Abstract
Progressive familial intrahepatic cholestasis (PFIC) is a rare childhood manifested disease associated with impaired bile secretion with severe pruritus yellow stool, and sometimes hepatosplenomegaly. PFIC is caused by mutations in ATP8B1, ABCB11, ABCB4, TJP2, NR1H4, SLC51A, USP53, KIF12, ZFYVE19, and MYO5B genes depending on its type. ABCB11 mutations lead to PFIC2 that encodes the bile salt export pump (BSEP). Different mutations of ABCB11 have been reported in different population groups but no data available in Pakistani population being a consanguineous one. We sequenced coding exons of the ABCB11 gene along with its flanking regions in 66 unrelated Pakistani children along with parents with PFIC2 phenotype. We identified 20 variations of ABCB11: 12 in homozygous form, one compound heterozygous, and seven heterozygous. These variants include 11 missenses, two frameshifts, two nonsense mutations, and five splicing variants. Seven variants are novel candidate variants and are not detected in any of the 120 chromosomes from normal ethnically matched individuals. Insilico analysis revealed that four homozygous missense variations have high pathogenic scores. Minigene analysis of splicing variants showed exon skipping and the addition of exon. This data is a useful addition to the disease variants genomic database and would be used in the future to build a diagnostic algorithm.
Similar content being viewed by others
Introduction
Bile formation is a significant function of the liver. Bile salts play various important physiological roles. They facilitate intestinal emulsification and absorption of fat and fat-like molecules. Bile salts also function as signaling molecules via acting as ligands for various proteins such as the nuclear hormone receptor farnesoid X receptor (FXR)1. The liver also excretes various molecules, such as bilirubin, cholesterol, and xenobiotics, via the bile. However, excessive intracellular bile salts are toxic to cells mainly due to the detergent properties of bile salts2,3,4.
Impaired bile secretion followed by subsequent accumulation of high bile salt content in the liver and blood is commonly studied in cholestasis. Cholestasis is one of the most common childhood liver disorders and one of the leading causes of hepatic failure. Despite immense investigations, the etiology of cholestasis remains unknown in a notable proportion of newborns5,6,7,8.
Progressive familial intrahepatic cholestasis (PFIC) is a rare childhood-onset disease characterized by impaired bile secretion. PFIC can be attributed to mutations in the ATP8B1, ABCB11, ABCB4, TJP2, NR1H4, SLC51A, USP53, KIF12, ZFYVE19, and MYO5B genes, depending on the specific type PFIC1, PFIC 2, PFIC 3, PFIC 4, PFIC 5, PFIC 6, PFIC 7, PFIC 8, PFIC 9 AND PFIC 10 respectively. PFIC type 1 and 2 are characterized by low to normal serum gamma-glutamyltransferase (GGT) levels while PFIC3 is characterized by high serum GGT levels9,10,11,12. Besides GGT level clinical symptoms of PFIC2 patients include pruritus, jaundice, splenomegaly, hepatomegaly, and elevated serum bilirubin13,14. Liver histology can show inconstant features, such as lobular and portal fibrosis, giant cell transformation, and inflammation. However, these symptoms are not exclusive to PFIC2 and tend to overlap with other types of cholestasis15,16. Although the actual prevalence of PFIC is still unknown, the approximate incidence is 1/50,000–100,000 births worldwide10.
PFIC2 is an autosomal recessive disorder resulting due to ABCB11 mutations. ABCB11 is located on chromosome 2 at position 3111,17. ABCB11 encodes bile salt export pump (BSEP) that belongs to a group of transporters called ABC (ATP binding cassette) transporter which consist of Transmembrane Domains (TMDs). Being a transporter, BSEP transports bile salts. BSEP is expressed in the canalicular membrane of hepatocytes. The main function of BSEP is to facilitate the secretion of bile salts from the liver into bile canaliculi (3, 14).
Defects in BSEP can cause severe consequences due to accelerated levels of bile salt in serum. Mutations in the ABCB11 gene can affect the function of BSEP leading to decreased bile salt secretion and decreased bile flow which ultimately increases bile inside hepatocytes resulting in cholestasis (6). High levels of bile in liver cells can enhance chances of hepatocellular carcinoma. Though BSEP is expressed in hepatocytes its mild expression is also detected in the testis and adrenal gland (7,12).
In vitro studies revealed that protein processing and trafficking is affected by some missense mutations that cause retention of protein in the Endoplasmic Reticulum (11,13).
Mutations in the ABCB11 gene have been found in some populations, including European, American, African, and Taiwan Chinese18,19,20 but no work has been done in Pakistani patients despite that the Pakistani population has high consanguinity. We sequenced the ABCB11 gene in Pakistani patients with PFIC2 symptoms and identified twenty variations.
Materials and methods
Sample collection
We conducted a study involving sixty-six patients who presented at the Children’s Hospital Lahore & Institute of Child Health Lahore. Prior to sample collection, informed consent was obtained from all legal guardians of the patients. These patients hailed from various regions of Pakistan. Blood samples were collected once clinical and biochemical diagnoses indicated PFIC2. Additionally, thorough clinical assessments were conducted to rule out the possibility of any bacterial or viral infections. We collected blood samples from healthy parents, wherever feasible. The age range of the patients varied from a few weeks to 5 years old, with the majority being only a few months old when they were admitted to the hospital. Serum GGT levels were measured in all patients. Patients with elevated GGT levels were considered to have PFIC3, while those with low or normal GGT levels were diagnosed with PFIC1 or PFIC2 based on clinical criteria, including the presence of cholestasis, pruritus, and elevated bilirubin levels. Patients were selected having low or normal GGT level as they are considered to having PFIC1 and PFIC2. ABCB11 gene responsible for PFIC2 has higher incidence of mutations reported in various populations. ABCB11 was further analyzed.
This study received approval from the Institutional Independent Ethical Committee of the University of Veterinary and Animal Sciences in Lahore, Pakistan. All methods employed in this study were conducted in strict accordance with the relevant guidelines and regulations.
Gene amplification
DNA was extracted from peripheral whole blood samples utilizing the QIAamp DNA Blood Mini Kit in accordance with the manufacturer’s instructions. Subsequently, we conducted amplification of all exon sequences, excluding the first exon, which does not participate in protein coding, as well as the intron–exon boundaries of ABCB11 (Accession ENSG00000073734) through polymerase chain reaction (PCR) using primers that were specifically designed with Primer Premier 5.0. Following the PCR, the resulting products were purified and subjected to direct sequencing using Big Dye Terminator (Applied Biosystems, Foster City, CA, USA) on an ABI 3130 genetic analyzer (Applied Biosystems).
Mutational analysis
To predict effect of missense mutations, we conducted in silico analyses using different tools, including PolyPhen-2 (Polymorphism Phenotyping v2, http://genetics.bwh.harvard.edu/pph2/), SIFT (http://sift.jcvi.org/), and ConSurf (http://consurftest.tau.ac.il). The Sorting Intolerant from Tolerant (SIFT) analysis involved the examination of an alignment of orthologous sequences to predict whether an amino acid substitution would impact protein function (http://sift.jcvi.org/)21. Additionally, we utilized PolyPhen-2 to predict the potential impact of an amino acid substitution on both the structure and function of the protein (http://genetics.bwh.harvard.edu/pph2/)22. The evolutionary conservation scores of mutated residues in the BSEP protein were calculated using the ConSeq web server reflecting the full alignment of all homologues from different species, including mammals, lower vertebrates, invertebrates, and lower eukaryotes, as obtained from the Uniref90 database. In silico analysis was performed for intronic variants.
Minigene reporter assay
We employed the pSPL3 vector, generously provided by Dr. Irene Bottillo (University of Sapienza, Italy) and Dr. Leping Shao (University of Qingdao, China) for this study. We focused on the analysis of three variations: IVS21-2A>T in exon 22, IVS14+1G>C in exon 14, and IVS6+5G>A in exon 6, with primer sequences detailed in Supplementary Table 1. Patient DNA carrying each of these mutations was utilized to generate genomic fragments of ABCB11, encompassing exon 22, exon 14, and exon 6, along with approximately ± 150 bp of their upstream and downstream intronic sequences. The design of both ends of the shortened introns was carefully carried out using the Human Splicing Finder program to prevent the activation of cryptic splicing. The primers used in this study were edited with Primer Premier 5.0 software (https://www.premierbiosoft.com/primerdesign/index.html) and are listed in the Table S1. All the specified fragments were cloned into a pSPL3 vector using XhoI and BamHI restriction sites, and the ClonExpressTM II One Step Cloning Kit from Vazyme Biotech Co., Ltd. was employed for this purpose. To validate the presence of distinct mutations and ensure the absence of off-target mutations in the constructs, all constructs underwent thorough sequencing.
Transfection and RT-PCR
HEK293T cells, obtained from the Cell Bank of the Chinese Academy of Sciences, were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) at 37 °C with 5% CO2. Twenty-four hours after seeding in 12-well plates, the cells were transfected with 1 μg of DNA constructs containing either wild-type or mutant alleles using Lipofectamine 2000 (Invitrogen). Following transient transfection, total RNA was extracted from the cells using Trizol Reagent (Takara, Japan) after another 24 h. The first cDNA strand was synthesized in a reverse transcription reaction using SuperScript III transcriptase (Invitrogen) along with random hexanucleotide primers. Two microliters of each cDNA sample were then subjected to amplification using TaKaRa Ex Taq polymerase (Takara, Japan) with primers located in the two cassette exons of the pSPL3 vector. The primer sequences were as follows: Forward SD 5′-TCTGAGTCACCTGGACAACC-3′ and reverse SA 5′-ATCTCAGTGGTATTTGTGAGC-3′. The PCR products were analyzed by agarose gel electrophoresis and confirmed by sequencing of the extracted DNA. The gel contained 1.5% agarose in Tris–acetate–EDTA buffer.
Results
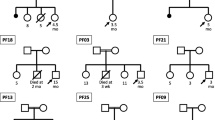
Sanger sequencing unveiled both known and several novel mutations within the Pakistani population (Fig. 1). We analyzed sixty-six patients suspected of having PFIC2, along with available parents, for mutations in ABCB11. This analysis led to the identification of twenty mutations in twenty patients, comprising 12 variants in a homozygous state, one in compound heterozygosity and seven heterozygotes, all listed in Table 1. Among these variants, 11 were missense mutations, two were frameshift mutations, and two were nonsense mutations and five splicing alterations. Notably, seven of these variants were candidate novel and were not detected in any of the 120 chromosomes from normal, ethnically matched individuals. Interestingly, we did not identify any mutations of ABCB11 in the 46 subjects initially diagnosed with PFIC2. This suggests the potential involvement of other identified genes in these cases. However, it is important to acknowledge that clinical evaluations may not always precisely differentiate between the various types of PFICs. This emphasizes the significance and necessity of genetic screening for early and accurate disease management.
Chromatograms of mutations identified in this study (This figure displays chromatograms representing the genetic variations identified in Pakistani population. The study comprised sixty-six patients from various regions of Pakistan, all exhibiting symptoms consistent with Progressive Familial Intrahepatic Cholestasis Type 2 (PFIC2). But only twenty were fond positive for variation in ABCB11 gene).
To assess the impact of the eleven missense variations, we conducted in silico analysis using two tools: PolyPhen-2 (Polymorphism Phenotyping v2, available at (http://genetics.bwh.harvard.edu.pph2) and SIFT (http://sift.jcvi.org). Additionally, we considered evolutionary conservation data obtained from the ConSurf Tests, as presented in Table 2. The scores for residues containing these missense variations in BSEP were based on a comprehensive alignment of BSEP homologs from various species sourced from the Uniref90 database. This alignment encompassed mammals, lower vertebrates, invertebrates, and lower eukaryotes. In silico analysis of the splicing effect of intronic variations was also performed using SpliceAI (https://spliceailookup.broadinstitute.org/#) and MaxEntScan (https://www.genes.mit.edu/burgelab/maxent/Xmaxentscan_scoreseq.html) (Table 3).
Splicing minigene reporter assay
We proceeded to conduct an in vitro minigene splicing assay, utilizing the pSPL3 exon trapping vector, to assess the consequences of the three splicing variations. Minigenes are designed with a conventional expression system, featuring two cassette exons, which allows for the analysis of the resulting mRNA transcripts (Fig. 2A). Following the transfection of minigenes containing the IVS21-2A>T, IVS6+5G>A, and IVS14+1G>C fragments into human kidney HEK293T cells, we extracted total RNA and converted it into cDNA. Subsequently, we performed PCR using flanking primers (arrows in Fig. 2A) and visualized the results on an agarose gel.
Minigene splicing assay (This figure depicts the Minigene Splicing Assay conducted in our study. The assay involved the use of pSPL3 vectors and the analysis of three specific mutations: IVS21-2A>T, IVS14+1G>C, and IVS6+5G>A in the ABCB11 gene. These variations were incorporated into genomic fragments and transfected into human kidney HEK293T cells. The resulting RNA transcripts were visualized through agarose gel electrophoresis for analysis).
When comparing the IVS14+1G>C mutant constructs with the wild-type plasmids, we observed that the mutant constructs produced only a single, smaller RT-PCR product, which corresponded to exon exclusion. This was confirmed through sequencing analysis (Fig. 2B). The results strongly indicate that the splice-donor site variation of IVS14+1G>C disrupts normal splicing, leading to exon skipping in vitro. In the case of IVS21-2A>T, we noticed the presence of a smaller RNA species compared to the product of the wild-type allele. Sequencing analysis of the RT-PCR fragment revealed the generation of a new acceptor site at + 80 bp in Exon22, resulting in the formation of a new exon containing 4 bp (Fig. 2C). The same results were obtained with IVS6+5G>A (Fig. 2D) as for IVS14+1G>C.
Evolutionary conservation analysis of eleven BSEP missense variations (This figure displays the results of an evolutionary conservation analysis conducted on eleven missense variations within the BSEP gene. It highlights the highly conserved amino acid residues across various species, emphasizing the significance of these variations in evolutionary conservation and their potential impact on BSEP function).
Discussion
This study marks the first genetic analysis of the ABCB11 gene associated with PFIC2 within the Pakistani population. PFIC is a hereditary disorder, and children born into families with a history of intermarriage face an increased risk of inheriting such disorders30. In Pakistan, a staggering 73% of marriages are consanguineous, with more than 50% occurring between first and second cousins. This high rate of consanguinity contributes to an elevated incidence of metabolic disorders among children31. The discovery of seven novel candidate ABCB11 variations in the Pakistani population underscores the possibility that its genetic diversity may differ from the data on PFIC reported in other populations.
We identified a nonsense mutation, c.989G>A (W330X), in a 4-month-old girl from a consanguineous marriage. Her liver profile revealed a slight elevation in ALT levels, while there was a significant increase in AST and ALP levels (Table S2). Her GGT level was recorded that was 13 IU. In her family history, cousin marriages were prevalent for the second generation, yet she was the sole individual to inherit this disease. Nonsense mutation was identified by Goto et al. at amino acid position 330 in a Japanese patient which led to PFIC due to termination codon23.
In another case, a deletion, c.1366_1366delC (S457VfsX23), was observed in a male patient who presented at the hospital at just three weeks old, showing signs of jaundice from birth. Both of his parents were found to be heterozygous for this variation, and sadly, one male sibling had already passed away. The patient hails from the province of Khyber Pakhtunkhwa and belongs to the Pakhtoon descendants. Notably, there is no record of this variation in the Human Gene Mutation Database (HGMD, http://www.hgmd.cf.ac.uk) nor in any other database or literature therefore, this represents a novel variation found within the Pakistani population.
A baby boy patient presented with jaundice and signs of pruritus. Blood analysis indicated a likelihood of PFIC2, as shown in the S2 Table. Genetic analysis revealed a novel variation at c.2144_2145delTA (V715GfsX3) in exon 18. He hailed from Lahore, Punjab Province, and while treatment was initiated, unfortunately, he did not survive. This is candidate novel variation. We couldn’t find any data from literature regarding this variation in any other population to make any comparison.
In another case, a nonsense variation, c.1156G>T (G386X), was identified in a patient from a family with a history of intermarriages spanning two generations. This patient presented at the hospital at the age of 8 months with significant jaundice. Clinical evaluation strongly suggested the possibility of PFIC2. She belonged to a Baloch ancestry but resided in Sargodha city, Punjab Province. Regrettably, the patient passed away a few months later. Similar case was reported by Shah. They reported an Indian child whose mutation analysis showed a homozygous nonsense pathogenic variant in the ABCB11 gene (c.1156G>T; p.Gly386*). He underwent a biliary diversion at 3½ years of age but subsequently died secondary to massive hematemesis28. Both patients belonged to same ethnic region and developed same mutation and passed away. This mutation might have serious pathogenic effect in Indian population.
Additionally, a missense variation, c.3382C>T (R1128C), was identified in three unrelated families from different geographic regions of Pakistan. This variation had previously been reported in the South Asian population17. Among these patients, one was a 3-month-old male infant from central Punjab province who, unfortunately, did not survive. His parents had a history of consanguineous marriages spanning three generations.
The second patient with this variation was a one-week-old male infant who was born healthy with a weight of 4 kg. He developed severe jaundice, and clinical studies revealed his GGT level at 27 IU/L indicated the possibility of PFIC2. Unlike the majority of patients, there was no history of cousin marriage in this family, although both parents were found to be heterozygous.
Strautnieks et al. reported this variation in south Asian population. Presence of this variation in multiple Pakistani families and support from literature indicates that this variation has high prevalence in south Asia24.
The third patient was a compound heterozygote, carrying not only c.3382C>T (R1128C) but also another variation, c.968G>A (G323E). She was born with a weight of 2.8 kg and was 5 months old when reported to the hospital with GGT level at 23 IU/L. Her family had a history of cousin marriages spanning three generations.
Though R1128C is already reported but G323E is only reported in gnomAD. There is no PMID and no literature related this variation. We can’t make any comparison to any other population due to lack of information.
In an individual, two missense variations, c.2633T>A and c.2636T>G, were found in a compound heterozygous form. Their proximity to each other raised the likelihood of pathogenicity, given the alteration at the adjacent amino acid residues. The patient was presented at the hospital at six months of age and received treatment for two years. Despite medication, her condition continued to deteriorate. It’s worth noting that her parents were first cousins, with the mother being heterozygous for the variations. Unfortunately, this patient, hailing from central Punjab, later passed away. While Droge et al. reported c.2636T>G in German population. This variation was reported in PFIC2 suspected patient25. c.2633T>A is reported in genomAD without any literature available.
We identified another variation, c.3164T>C (L1055P), in a 1-month-old patient. This deleterious variation had been previously reported in the American population32. Unfortunately, the mother’s sample was not available for analysis. The father brought the child to the hospital and was found to be heterozygous for the variation. There was a history of cousin marriage in the family spanning the previous two generations. The family hailed from Jhang city in the Punjab province. Evason et al.27 reported this variation in American population. Although Asian and American populations have different genetics but ABCB11 variations do not look region dependent.
In addition, we detected a missense variation, c.593T>C (L198P), in a 2-year-old male patient who also belonged to Pakhtoon ancestors. This identified variation was present in a homozygous form, but blood samples from both parents were unavailable for analysis. Byrne et al. studies revealed this variation significantly reduced mature protein level thus highlighting its importance in proper functioning of BSEP26.
In seven patients (as listed in Table 1), a single mutated heterozygous allele was identified. One such case involved a missense variation, c.3382C>G (R1128G), in a patient from Lahore, Punjab, who was admitted to the hospital at 5 months of age due to severe itching and cholestasis. Notably, this variation had not been previously reported. Previous studies have noted both transition and transversion variations occurring at this position. Mutations at the same positions, such as p.Arg1128Pro, p.Arg1128His, and p.Arg1128Cys have been reported in the database but p.Arg1128Gly has not been reported17. So no relevant data available. A female patient was presented at 8 months with mild pruritus and cholestasis. She exhibited heterozygous variation c.3149G>A (p.Arg1050His). She did not survive. p. Arg1050His. is only available at gnomAD. There is no publication reporting this variation. It is not even reported in ClinVar. Another patient found positive for variation c.1664G>C (p.Gly555Ala) was brought to hospital ate age of 6 months with pruritus and cholestasis. He passed away soon. Wang et al. (2016) reported p. Gly555Ala in Chinese population. While variations in a heterozygous form may not have as severe effects as homozygous variations, they remain potential candidates for future research. Furthermore, two additional patients were identified with heterozygous variations: c.3568G>A (p.Ala1190Thr) and c.3691C>T (p.Arg1231Trp). While the later reported by Alvarez et al.29 p.Ala1190Thr is only available at gnomAD. There is no publication related to this variation and it’s not reported in ClinVar.
In addition to missense variations, we also identified splicing variations in our study. Two of these splicing variations were novel and were found in a homozygous form, while two other novel splicing variations, IVS6+8G>T and IVS16-51T>C, were identified in a heterozygous form. These patients were suspected for PFIC2. Intronic variants might have a role in diseases but as these variants were identified in heterozygous form, variants in other PFIC- related genes maybe be causative. Further research is required in this regard.
One patient carrying IVS21-2A>T was admitted to the hospital at the age of twelve months. He resided in Lahore, and his family had a history of cousin marriages. Despite developing symptoms earlier, he was brought to the hospital a bit late and unfortunately did not survive. Both of his parents tested negative for the variation. This variation was reported by Strautnieks et al. in Canadian population24.
Another patient, who had the splicing variant IVS6+5G>A, initially received treatment with medication but later underwent a liver transplant. Fortunately, he started to recover. He belonged to the Narowal district of Punjab. We cannot compare effect of this variation with other population as this is not reported anywhere in literature or databases.
A female patient from the Shakargarh district was brought to the hospital and was identified with IVS14+1G>C. Both of her parents were found to be heterozygous for this variant, and they had a history of cousin marriages. This is another novel candidate variation found in Pakistani population.
Out of the 66 enrolled patients, 12 exhibited homozygous variations, while seven were identified with heterozygous variations in the ABCB11 gene and one compound heterozygote. However, some patients did not show variations in the ABCB11 gene, and some with deteriorating conditions despite being heterozygous. These cases suggest the possible involvement of other candidate genes within the PFIC family. Given that many symptoms of PFIC2 overlap with those of other types of PFIC, it is plausible that other genes might be responsible for these cases. Further investigation is warranted to identify these potential candidate genes and their role in the development of the disorder.
Protein alignments have revealed that some of the identified variations (L198P, G323E, R1128C/G, and R1231W) occur at highly conserved amino acid residues among different species (Fig. 4). We utilized the TOPO2 model to predict the transmembrane topology of membrane proteins. Variations within the transmembrane domains can potentially alter the predicted topology, whereas variations in the nucleotide-binding domain (NBD1 and NBD2) may not directly affect topology predictions but can have an impact on protein function.
TOPO2 diagram of identified variations in the ABCB11 gene (Schematic view of BSEP (NCBI reference sequence, NP_003733.2) with the location of the variants identified in the study using the TOPO2 program http://www.sacs.ucsf.edu/TOPO2/).
Based on the Cryo-EM structure of BSEP, G323E and W330X are located in the transmembrane domain (TMD) 5, M878K and I879R reside within TMD9, L189P is found in NBD 1, and R1128C/G and R1231W are located within NBD233,34, as depicted in Fig. 4. The precise mechanisms through which these variations affect BSEP-mediated bile salt export are not yet completely understood. Further experiments at the cellular level will be required to provide a more detailed mechanistic explanation.
Conclusion
This study underscores the necessity for an expanded newborn genetic screening program focused on metabolic disorders in Pakistan, alongside the establishment of comprehensive metabolic healthcare services. The early and precise diagnosis of inborn errors of metabolism (IEM) can facilitate the timely implementation of appropriate disease management strategies. While larger trials are essential, our results suggest that these variants could serve as valuable molecular markers for the initial screening of affected families and patients. The identification of candidate genetic markers for these disorders would empower affected families to explore options such as carrier and prenatal testing, preimplantation genetic diagnosis, and informed decision-making regarding within-family marriages.
Data availability
DNA of patients used for this study is available upon request to Hafsa Riaz via email 2014-phd-1069@uvas.edu.pk. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables. The supplementary file comprises Supplementary Table 1 describing Primers for exon-specific sequencing of the ABCB11 gene and Supplementary Table 2 showing available data of patients with mutations in ABCB11 and Supplementary Fig. 1 representing plasmid card of pSPL3.
References
Camilleri, M. & Gores, G. J. Therapeutic targeting of bile acids. Am. J. Physiol. Gastrointest. Liver Physiol. 309(4), G209–G215 (2015).
Jansen, P. L. et al. Hepatocanalicular bile salt export pump deficiency in patients with progressive familial intrahepatic cholestasis. Gastroenterology 117, 1370–1379 (1999).
Kato, T., Hayashi, H. & Sugiyama, Y. Short-and medium-chain fatty acids enhance the cell surface expression and transport capacity of the bile salt export pump (BSEP/ABCB11). Biochim. Biophys. Acta Mol. Cell 1801(9), 1005–1012 (2010).
Hu, G. et al. Diagnosis of ABCB11 gene mutations in children with intrahepatic cholestasis using high resolution melting analysis and direct sequencing. Mol. Med. Rep. 10(3), 1264–1274 (2014).
Jackson, J. P. et al. Basolateral efflux transporters: A potentially important pathway for the prevention of cholestatic hepatotoxicity. Appl. Vitro Toxicol. 2, 207–216 (2016).
Knisely, A. S. Perspectives in pediatric pathology: Progressive familial intrahepatic cholestasis: A personal perspective. Pediatr. Dev. Pathol. 3, 113–125 (2000).
Kubitz, R., Dröge, C., Stindt, J., Weissenberger, K. & Häussinger, D. The bile salt export pump (BSEP) in health and disease. Clin. Res. Hepatol. Gastroenterol. 36, 536–553 (2012).
Kubitz, R., Keitel, V. & Scheuring, S. Benign recurrent intrahepatic cholestasis associated with mutations of the bile salt export pump. J. Clin. Gastroenterol. 40, 171–175 (2006).
Lang, C. et al. Mutations and polymorphisms in the bile salt export pump and the multidrug resistance protein 3 associated with drug-induced liver injury. Pharmacol. Genet. 17, 47–60 (2007).
Baker, A. et al. A systematic review of progressive familial intrahepatic cholestasis. Clin. Res. Hepatol. Gastroenterol. 43(1), 20–36 (2019).
Strautnieks, S. S. et al. Identification of a locus for progressive familial intrahepatic cholestasis PFIC2 on chromosome 2q24. Am. J. Hum. Genet. 61(3), 630–633 (1997).
Davit-Spraul, A. et al. ATP8B1 and ABCB11 analysis in 62 children with normal gamma-glutamyl transferase progressive familial intrahepatic cholestasis (PFIC): Phenotypic differences between PFIC1 and PFIC2 and natural history. Hepatology 5, 1645–1655 (2010).
Hori, T., Nguyen, J. H. & Uemoto, S. Progressive familial intrahepatic cholestasis. Hepatobil. Pancreatic Dis. Int. 9, 570–578 (2010).
Liu, L. Y., Wang, Z. L., Wang, X. H., Zhu, Q. R. & Wang, J. S. ABCB11 gene mutations in Chinese children with progressive intrahepatic cholestasis and low γ glutamyltransferase. Liver Int. 30, 809–815 (2016).
Misawa, T., Hayashi, H., Makishima, M., Sugiyama, Y. & Hashimoto, Y. E297G mutated bile salt export pump (BSEP) function enhancers derived from GW4064: Structural development study and separation from farnesoid X receptor-agonistic activity. Bioorg. Med. Chem. Lett 15, 3962–3966 (2012).
Byrne, J. A. et al. Missense mutations and single nucleotide polymorphisms in ABCB11 impair bile salt export pump processing and function or disrupt pre-messenger RNA splicing. Hepatology 49, 553–567 (2009).
Strautnieks, S. S. et al. Severe bile salt export pump deficiency: 82 different ABCB11 mutations in 109 families. Gastroenterology 134, 1203–1214 (2008).
Bull, L. N. et al. A gene encoding a P-type ATPase mutated in two forms of hereditary cholestasis. Nat. Genet. 18, 219–224 (1998).
Chen, H. L. et al. Diagnosis of BSEP/ABCB11 mutations in Asian patients with cholestasis using denaturing high performance liquid chromatography. J. Pediatr. 153, 825–832 (2008).
Li-Yan, L., Zhong, L. W., Xiao, H. W. & Qi-Rong, Z. ABCB11 gene mutations in Chinese children with progressive intrahepatic cholestasis and low γ glutamyltransferase. Liver Int. 30(6), 809–815 (2009).
Ng, P. C. & Henikoff, S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 31, 3812–3814 (2003).
Adzhubei, I., Jordan, D. M. & Sunyaev, S. R. Predicting functional effect of human missense mutations using PolyPhen-2. Curr. Protoc. Hum. Genet. 76, 7–20 (2013).
Goto, K. et al. Bile salt export pump gene mutations in two Japanese patients with progressive familial intrahepatic cholestasis. J. Pediatr. Gastroenterol. Nutr. 36(5), 647–650 (2003).
Strautnieks, S. S. et al. Severe bile salt export pump deficiency: 82 different ABCB11 mutations in 109 families. Gastroenterology 134(4), 1203–1214 (2008).
Dröge, C. et al. Sequencing of FIC1, BSEP, and MDR3 in a large cohort of patients with cholestasis revealed a high number of different genetic variants. J. Hepatol. 67(6), 1253–1264 (2017).
Byrne, J. A. et al. Missense mutations and single nucleotide polymorphisms in ABCB11 impair bile salt export pump processing and function or disrupt pre-messenger RNA splicing. Hepatology 49(2), 553–567 (2009).
Evason, K. et al. Morphologic findings in progressive familial intrahepatic cholestasis 2 (PFIC2): Correlation with genetic and immunohistochemical studies. Am. J. Surg. Pathol. 35(5), 687–696 (2011).
Shah, I. & Chilkar, S. Progressive familial intrahepatic cholestasis type 2 in an Indian child. Case reports. J. Pediatr. Genet. 6(2), 126–127 (2017).
Alvarez, L. et al. Reduced hepatic expression of farnesoid X receptor in hereditary cholestasis associated with a mutation in ATP8B1. Hum. Mol. Genet. 13(20), 2451–2460 (2004).
Afzal, M., Ali, S. M., Siyal, H. B. & Hakim, A. Consanguineous marriages in Pakistan. Pak. Dev. Rev. 33, 663–676 (1994).
Zahoor, Y. M., Cheema, H. A., Ijaz, S. & Fayyaz, Z. Genetic analysis of tyrosinemia type 1 and fructose-1, 6 bisphosphatase deficiency affected in Pakistani cohorts. Fetal Pediatr. Pathol. 39, 430–440 (2020).
Evason, K. et al. Morphologic findings in progressive familial intrahepatic cholestasis 2 (PFIC2): Correlation with genetic and immunohistochemical studies. Am. J. Surg. Pathol. 35, 687 (2011).
Leabman, M. K. et al. Natural variation in human membrane transporter genes reveals evolutionary and functional constraints. Proc. Natl. Acad. Sci. 10, 5896–5901 (2003).
Wang, L. et al. Cryo-EM structure of human bile salts exporter ABCB11. Cell Res. 30, 623–625 (2020).
Acknowledgements
The authors are thankful to the patients who participated in the study and to the Department of Pediatric Gastroenterology and Hepatology, Children’s Hospital Lahore, Pakistan. They are thankful to the Higher Education Commission of Pakistan for a Ph.D. indigenous fellowship to Hafsa Riaz and for awarding a visiting fellowship at the University of Alberta Canada.
Funding
The funding was provided by Higher Education Commision, Pakistan (213-67920-2MB2-106).
Author information
Authors and Affiliations
Contributions
This is a research work of Hafsa Riaz for her Ph.D. dissertation under the major supervision of Da-wei Zhang and Muhammad Yasir Zahoor. All authors made substantial contributions to the study’s conception and design and/or acquisition of the data and/or analysis and interpretation of the data. Each author participated in drafting the article or revising it critically for important intellectual content and gave the version’s final approval and any revised version.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Riaz, H., Zheng, B., Zheng, Y. et al. The spectrum of novel ABCB11 gene variations in children with progressive familial intrahepatic cholestasis type 2 in Pakistani cohorts. Sci Rep 14, 18876 (2024). https://doi.org/10.1038/s41598-024-59945-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-59945-0
- Springer Nature Limited