1038/srep19022 (2016).
Wang, Y. et al. Synergistic roles of LAX1 and FZP in the development of rice sterile lemma. Crop J. 8, 16–25 (2020).
Dai, D. et al. Panicle apical abortion 7 regulates panicle development in rice (Oryza sativa L.). Int. J. Mol. Sci. 23, 9487. https://doi.org/10.3390/ijms23169487 (2022).
Wang, W., Chen, W. & Wang, J. FRIZZLE PANICLE (FZP) regulates rice spikelets development through modulating cytokinin metabolism. BMC Plant Biol. 23, 650. https://doi.org/10.1186/s12870-023-04671-4 (2023).
Bai, X. et al. Duplicationof an upstream silencer of FZP increases grain yield in rice. Nat. Plants 3, 885–893 (2017).
Fujishiro, Y. et al. Comprehensive panicle phenotyping reveals that qSrn7/FZP influences higher-order branching. Sci. Rep. 8, 12511. https://doi.org/10.1038/s41598-018-30395-9 (2018).
Yi, G. et al. Morphological and molecular characterization of a new frizzy panicle mutant, “fzp-9(t)”, in rice (Oryza sativa L.). Hereditas 142, 92–97 (2005).
Kato, T. & Horibata, A. A novel frameshift mutant allele, fzp-10, affecting the panicle architecture of rice. Euphytica 184, 65–72 (2012).
Huang, Y. et al. Variation in the regulatory region of FZP causes increases in secondary inflorescence branching and grain yield in rice domestication. Plant J. 96, 716–733 (2018).
Ren, D. et al. FZP determines grain size and sterile lemma fate in rice. J. Exp. Bot. 69, 4853–4866 (2018).
Chen, H. et al. Identification of a novel mutant allele, fzp-15, involved in panicle branch pattern of rice (Oryza sativa). Plant Breed. 140, 595–602 (2021).
Ookawa, T. et al. New approach for rice improvement using a pleiotropic QTL gene for lodging resistance and yield. Nat. Commun. 1, 132. https://doi.org/10.1038/ncomms1132 (2010).
Yano, K. et al. Isolation of a novel lodging resistance QTL gene involved in strigolactone signaling and its pyramiding with a QTL gene involved in another mechanism. Mol. Plant 8, 303–314 (2015).
Ookawa, T. et al. Pyramiding of multiple strong-culm genes originating from indica and tropical japonica to the temperate japonica rice. Sci. Rep. 12, 15400. https://doi.org/10.1038/s41598-022-19768-3 (2022).
Yang, X. et al. Isolation of a novel QTL, qSCM4, associated with strong culm affects lodging resistance and panicle branch number in Rice. Int. J. Mol. Sci. 24, 812. https://doi.org/10.3390/ijms24010812 (2023).
Chigira, K., Yamasaki, M., Adachi, S., Nagano, A. J. & Ookawa, T. Identification of novel quantitative trait loci for culm thickness of rice derived from strong-culm landrace in Japan. Omachi. Rice 16, 4 (2023).
Panigrahi, R., Kariali, E., Panda, B. B., Lafarge, T. & Mohapatra, P. K. Controlling the trade-off between spikelet number and grain filling: The hierarchy of starch synthesis in spikelets of rice panicle in relation to hormone dynamics. Funct. Plant Biol. 46, 507–523 (2019).
Panigrahi, S., Kariali, E., Dash, S. K., Sahu, B. B. & Mohapatra, P. K. Ethylene sensitivity underscores the yield advantage of high-grain numbers in cylinder-shaped rice panicles. Environ. Exp. Bot. 214, 105466 (2023).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Kawahara, Y. et al. Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice 6, 4 (2013).
Li, H. & Durbin, R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26, 589–595 (2010).
Li, H. A Statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27, 2987–2993 (2011).
Acknowledgements
This manuscript was edited by Wallace Academic Editing.
Funding
This work was supported by Tainan District Agricultural Research and Extension Station, Ministry of Agriculture, Taiwan (108AS-7.6.3-NS-N2).
Author information
Authors and Affiliations
Contributions
S-SW and K-YC designed research; R-KC performed field experiments; S-SW, P-HT and S-FC conducted phenotyping, genotyping and the data analysis; S-SW and K-YC wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Wang, SS., Tsai, PH., Cheng, SF. et al. Identification of genomic regions controlling spikelet degeneration under FRIZZLE PANICLE (FZP) defect genetic background in rice. Sci Rep 14, 12451 (2024). https://doi.org/10.1038/s41598-024-63362-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-63362-8
- Springer Nature Limited