Abstract
Improved phenotyping in pneumonia is necessary to strengthen risk assessment. Via a feasible and multidimensional approach with basic parameters, we aimed to evaluate the effect of host response at admission on severity stratification in COVID-19 and community-acquired pneumonia (CAP). Three COVID-19 and one CAP multicenter cohorts including hospitalized patients were recruited. Three easily available variables reflecting different pathophysiologic mechanisms—immune, inflammation, and respiratory—were selected (absolute lymphocyte count [ALC], C-reactive protein [CRP] and, SpO2/FiO2). In-hospital mortality and intensive care unit (ICU) admission were analyzed as outcomes. A multivariable, penalized maximum likelihood logistic regression was performed with ALC (< 724 lymphocytes/mm3), CRP (> 60 mg/L), and, SpO2/FiO2 (< 450). A total of 1452, 1222 and 462 patients were included in the three COVID-19 and 1292 in the CAP cohort for the analysis. Mortality ranged between 4 and 32% (0 to 3 abnormal biomarkers) and 0–9% in SARS-CoV-2 pneumonia and CAP, respectively. In the first COVID-19 cohort, adjusted for age and sex, we observed an increased odds ratio for in-hospital mortality in COVID-19 with elevated biomarkers altered (OR 1.8, 3, and 6.3 with 1, 2, and 3 abnormal biomarkers, respectively). The model had an AUROC of 0.83. Comparable findings were found for ICU admission, with an AUROC of 0.76. These results were confirmed in the other COVID-19 cohorts Similar OR trends were reported in the CAP cohort; however, results were not statistically significant. Assessing the host response via accessible biomarkers is a simple and rapidly applicable approach for pneumonia.
Similar content being viewed by others
Introduction
The COVID-19 pandemic excessively burdened health systems worldwide, affecting millions of people. The scientific community, in response, shared an objective to develop models that would recognize disease subtypes or endotypes in patients with varying susceptibility to determine clinical evolution1. To date, several sophisticated omics-based studies have been done to provide undeniably informative knowledge about both the disease’s complex pathogenic mechanisms and the intricate host response2,3,4. Furthermore, multiple scores incorporating many clinical variables have been proposed; however, most require an on-line calculator5. The difficulty in stratifying patient subtypes depends on causal microorganism factors, such as SARS-CoV-2 or others; the heterogeneity of the host; and the eventual outcome of the interplay occurring between the aforementioned elements.
In pneumonia, improved phenotyping considering host response has been encouraged. Innate and adaptive responses could provide insights that contribute to individualized severity assessment and management6. In host response, there are three main basic components to determine, two of which are common in all infections—immune cells and inflammatory response—and the third is related to the lung—gas exchange function. From a very practical perspective, these components are measurable by quantifiable biomarkers/parameters that reflect a specific pathophysiologic area: absolute lymphocyte count (ALC), C-reactive protein (CRP), and peripheral blood oxygen saturation (SpO2)/fraction of inspired oxygen (FiO2). Indeed, lymphopenia was consistently reported in severe episodes during the pandemic. Similarly, in community-acquired pneumonia (CAP), lymphopenia at admission has been shown to double the risk of mortality7,8. CRP is one of the most frequent acute phase reactants available in the emergency department (ED) in hospitals and has also been studied in SARS-CoV-2 pneumonia. Oxygen saturation that indicates respiratory failure is considered one of the strongest predictors of deterioration9.
We hypothesized that a simple and feasible multidimensional approach that would require blood analyses and oxygen saturation, and include assessing three physiologic host response areas—inflammation, immune, and functional—could offer useful information during an initial evaluation of patients with either SARS-CoV-2 pneumonia or possibly CAP due to other causal microorganisms. In that case, patients could be classifiable per risk of mortality and clinical deterioration regardless causal microorganism.
The aim of the study was to estimate the effect of a basic and rapidly applicable tool based on ALC, CRP, and SpO2/FiO2 in addressing patient stratification per mortality risk or disease progression. A secondary aim was to evaluate this tool in CAP not caused by SARS-CoV-2.
Methods
Design and patients
A retrospective and multicenter study was designed. The COVID-19 cohort included patients admitted across four Spanish hospitals (La Fe University and Polytechnic Hospital in Valencia; Cruces University Hospital in Barakaldo; Galdakao-Usansolo Hospital in Galdacano; and Hospital Clinic of Barcelona) between March and May 2020. Inclusion criteria comprised clinical symptoms and a confirmed microbiologic diagnosis of SARS-CoV-2 by reverse transcription polymerase chain reaction (RT-PCR) testing on nasopharyngeal swabs. Exclusion criteria included those patients transferred from other medical facilities and under the age of 18 years. This study was conducted in accordance with the amended Declaration of Helsinki. The study was approved by the Biomedical Research Ethics Committee of La Fe University and Polytechnic Hospital (2020-122-1). Written informed consent was waived given the non-interventional nature of the study.
A second multicenter cohort with COVID-19 patients was included (hereafter referred to as the COVID-19 2020 validation cohort). This cohort comprised patients from across eight hospitals in Galicia who were diagnosed and admitted to hospital between March 1st and April 24th 2020. Inclusion and exclusion criteria were the same as in the first cohort. The Ethics Committee of Galicia (Cod. 2020/239) approved the study for the validation cohort.
A third cohort with COVID-19 patients admitted between 2022 and 2023 was included (hereafter referred to as the COVID-19 2022–23 validation cohort). This cohort comprised patients from La Fe University and Polytechnic Hospital who were diagnosed and admitted to hospital between January 1st 2022 and December 31st 2023 when omicron variant was circulating and population was highly vaccinated. Inclusion and exclusion criteria were the same as in the first cohort.
A fourth cohort (hereafter referred to as the CAP cohort) was considered in order to assess the generalization of the biomarkers’ effect on patients without COVID-19 yet presenting pneumonia. The cohort included those patients admitted to the same Spanish hospitals for CAP before December 2019. Patients recruited for this cohort met inclusion criteria if they received a diagnosis for pneumonia based on a new radiologic infiltrate and the presence of at least two compatible clinical symptoms. Exclusion criteria included admission within the previous 15 days, nursing home residency, and immunosuppressed patients. The study was approved by the ethics committee of each hospital, and patients signed an informed consent (2013/0204).
Demographics, comorbidities, treatments, and laboratory data were collected at baseline (emergency department admission), including those variables of interest for evaluating host response areas at admission: ALC (immune); CRP (inflammatory); and SpO2/FiO2 (respiratory). The main outcome was in-hospital mortality due to any cause. The secondary outcome analyzed was intensive care unit (ICU) admission.
Statistical analysis
Patients were classified according to in-hospital mortality (alive or dead). Categorical variables under study were presented as absolute frequency and percentage, whilst quantitative variables as median and interquartile range (IQR). Differences between the two analysis groups were evaluated using the Wilcoxon rank-sum and Chi-square tests, as appropriate. Tables of association between biomarkers and in-hospital mortality were made, represented by absolute frequency, percentage and a 95% confidence interval (CI). There were 93.8% of cases with complete data, so multiple imputation was not performed.
A multivariable, penalized maximum likelihood logistic regression was performed10. Lymphocyte count, CRP, and SpO2/FiO2 were selected a priori as predictors of in-hospital mortality, as they had been identified as clinically relevant in the literature. A cut-off point was selected for each of the biomarkers based on former studies. The cut-off value for lymphocyte count was 724 cells/mm3. Patients with CAP presenting < 724 lymphocytes/mm3 at admission had been identified with having a higher risk of mortality8,11. Cut-off values for CRP and SpO2/FiO2 were 60 mg/L and 450, respectively. The latter is equivalent to an SpO2 < 94% in room air per the definition of severe COVID-19 by the Infectious Diseases Society of America (IDSA)12, and the cut-off point for CRP was selected from a prior study as we have found that < 60 mg/L was independently associated with low-risk for mortality13. Patients were grouped into four risk categories per the number of altered biomarkers in accordance with cut-off values (normal, or one, two or all abnormal markers). The estimate effect of each risk group was adjusted by sex and age, and represented as odds ratios (OR) and CI 95%. The reference was the normal marker group.
For the different biomarkers, survival curves of in-hospital mortality and ICU admission were estimated using the Kaplan–Meier method and compared by the log-rank test. All analyses were performed using STATA/IC 16.1 software (Stata Corporation, College Station, TX, USA).
Ethics approval and consent to participate
This study was conducted in accordance with the amended Declaration of Helsinki. The study of the first cohort was approved by the Biomedical Research Ethics Committee of La Fe University and Polytechnic Hospital (2020-122-1). Written informed consent was waived given the non-interventional nature of the study. The Ethics Committee of Galicia (Cod. 2020/239) approved the study for the validation cohort. The CAP cohort study was approved by the ethics committee of each participant hospital, and patients signed an informed consent (Biomedical Research Ethics Committee of La Fe University and Polytechnic Hospital 2013/0204).
Results
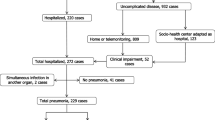
A total of 1548 patients were included in the COVID-19 cohort. Of these, 1452 (93.8%) had complete data for the selected variables (age, sex, ALC, CRP, and SpO2/FiO2) and main outcome under study (in-hospital mortality) (Fig. 1). Table 1 details baseline characteristics according to in-hospital mortality. Briefly, the patients who died were older, predominantly male, and with more comorbidities. In addition, they had lower levels of ALC and SpO2/FiO2, and higher levels of CRP. Supplementary Table 1, baseline characteristics are shown according to the number of markers (ALC, CRP, and SpO2/FiO2) altered.
A total of 1292 patients were included in the COVID-19 2020 validation cohort. Of these, 1222 had complete data for the selected variables (Supplementary Fig. 1). Supplementary Table 2 shows baseline characteristics of the validation cohort. Moreover, baseline characteristics according to the number of markers (ALC, CRP, and SpO2/FiO2) altered can be found in supplementary Table 3. A total of 462 patients were included in the COVID-19 2022–23 validation cohort.
A total of 1601 patients were included in the CAP cohort. Of these, 1292 had complete data for the selected variables and outcome under study (Supplementary Fig. 2). Supplementary Table 4 shows baseline characteristics according to in-hospital mortality. In addition, supplementary Table 5 shows baseline characteristics according to the number of markers (ALC, CRP, and SpO2/FiO2) altered.
Primary outcome: in-hospital mortality
In-hospital mortality was 14.8% (215 of 1452), 17.1% (221 of 1292), 13.6% (63 of 462), and 3.6% (47 of 1292) for the three COVID-19 (derivation, validation 2020 and 2022–23 validation cohorts, respectively) and CAP cohorts, respectively. Depending on whether all markers were normal or altered, mortality ranged from 4–32%, 4–40%, 0–32%, and 0–9% in the COVID-19, COVID-19 2020 validation, COVID-19 2022–23 validation and CAP cohorts, respectively (Table 2 and Fig. 2). The progressive increase in odds for in-hospital mortality in COVID-19 per the number of biomarkers altered was confirmed after we adjusted for age and sex (Table 2). The area under the receiver operating curve (AUROC) was 0.83 (0.80;0.86) and 0.83 (0.80;0.86) for the COVID-19 and the COVID-19 validation 2020 cohorts, respectively. Again, adding CURB65 to the model, similar results were found after adjusting for age and sex (supplementary Table 6). This analysis with CURB65 could not be performed in the validation COVID-19 cohorts due to the high number of missing data. To assess the influence of time since symptom onset, a subgroup analysis was performed according to whether the patients had had symptoms for less than 7 days or not (Table 3). This analysis showed a similar increase in odds of death according to the number of markers altered. The COVID-19 and CAP cohorts presented a comparable trend, with the latter presenting an AUROC of 0.75 (0.68;0.82) (Table 2). However, due to the low number of events, CIs were very wide and results were not statistically significant. As shown in Fig. 3, the number of altered markers allows various groups to already be differentiated within the first days per the risk of mortality.
A net reclassification analysis (NRI) for the COVID-19 derivation cohort was calculated to evaluate the effect of combining the CURB65 score with the 3 biomarkers model (Fig. 4).
Secondary outcome: ICU admission
ICU admission was necessary for 282 (19.4%), 232 (16.9%) and 89 (6.9%) patients in the COVID-19, COVID-19 validation and CAP cohorts, respectively. The proportion of ICU admissions rose in both cohorts as the number of altered markers grew (Table 4 and Fig. 5). After adjusting for age and sex, we observed that involvement of the immune, inflammatory and respiratory responses was related to increased odds for ICU admission in COVID-19 (Table 3). The AUROC was 0.76 (0.73;0.79) and 0.76 (0.73;0.79) for the COVID-19 and COVID-19 validation cohorts. Table 3 shows the logistic regression analysis for ICU admission with similar findings according to time since symptom onset. In the CAP cohort, we found a comparable trend for ICU admission to that of the COVID-19 cohort; AUROC was 0.76 (0.71;0.82) (Table 3). However, due to the low number of events, the CIs were very wide, and the results were not statistically significant. As with mortality, the number of altered markers differentiated the groups within the first days, according to the risk of ICU admission (Fig. 6).
Discussion
The main findings of the study were as follows: (i) three feasible-to-obtain parameters at pneumonia diagnosis –CRP > 60 mg/L, lymphocyte count < 724 cells/mm3 and, SpO2/FiO2 < 450– were able to stratify the risk of mortality into four groups; (ii) the mortality rate ranged between 4 and 32% (from 0 to 3 parameters altered) in COVID-19 and 0–9% in CAP; (iii) that simple tool showed usefulness in two different microbiological pneumonia scenario: COVID-19 and CAP although with worse performance in CAP.From the start of the COVID-19 pandemic and like in CAP, the search for useful prognostic scales in routine practice has generated great interest. Many studies have received wide acceptance whilst others have presented limitations14. Multiple scales and scores have shown their usefulness for short- and long-term mortality risk. However, none of them address the host's response with basic parameters, measured in emergency rooms, in such a simple and practical way and this represents a different personalized approach. Our study explored a basic and three-dimensional approach to host response that included performing a blood analysis that provided lymphocyte count and CRP (surrogate of inflammation) and measuring oxygen saturation (surrogate of lung function). This approach allowed for patient stratification into four subgroups of increasing severity similar to other strategies in different conditions and settings15. This information offers additional utility to well-known and wide used scores as CURB65 to discern risk more accurately because if the three biomarkers are abnormal, mortality increases despite a low CURB65 punctuation and vice versa. We selected easy parameters available in all hospitals and many facilities, and attained thresholds previously published in other studies. In CAP, a lymphopenia < 724 cells/mm3 had been reported to be associated with increased mortality after adjusting for CURB65 scale whilst, in COVID-19, lymphopenia was a constant finding in the most severe episodes7,8,16. If an infection is present, lymphopenia could indicate poor control of the pathogen. For example, it may lead to persistent hypercytokinemia in cases of CAP8. Although several biomarkers have been related to outcomes in COVID-19 and CAP17,18,19, CRP has been used in clinical scores and trials exploring the use of immunomodulators5,20,21. Similarly, oxygenation level—representing the main physiological function of the lung—was computed through SpO2/FiO2 ratio. It corresponded to an SpO2 94% at room air and which was widely used in many publications and trials for COVID-1922.
Our approach is easy to implement because it depends on only three parameters. The alteration of a single factor–inflammation, lymphocyte count or oxygenation level–approximately doubles the odds of death. When all three factors are altered, there is a more-than-sixfold increase in such risk. This information is valuable, as it can influence resource optimization, patient allocation and mortality risk calculations. Adjusted for age and sex, our analysis had an AUROC of 0.83 for the mortality risk. This is similar to the most widespread, and complex, COVID-19-specific scores: the 4C Mortality Score (8 items) with an AUROC of 0.79 for in-hospital mortality and the ISARIC 4C Deterioration model (11 items) for clinical deterioration5,23. CAP scores evaluated for COVID-19 showed AUROC of less than 0.8 in all cases as well24.
As time course is an important aspect in pathogenesis–the initial viral replication and inflammation phases–to consider for stratifying risk, our model was probed in both stages. It performed similarly within the first seven days (AUROC 0.74 [0.69;0.79]) since symptom onset and thereafter (AUROC 0.78 [0.74;0.81]). Our multidimensional approach was reproduced in both a different cohort of SARS-CoV-2 pneumonia and in patients with CAP.
There is no certainty about how new variants would modify the epidemiology of CAP and COVID-19, though SARS-CoV-2 became an endemic virus, like other coronaviruses (HCoV-229E, HCoV-OC43, HCoV-NL63, and HCoV-HKU1)25. In this new scenario, clinicians will have to face pneumonia of different or combined etiologies, including SARS-CoV-2, other viruses, and common bacteria. Basic tools will be necessary to assess and characterize clinical profiles of host response, irrespective of microbiologic etiology. Our present study aimed to address this aspect. The need for such an intervention reinforces the use of the model application and its extensibility to any pneumonia. The strength of our approach is its simplicity in quickly and pragmatically identifying patients with higher risk of worse outcomes, especially in the absence of sophisticated studies in some hospitals or other facilities. Other advantages include the use of objective data and adjustments in our analysis by age and sex.
The study has several limitations, though. The COVID-19 cohort had an in-hospital mortality four times higher than that of the CAP cohort. Nonetheless, the model behaved identically when evaluating the host response; there was an increase in risk per the number of markers in both cohorts. The COVID-19 cohort was recruited when there were no specific treatments. The COVID cohorts were established in the early phase of the pandemic when the healthcare system was overloaded. The scarcity of health care resources may introduce bias into the analysis. The potential impact of vaccination with induced immunity and the appearance of variants with varying virulence may have affected the course and outcomes of the disease. It is, therefore, necessary to confirm these data in the future. The strengths of our study are multi-site cohorts and the evaluation of interplay between microorganism and host by the approach. This latter detail is important, as it allows personalized risk assessment. Quoting DeMerle et al.1, “not all hosts nor all host responses are the same”. A step towards more personalized medicine in COVID-19 and CAP does not always imply the necessary use of complex techniques. CRP, ALC, and SpO2/FiO2 are routinely available at both low- and high-resource settings.
In conclusion, we presented a feasible and streamlined multidimensional approach to assess pneumonia severity per the host response and regardless of age or sex. Three simple biomarkers mirroring inflammatory, immune, and respiratory function responses allow for an easy on-site or point-of-care strategy that facilitates severity assessment in varying hospitals. In the future, tools capable of identifying different severity profiles based on the characteristics of the host will be necessary to improve overall care and optimize patient and resource allocation.
Data availability
The data that support the findings of this study are available from the corresponding author, RM, upon reasonable request.
References
Demerle, K., Angus, D. C. & Seymour, C. W. Precision medicine for COVID-19: Phenotype anarchy or promise realized?. JAMA J. Am. Med. Assoc. 325(20), 2041–2042 (2021).
Wu, P. et al. The trans-omics landscape of COVID-19. Nat. Commun. https://doi.org/10.1038/s41467-021-24482-1 (2021).
Stephenson, E. et al. Single-cell multi-omics analysis of the immune response in COVID-19. Nat. Med. 27(5), 904–916 (2021).
Su, Y. et al. Multi-omics resolves a sharp disease-state shift between mild and moderate COVID-19. Cell 183(6), 1479–1495 (2020).
Knight, S. R. et al. Risk stratification of patients admitted to hospital with covid-19 using the ISARIC WHO clinical characterisation protocol: Development and validation of the 4C mortality score. BMJ https://doi.org/10.1136/bmj.m3339 (2020).
Dela Cruz, C. S. et al. Understanding the host in the management of pneumonia: An official American thoracic society workshop report. Ann. Am. Thorac. Soc. 18(7), 1087–1097 (2021).
Bermejo-Martin, J.F., Almansa, R., Menéndez, R., Mendez, R., Kelvin, D.J. & Torres, A. Lymphopenic community acquired pneumonia as signature of severe COVID-19 infection. J. Infect. Available from: http://www.ncbi.nlm.nih.gov/pubmed/32145214 (2020).
Méndez, R. et al. Lymphopenic community-acquired pneumonia is associated with a dysregulated immune response and increased severity and mortality. J. Infect. 78, 423–431 (2019).
Gupta, R. K. et al. Systematic evaluation and external validation of 22 prognostic models among hospitalised adults with COVID-19: An observational cohort study. Eur. Respir. J. https://doi.org/10.1183/13993003.03498-2020 (2020).
Heinze, G. & Schemper, M. A solution to the problem of separation in logistic regression. Stat. Med. 21(16), 2409–2419 (2002).
Bermejo-Martin, J. F. et al. Lymphopenic community acquired pneumonia (L-CAP), an immunological phenotype associated with higher risk of mortality. EBioMedicine 24, 231–236 (2017).
Rice, T. W. et al. Comparison of the SpO2/FiO2 ratio and the PaO2/FIO2 ratio in patients with acute lung injury or ARDS. Chest 132(2), 410–417 (2007).
Menéndez, R., Méndez, R., González-Jiménez, P., Zalacain, R., Ruiz, L.A. & Serrano. L., et al. Early recognition of low-risk SARS-CoV-2 pneumonia: A model validated with initial data and IDSA/ATS minor criteria. Chest. Published by Elsevier Inc under license from the American College of Chest Physicians. S0012-3692(22)01007-8. Available from: https://pubmed.ncbi.nlm.nih.gov/35609674 (2022).
Wynants, L. et al. Prediction models for diagnosis and prognosis of covid-19: Systematic review and critical appraisal. BMJ https://doi.org/10.1136/bmj.m1328 (2020).
Kutz, A. et al. The TRIAGE-ProADM score for an early risk stratification of medical patients in the emergency department - development based on a multi-national, prospective, observational study. PLoS One 11(12), e0168076 (2016).
Wang, D., Hu, B., Hu, C., Zhu, F., Liu, X. & Zhang, J. et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus–infected pneumonia in Wuhan, China. JAMA. Available from: https://jamanetwork.com/journals/jama/fullarticle/2761044 (2020).
Méndez, R. et al. Acute and sustained increase in endothelial biomarkers in COVID-19. Thorax https://doi.org/10.1136/thoraxjnl-2020-216797 (2021).
Méndez, R., Aldás, I. & Menéndez, R. Biomarkers in community-acquired pneumonia (Cardiac and Non-Cardiac). J. Clin. Med. 9. Available from: http://www.ncbi.nlm.nih.gov/pubmed/32085380 (2020).
Copaescu, A. et al. The role of immunological and clinical biomarkers to predict clinical COVID-19 severity and response to therapy—a prospective longitudinal study. Front. Immunol. 12, 646095 (2021).
Abani, O. et al. Tocilizumab in patients admitted to hospital with COVID-19 (RECOVERY): A randomised, controlled, open-label, platform trial. Lancet 397(10285), 1637–1645 (2021).
Marconi, V. C. et al. Efficacy and safety of baricitinib for the treatment of hospitalised adults with COVID-19 (COV-BARRIER): A randomised, double-blind, parallel-group, placebo-controlled phase 3 trial. Lancet Respir. Med. 9(12), 1407–1418 (2021).
Diaz, G. A. et al. Remdesivir and mortality in patients with coronavirus disease 2019. Clin. Infect. Dis. 74, 1812–1820. https://doi.org/10.1093/cid/ciab698 (2021).
Gupta, R. K. et al. Development and validation of the ISARIC 4C deterioration model for adults hospitalised with COVID-19: A prospective cohort study. Lancet Respir. Med. 3, 18 (2021).
Lazar Neto, F. et al. Community-acquired pneumonia severity assessment tools in patients hospitalized with COVID-19: A validation and clinical applicability study. Clin. Microbiol. Infect. 27(7), 1037-e1 (2021).
Jo, W. K., Drosten, C. & Drexler, J. F. The evolutionary dynamics of endemic human coronaviruses. Virus Evol. https://doi.org/10.1093/ve/veab020 (2021).
Acknowledgements
Integrate Research Program (PII) of Respiratory Infections of Sociedad Española de Neumología y Cirugía Torácica (SEPAR).
Funding
This study was supported by: El Instituto de Salud Carlos III (ISCIII) through Project [PI17/01421] and Project [COV20/00385] (Co-funded by the European Regional Development Fund/European Social Fund. “Investing in your future”). Sociedad Española de Neumología y Cirugía Torácica (SEPAR): 1078/2020. Sociedad Española de Neumología y Cirugía Torácica (SEPAR): Convocatoria extraordinaria PII Infecciones Respiratorias 2011. Raúl Méndez is the recipient of a Juan Rodés grant, supported by the Instituto de Salud Carlos III (ISCIII [JR21/00051]). Paula González-Jiménez is the recipient of a Río Hortega grant, supported by the Instituto de Salud Carlos III (ISCIII [CM23/00062]).
Author information
Authors and Affiliations
Contributions
Study design: R. Menéndez and R. Méndez. Patient enrolment: R. Méndez, P. González-Jiménez, A. Latorre, S. Reyes, R. Zalacaín, LA Ruiz, L. Serrano, PP. España, A. Uranga, C. Cillóniz, L. Pérez-de-Llano, R. Golpe, and A. Torres. Statistical analysis: A. Gaetano-Gil and BM Fernández-Félix. Drafting the manuscript: R. Menéndez, R. Méndez, and A. Torres. Revision of manuscript and approval of the final version: all authors. R. Menéndez, R. Méndez and Antoni Torres are the guarantors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Menéndez, R., Méndez, R., González-Jiménez, P. et al. Basic host response parameters to classify mortality risk in COVID-19 and community-acquired pneumonia. Sci Rep 14, 12726 (2024). https://doi.org/10.1038/s41598-024-62718-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-62718-4
- Springer Nature Limited