Abstract
This study aimed to identify the teleost fish species sold in Bragança, a major fishing hub on the north coast of Brazil. The COI gene analysis was performed for the identification of fish species. The local market uses common names that are not accurate and do not reflect the diversity of the species. 204 sequences were obtained, with 119 haplotypes. 83 species were identified by comparing with public databases and constructing phylogenetic trees, with Carangidae being the most prevalent family. The study also found Haemulon atlanticus, Menticirrhus cuiaranensis and Hoplias misioneira, a newly described species from the Amazon basin, among the samples. Additionally, 73 commercial names were recorded, including 10 categories, and the illegal trade of Epinephelus itajara was detected. The DNA Barcode method proved to be effective for discriminating the species. The study highlights that common and commercial names are vague and underestimate the fish diversity, and that Brazil needs to revise its regulations for commercial and scientific names.
Similar content being viewed by others
Introduction
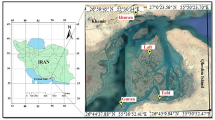
The North Brazilian coast spans the states of Amapá, Pará and Maranhão and is about 2500 km long1. This region is home to a rich diversity of fish, both marine and estuarine, that have high economic and social importance2,3,4,5. Various fishing fleets operate in this region and bring their catch to state of Pará6, where Bragança is one of the main fish markets7. Bragança is a strategic location for studying the coastal ecosystem, as it has a large area of mangroves, estuaries and rivers nearby, such as the Caeté River, which provides a constant flow of organic matter and nutrients to the marine environment,8,9, resulting in high productivity and a rich biodiversity in the region8,9. The latest survey conducted in the Caeté estuary recorded 120 fish species belonging to 48 families, of which 19 species were new records for the north coast of Brazil10.
The fishing activity is influenced by the great diversity of resources available. In the municipality, fish landings happen daily6 and the fish originate from two different fleets, the large-scale (industrial), which targets fish in the deeper areas of the continental shelf, and the small-scale (artisanal) fleet, which operates in the areas closer to the shore9. Both catch a high diversity of species that supply the regional, national, and international market6,11. A lot of fish species from different regions come to the Municipal market and the City Fair, where they are available for sale all year round. According to a recent survey of the fish trade in Bragança, there were 98 species sold under 103 different trade names12. However, the survey only used morphology to identify the species. The use of trade names makes it hard to know the true diversity of the fish being sold, because these names can change within and between regions13. Moreover, sometimes multiple species are sold under one name as a category14, or one species may have different trade names depending on the region15. This lack of precision can cause confusion about the identity of the fish being sold, as well as enable the trade of endangered species16,17 and even harm the consumer, since it may increase the chance of commercial fraud18.
The challenge of accurately identifying species is a barrier that hinders the assessment and monitoring of the status of exploited resources, because there is no consistency in the use of the common name by fishermen19 and, therefore, by traders and consumers. To address the difficulty of standardizing trade names, Normative Instruction No. 53 of September 1, 2020 (MAPA), established a correlation between common and scientific names for the main species of fish with commercial importance in Brazil. However, this list still has many generic terms and ambiguities20. In this way, the precise identification of commercialized fish is necessary, to know the real diversity of fish sold, a primary measure for the effective management of resources to promote sustainable fishing. Traditionally, the identification of species is performed based on morphological characters, however, the small number of specialists in various groups has made it difficult to register biodiversity21. In addition, morphological approaches began to present some limitations, such as identification errors due to phenotypic plasticity, cryptic species, and individuals in the early stages of life, which are generally not contemplated in identification keys22, problems are even more evident in groups where species are highly similar and/or in processed fish, where diagnostic characters are removed18,23.
In view of this, the use of alternative methodologies is essential, such as molecular tools that use DNA for species-specific identification, especially the initial portion of the mitochondrial gene Cytochrome Oxidase C—subunit I (COI), a tool widely used as DNA Barcoding22. Since its proposal, several works have been reported in the literature for fish identification, demonstrating its efficiency for species discrimination and understanding of the diversity offered in categories14,16.
Among Brazilian examples, we highlight14, who were able to identify seven different species of fish sold together under the name Acará, in the Amazon, and16, who identified fish from markets in southern Brazil and found, in addition to replacement cases, trade in endangered species. DNA Barcoding has been shown to be a powerful tool for identifying species and revealing the hidden diversity that may be overlooked by traditional methods16,17,24. In this study, we applied DNA Barcoding to investigate the diversity of Teleost fish sold in Bragança, a coastal city in northeastern Pará, Amazon region. We compared the common names used in the market with the actual species identified by DNA Barcoding. We also detected the presence of endangered and newly described Teleost fish species in the trade. Our findings provide valuable information for the conservation and management of the fishery resources, as well as for the consumers' awareness and education.
Results
Teleost diversity traded using DNA barcoding
We analyzed 500 base pairs of DNA from 204 fish samples and found 258 polymorphic sites. The final alignment did not contain any deletions, insertions or stop codons. We obtained 119 haplotypes and compared their sequences to public databases. Table 1 shows the haplotypes, their molecular identification based on genetic similarity, and the commercial names of the fish samples. The sequences are publicly available, codes OR459502-OR459617 and OR515260-OR515262.
We identified 82 species of teleost fish, belonging to 15 orders, 31 families and 58 genera, from the 73 commercial names previously recorded. We used both genetic similarity and phylogeny for molecular identification (Fig. 1). Many taxa did not match the commercial name assigned to them. The most diverse families were: Carangidae (12 species), Sciaenidae (10 species) and Ariidae (9 species) (Supplementary Table S1).
Most of the recorded species (68) were from marine and/or estuarine habitats (Fig. 2), while 14 were from freshwater (Fig. 3). We also report the first record of commercialization of Haemulon atlanticus (formerly known as H. steindachneri) (Family Haemulidae), Menticirrhus cuiaranensis (Sciaenidae) and Hoplias misioneira (Erythrinidae) species on the north coast of Brazil.
Molecular identification
Using DNA Barcoding, we discriminated 82 species of fish from the Amazon region. We compared their genetic sequences with public databases and found 81 matches at the species level. The exceptions were Aspistor quadriscutis and Batrachoides surinamensis, which we identified by morphology and phylogenetic analysis. These two species had no reference sequences in public databases, so our sequences will serve as the first references for them (Table 1; Figs. 2, 3).
We also identified eight species by similarity only using the BOLD platform, but we did not include their sequences in our analyses because they came from a private source. They were Trachinotus cayennensis, Cetengraulis edentulus, Hoplerythrinus unitaeniatus, Astyanax bimaculatus, Notarius grandicassis, Sciades parkeri, Sciades proops and Sciades herzbergii.
Some samples had ambiguous identification results, such as the ones labeled as “cangatã”, which matched Aspistor luniscutis by similarity and A. quadriscutis by morphology. Other samples had high similarity with more than one species, such as “caica” 01 with Mugil curema and Mugil rubrioculus, “caica” 02 with Mugil hospes and Mugil brevirostris, “caica” 03 with Mugil curema and Mugil trichodon; the “gurijuba” sample with Netuma sp. and S. parkeri, “urubaiana” with Elops smithi and Elops saurus (Table 1). Another controversial case was found for the sequences of “bragalhão”, “bagre” and “uricica branca”, which the comparisons returned them ash Sciades couma, based on different sequences (ITAPE024 and ITAPE351). The clusters resulting from the NJ tree showed that “bragalhão” and the ITAPE024 sequence form a cluster and that the “bagre” and “uricica branca” form another clustering with the ITAPE351 sequence (Fig. 2). The two groups differ from each other with a divergence of 5.20%.
Mean genetic distances increased according to taxonomic level, with average of 0.13% within species, 11.55% within genera (between species) and 18.28% within families (between genera) respectively (Table 2). Intraspecific values ranged from 0.0% to 1.42%. The species showed barcode gaps, with a minimum distance between congeners of 4.16%, between S. couma and S. props (Table 2).
Trade names and endangered species
73 trade names were identified in this study, corresponding to 82 species (Table 1). Among the designations, 10 were considered categories (Fig. 4), as they presented more than one species being sold by the same name. As an example, we have the “pampo” category, which had the highest number of species (n = 5), including Chloroscombrus chrysurus, Hemicaranx amblyrhynchus, Trachinotus carolinus, Trachinotus goodei and Peprilus crenulatus (Fig. 4).
Beside from this, 10 cases in which different trade names were used for the same species were also observed, such as Cynoscion acoupa, which has been sold as “pescada amarela”, “pescada branca” and “garoupa” (Table 1). On the other hand, the species Haemulon parra, H. atlanticus and M. cuiaranensis were found being sold without presenting a commercial name, therefore they were called “without commercial designation” (SDC).
The commercial designations, when compared with the correlation of common names and respective scientific names provided by Normative Instruction No. 53 of September 1, 2020 (MAPA), showed that there is compatibility for many of the identified species (47.6%), however, for other species, the names are different (23.4%), in addition to species found being commercialized, but which do not have a name on the MAPA list (28%) (Supplementary Table S1).
The study revealed five species with some level of threat, according to IUCN list (2023) and by Ordinance MMA nº 14825, including: the “mero” Epinephelus itajara (vulnerable—VU), “gurijuba” S. parkeri (vulnerable—VU), “pirapema” Megalops atlanticus (vulnerable—VU) and “ariacó” Lutjanus synagris (near threatened -NT)26. By MMA Ordinance No. 148, of June 7, 2022, Epinephelus itajara was considered (critically endangered-CR), S. parkeri (vulnerable—VU), Megalops atlanticus (vulnerable—VU) and “pargo” Lutjanus purpureus (vulnerable—VU)25.
Discussion
This work represents the most comprehensive molecular analysis with Teleost fish traded in the coastal Amazon, with more than 200 individuals collected over four years of study. DNA barcoding tool was used to identify and validated the real commercialized diversity, masked due to the use of categories, and revealed an important trade of threatened species, in addition to species that its commercialization was first recorded in this study.
DNA barcoding for ichthyodiversity identification
The DNA Barcoding tool was used to identify the fish diversity traded in Braganca. Comparisons were made with public database and phylogenetic trees, comprising 82 fish species, corresponding to the highest ichthyodiversity recorded to date, which was higher than those found by Braga et al.9 and Freire et al.29 and like the ones found by Martins et al.12, when considering only teleost fish. The identifications carried out in previous studies were based on vernacular nomenclatures and taxonomic keys, while in this study, we identified a large number of species commercialized in Bragança and in the North region through the DNA Barcoding approach, confirming the efficiency of the molecular tool to discriminate taxa, as observed in other studies with ichthyodiversity24,27, as well as to identification of processed products by the fishing industry28.
Within the identified species, the family Carangidae was the most representative with 12 species, contrary with previous studies that positioned the Sciaenidae family as the most representative9,12,29. Carangidae is constantly identified as one of the main families that composes the ichthyofauna of the Brazilian north coast3,30.
The entry of large number of Carangidae family species into local trade can be attributed to the emergence of a new market, during the closed season for “pargo” (L. purpureus) and other species of greater commercial value from the north coast of Brazil where the vessels are licensed for various fish species and many carangids, popularly known as 'black fish', are caught (personal communication). Another important fact is that this study is the first implementing the molecular approach on the diversity of teleost fish commercialized in Bragança, when compared to previous research that only used taxonomic keys9,12,29, leading the authors not to reach the identity of the evaluated species, thereby underestimating the group of Carangidae species sold.
In this study, the commercialization of H. misioneira in the North of Brazil was also observed for the first time. This species was described from the Hoplias malabaricus species complex, in the Uruguay, Paraguay and Paraná basins31 and according to Guimaraes et al.24, this species has a disjunct distribution, also occurring in the Amazon Basin. This is the second record outside its natural range, which shows that this species is probably distributed in other areas, since the H. malabaricus complex has a wide distribution32.
In addition, we recorded the trade of newly described species already being sold, and without having a commercial name, such as M. cuiaranensis, and H. atlanticus. This scenarios shows how the diversity of fish in the coastal Amazon is underestimated and misunderstood, since the capture and commercialization of this species was already happening even before we were aware of its presence. This is worrying, because while part of the biodiversity remains unknown, natural resources are being exploited at an increasingly accelerated pace5.
Probably, when it comes to fish, many taxa can be extinct even before being formally described due to the intense dynamics of capture and commercialization, with diverse and non-standard nomenclatures, associated with inefficient and/or non-existent inspection which strongly collaborate to reduce the biodiversity.
Inconsistencies between morphological and molecular identifications
Ambiguities in the identifications were found for the “cangatã” fish, which molecular identification presented them as A. luniscutis, however, the species that is found in the Brazilian north region is A. quadriscutis. In addition, a study with a molecular and morphological approach showed that Aspistor species found on the Brazilian coast have morphological differences, but do not present significant genetic distances in mitochondrial genes such as cytochrome b (Cyt b) and subunits 8 and 6 of ATP synthase (ATPase 8/6)33. It may be that the same happens for the COI gene, causing the samples of A. quadriscutis to show great similarity with A. luniscutis and forming a clade in the phylogenetic tree (NJ).
For some species of the Ariidae family, the inconsistencies in the identifications probably occurred due to identification errors and consequent erroneous deposits in public database, as observed for the designations “bragalhão”, “bagre” and “uricica branca”, identified as S. couma, but which formed two distinct groups with a genetic distance of 5.20%, in the NJ tree, being “bragalhão” (clade 1), bagre” and “uricica branca” (clade 2), that was identified based on morphology as S. couma and S. herzbergii respectively.
Comparisons with public database showed ambiguity in identification. Cases like these were observed for the Mugilidae family, the species referring to “caica” 02, was 100% similar to M. hospes and M. brevirostris in the NCBI and BOLD. However, in the South Atlantic only M. brevirostris occurs34, therefore, the sequences deposited in Brazil as M. hospes are, considered M. brevirostris35. For the other members of the Mugilidae family, the identification was confirmed from the study conducted by Durand et al.35, where a new identification of sequences from the Mugilidae family deposited in Genbank was carried out, correcting erroneous deposits, therefore we identified “caica” 01 as M. rubrioculus, and “caica” 03 as M. curema.
Another case of incongruity was observed for the “urubaiana” fish, identified as Elops smithi and Elops saurus in public banks. However, Sousa et al. revealed the occurrence of only E. smithi on the Brazilian coast. Thus, the sequence assigned to E. saurus in Brazil is possibly a taxonomic error, which has already been reported for these two species in the literature36,37.
The samples considered to be H. atlanticus were identified in public database as H. steindachneri, before the description of H. atlanticus for the Western Atlantic, both belonging to the H. steindachneri complex38. Despite the study by Carvalho et al.38 used the genetic tool to confirm the existence of the two species, these sequences could not be used in this work, as they are not available in public databases.
Records of incompatibilities due to inaccurate or erroneous deposits in public database have been reported in the literature for both the BOLD system27 and the NCBI34. Faced with these shortcomings, researchers must be carefully when carrying out identifications by consulting specialized literature or specialists in each group or using reference database to resolve ambiguous cases, so that identification errors are mitigated and not perpetuated and reliability in the data deposited in public database is maintained.
Despite some ambiguities in species identifications, the DNA Barcoding tool was efficient in discriminating most of the taxa found in this study, with expressive Barcode gap. We recovered as the greatest intraspecific distance, (1.42%), found for the species Mylossoma duriventre, and the smallest interspecific distance (4.16%) between the species Sciades couma and Sciades proops.
Commercial name and hidden diversity
82 species were found out of 73 trade names sampled, showing that there is no correspondence between the number of trade names and the number of traded species, since in some cases the designations act as a category and in others the same species receives different trade names. Marketing by category, as in Bragança and as it happens in most places, ends up with underestimating the fish diversity offered, mainly due to the difficulty of differentiating the taxa of some families with similar morphology, as observed for Centropomidae and its congeners sold as “camurim”, the C. undecimalis, C. parallelus and C. ensiferus; and Mugilidae sold as “caica”, M. rubrioculus, M. brevirostris, M. curema, and M. incilis.
An interesting case is the category “pampo” used for five species, including those from different genera (C. chrysurus, H. amblyrhynchus, T. carolinus, T. goodei and P. crenulatus) confirming that commercial designations do not offer precision about the commercialized species. Some commercial names describe large groups, configuring themselves as a category, this is already described for nomenclatures such as “pargo”, “bagre”, “sardinha”, “pescada”, “garoupa”, and “arraia”17,39.
Marketing through generalist names can pose a threat to fish conservation, as several species can be sold through categories, including endangered species14,16,17, as the case of E. itajara sold under the designation/category “garoupa”. It is important to note that the commercialization of E. itajara in Brazil has been prohibited since 200240, therefore, the commercialization of this species is taking place illegally. The trade of endangered species is worrying, basically when it is facilitated by the non-standardization of the commercial nomenclature that masks this market.
One case of substitution occurred for fish sold under the name “pescada branca”, which according to normative instruction MAPA Nº 53 of 2020, should only be used for the species Cynoscion leiarchus and Plagioscion squamosissimus. However, all samples collected under this designation were identified as Cynoscion acoupa, a species normally sold as “pescada amarela” and which is of great commercial importance7,12. This replacement probably occurred accidentally, since the individuals collected were juveniles and many species of the Sciaenidae family are morphologically similar in the early stages and have a sympatric distribution, which can lead to difficulties in the correct identification of taxa, as already reported in other works18.
For continental species, we found different taxa being marketed through the designation “traíra”, including different genera, they are: “traíra” 01, identified as Hoplerythrinus unitaeniatus and “traíra” 02 H. misioneira.
Although the MAPA normative tries to establish and standardize the relationship between the common names and respective scientific names for the main commercialized species, it still has redundancies, as it provides several common names for a single species and in some cases displays nomenclatures for fish down to the genus level, such as “canguiro” and “pampo” for Trachinotus sp., opening space for permanence of categories. When comparing the Feira Livre designations with Normative Instruction No. 53 of September 1, 2020, we noticed that a range of taxa does not have a similar name in the normative instruction, as well as several species are not present in the list. This reveals that we have a document that needs to be revised to establish in a coherent and specific way the trade name and corresponding species in Brazil. The alternative to reduce the gaps left by categorization is the creation of lists by region, since the nomenclatures vary a lot, even in nearby places.
The importance of knowing the diversity of fish in the trade
Trade in Bragança is predominantly carried out with marine species, but some freshwater species are also sold, including fish from fish farming such as “tilápia” Oreochromis niloticus and “Tambaqui” Colossoma macropomum.
We observed the trade of species that had not been previously registered in the Bragantina region9,12,29, including Katsuwonus pelamis (“atum”), C. ensiferus (“camurim”), T. cayennensis (“pampo”), T. goodei (“pampo”), Selene setapinnis (“peixe galo”), H. parra (SDC) and M. cuiaranensis (SDC). For freshwater fish we found Schizodon fasciatus (“aracu/piau”), H. missioneira (“traira”), M. duriventre (“pacu/paboca”) and Pygocentrus nattereri (“piranha”).
The results reveal that trade in Bragança is quite dynamic, with changes in the composition of species offered over the year (Table 3). Certainly, there are species that were not sampled, as the landings and commercialization of fish in Bragança occur daily6, however, this study presents the most complete data regarding the diversity of teleost fish commercialized in the Bragança region (Table 3).
Our results shows the commercialization of species that were hidden by popular nomenclature and imprecise taxonomic identification. These data raise an alert about the capture and sale of species that already have low stocks, allowing the competent authorities to manage and supervise this market.
The measures for conservation and fisheries management be effective, it is first necessary to know the really diversity. We present here a list (Supplementary Table S1) with the correspondence between the commercial and biological designation for the species commercialized in Bragança, Amazonian coastal region, the first obtained through molecular identification and which will be an important tool for ordering the commercialization of fish in the region, considering all fish collected and molecular identifications carried out.
Final considerations
In the present research, the DNA Barcoding tool proved to be extremely efficient for the discrimination and correct identification of the species sampled in Bragança. The results showed cases of replacement, trade of endangered species and unrecorded species diversity. Our results confirm that common and commercial names are inaccurate which underestimate ichthyodiversity and may favour replacements and trade of endangered species. Although we have a regulation to establish the relationship of commercial and specific names, it is incomplete, inefficient and needs a reformulation, which considers the diversity of names and the different Brazilian regions, to propose a standard name for each species. We therefore present a list of correspondence between trade name and referent species, considering the trade of Teleosts in the coastal portion of the Brazilian Amazon (Supplementary Table S1).
Methods
Ethics statement
All individuals were obtained from points of sale, they were already dead. There was no need to apply the guidelines of the Institution's Ethics Committee. In the same way, it was not necessary to obtain a collection license, since individuals were purchased during the commercialization process, or donated by traders.
Sampling
The Bragança Free Market (“Feira Livre”) is the main place for selling fish, and is divided into two distinct environments, Market (“Mercado”) and Fair (“Feirinha”)12. The “Mercado” is supplied mainly by industrial fishing, aimed at target species of great commercial value, while the “Feirinha” is supplied mainly by artisanal fishing and that capture fish with less selectivity and greater diversity9,12,19. In this way, sample collection took place in both environments.
The collections were carried out monthly from April 2016 to February 2019, adding up to a total of 35 months of sampling. Fish samples were obtained through the purchase of whole individuals or donations from merchants. After tipping over, a sample of biological tissue was taken from each specimen. Three different tissues were used, depending on availability, tongue, fin and/or whole fish muscle. In this work, we adopted the term “commercial designation” to refer to the names observed during commercialization. For all collected commercial designations, whole individuals were fixed/preserved and incorporated, as exemplary testimonies, in the Ichthyological Collection of the Laboratory of Applied Genetics, of the Instituto of Coastal Studies, UFPA, Bragança. In addition, the photographic record has also been included for most commercial designations.
The whole acquired fish Specimens were identified to the taxonomic level possible, using morphological characters, through specialized literature4,10,41.
The biological tissue samples were stored in Eppendorf-type microtubes (2.0 mL), with 70% commercial alcohol and in a freezer at -20º C, for molecular analysis.
Laboratory procedures
Genomic DNA was obtained using the commercial Wizard Genomic Kit (PROMEGA), following the manufacturer's instructions. After isolation, the samples were mixed with blue juice buffer solution and GelRed dye (2μL of the mixture and 2μL of DNA) and subjected to horizontal underwater electrophoresis in agarose gel (1%) for 30 min/60 V. After the electrophoretic run, the samples were visualized under ultraviolet light to verify the quality of the extracted DNA.
The COI gene target fragment was amplified by Polymerase Chain Reaction (PCR), using the FishF1 and FishR1 and FishF2 and FishR2 primers described by42. The reaction consisted of a final volume of 15μL, and the amplification conditions were those used by42, with modifications in the hybridization temperatures to 53º C and 54º C.
After PCR, the positive samples were purified with Polyethylene Glycol (PEG) according to the protocol by43 and submitted to the sequencing reaction, using the dideoxyterminal method44, with reagents from Big Dye Kit (ABI PrismTMDye Terminator Cycle Sequencing Reading Reaction—Thermo Fisher). The precipitated products were subjected to capillary electrophoresis in an ABI 3500XL automatic sequencer (Thermo Fisher).
Sequences database and genetic analysis
The generated sequences were inspected manually, edited in the BioEdit v. 7.1.3.045 and automatically aligned using the CLUSTAL W application46. The DNAsp v 6 program47 was used to generate a list of haplotypes, to assist in the sample identification process.
For the specimen identification process, each haplotype was initially compared to sequences available in public banks, GenBank Platforms, more specifically in the Basic Local Alignment Search Tool (BLAST), in the “nucleotide blast” field48 and BOLD (Barcoding of Life Database)49. The maximum divergence level adopted for individuals of the same species was 2%22.
Species reference sequences, available at NCBI and BOLD Systems, were added to the haplotype database generated in the work for the construction of the Neighbor Clustering tree, using the Kimura-2-parameters evolutionary model50, in MEGA 1151 program with the significance of the clusters estimated by Bootstrap analysis, 1000 pseudoreplicas52.
The maximum and average Barcode gap distances of the sequences were evaluated on the BOLD Systems platform (http://www.boldsystems.org/index.php/MAS_Management_DataConsole?codes=CA), through the Barcode Gap Analysis tool and the possible presence of stop codon were also checked in the same plataform. To complement the genetic distance data, we also used the MEGA 11 Program50, based on the K2P evolutionary model49.
The choice of the best evolutionary model for Bayesian inference trees was obtained on the CIPRES Science Gateway v3.3 platform53, using jModelTest2 in XSEDE54, the analysis recommended the HKY +|I + G evolutionary model for the marine and freshwater species bank, based on the Bayesian Information Criterion (BIC).
The construction of the Bayesian inference tree (BI) was performed using BEAST v. 1.8.455,56. In the construction of the trees, a strict clock and the Yule speciation process were used. The posterior probability was estimated with three million generations and 10% burn-in. The log files were checked in Tracer v1.7.2 tool57 to assess chain convergence and proper burn-in length. The convergence chains considered adequate showed a value greater than > 200 ESS (effective sample size). Trees generated in BEAST were summarized in TreeAnnotator, v1.10.4, to obtain the best tree. FigTree, v1.4.458 was used to visualize the resulting tree.
Comparison of trade designations and threat status
Based on species identification, we compared the list of commercial names and corresponding species with the list of common names and respective scientific names provided by Normative Instruction No. 53 of September 1, 2020 (MAPA). We verified the threat status of each species in the red list of the International Union for the Conservation of Nature (IUCN) and in the Official List of Threatened Species of Brazil, Ordinance of the Ministry of the Environment no. 148, of June 07, 2022 (MMA no. 148)25.
The graph used to illustrate the categories obtained in this study was created in RAWGraphs (https://rawgraphs.io/).
Data availability
The data sets generated during and analyzed during the present study are available in the repository of the National Center for Biotechnology Information, with the codes OR459502-OR459617 and OR515260-OR515262.
References
Ekau, W. & Knoppers, B. An introduction to the pelagic system of the North-East and East Brazilian shelf. Arch. Fish. Mar. Res. 47, 113–132 (1999).
Camargo, M. & Isaac, V. Os peixes estuarinos da região Norte do Brasil: lista de espécies e considerações sobre sua distribuição geográfica. Bol. Mus. Para. Emílio Goeldi Zoo. 17, 133–157 (2001).
Marceniuk, A. P. et al. The bony fishes (Teleostei) caught by industrial trawlers off the Brazilian North coast, with insights into its conservation. Neotrop. Ichthyol. 17, 2 (2019).
Marceniuk, A. P. et al. Teleostei fishes of the North Coast of Brazil. Revista CEPSUL-Biodiversidade e Conservação Marinha. 10, e2021006–e2021006 (2021).
Marceniuk, A. P., Caires, R. A., Wosiacki, W. B. & Dario, F. D. Conhecimento e conservação dos peixes marinhos e estuarinos (Chondrichthyes e Teleostei) da costa Norte do Brasil. Biota Neotrop. 13, 251–259 (2013).
Isaac, V. J., Espírito Santo, R. V. & Nunes, J. L. G. A estatística pesqueira no litoral do Pará: resultados divergentes. Pan-Am. J. Aquat. Sci. 3, 205–213 (2008).
Furtado Júnior, I., Tavares, M. C. S. & Brito C. S. F. Estatísticas das produções de pescado estuarino e marítimo do estado do Pará e políticas pesqueiras. Bol. Mus. Para. Emílio Goeldi Ciênc. hum. 1, 95–111 (2006).
Espírito Santo, R.D., Isaac. V. J., Silva, L. M. A., Martinelli, J. M., Higuchi, H., E Saint-Paul, U. Peixes e camarões do litoral bragantino, Pará, Brasil. Belem: Madam, 268p. (2005).
Braga, C. F., Espírito Santo, R. V., Silva, B. B., Giarrizzo, T. & Castro, E. R. Considerações sobre a comercialização do pescado no município de Bragança -PA. Bol. Téc. Cient. Cepnor. 6, 105–120 (2006).
Marceniuk, A. P., Caires, R. A., Rotundo, M. M., Alcântara, R. A. K. & Wosiacki, W. B. The icthyofauna (Teleostei) of the Rio Caeté estuary, northeast Pará, Brazil, with a species identification key from northern Brazilian coast. Pan-Am. J. Aquat. Sci. 12, 31–79 (2017).
Isaac, V. J. & Barthem, R. B. Os recursos pesqueiros da Amazônia Brasileira. Bol. Mus. Para. Emílio Goeldi Sér. Ant. 11, 295–339 (1995).
Martins, T. S. et al. Diversity and abundance of commercialized fish in northeastern Pará, coastal amazon: The case of the street market in Bragança–pa. Arq. Ciênc. Mar. 54, 27–43 (2021).
Keskin, E. & Atar, H. H. DNA barcoding commercially important fish species of Turkey. Mol. Ecol. Resour. 13, 788–797 (2013).
Ardura, A., Linde, A. R., Moreira, J. C. & Garcia-Vazquez, E. DNA barcoding for conservation and management of Amazonian commercial fish. Biol. Conserv. 143, 1438–1443 (2010).
Previero, M., Minte-Vera, C. V. & Moura, R. L. Fisheries monitoring in Babel: Fish ethnotaxonomy in a hotspot of common names. Neotrop. Ichthyol. 11, 467–476 (2013).
Carvalho, D. C., Palhares, R. M., Drummond, M. G. & Frigo, T. B. DNA Barcoding identification of commercialized seafood in South Brazil: A governmental regulatory forensic program. Food Control. 50, 784–788 (2015).
Martins, T. et al. Intensive commercialization of endangered sharks and rays (Elasmobranchii) along the coastal Amazon as revealed by DNA barcode. Front. Mar. Sci. 8, 769908 (2021).
Barbosa, A. J., Sampaio, I. & Santos, S. Re-visiting the occurrence of mislabeling in frozen “pescada-branca” (Cynoscion leiarchus and Plagioscion squamosissimus–Sciaenidae) sold in Brazil using DNA barcoding and octaplex PCR assay. Food Res. Int. 143, 110308 (2021).
Espírito-Santo, R. V. & Isaac, V. J. Desembarques da pesca de pequena escala no município de Bragança-PA, Brasil: Esforço e produção. Bol. Lab. Hidrobiol. 25, 31–48 (2012).
Freire, J. L., Sarmento, G. C., Lutz, Í., Bentes, B. & Isaac, V. J. New insight into the reproductive biology and catch of juveniles of the Lutjanus purpureus in a portion of the Great Amazon reef system off the Northern Brazilian Coast. Front. Mar. Sci. 9, 804648 (2022).
Pires, A. C. & Marinoni, L. DNA barcoding and traditional taxonomy unified through Integrative Taxonomy: A view that challenges the debate questioning both methodologies. Biota Neotropica 10, 339–346 (2010).
Hebert, P. D. N., Cywinska, A., Ball, S. L. & deWaard, J. R. Biological identifications through DNA Barcodes. Proc. R. Soc. B: Biol. Sci. 270, 313–332 (2003).
Gomes, G. et al. Forensic analysis reveals fraud in fillets from the “Gurijuba” Sciades parkeri (Ariidae-Siluriformes): A vulnerable fish in Brazilian Coastal Amazon. Mitochondrial DNA A. Seq. Anal. 30, 1–9 (2019).
Guimarães, K. L. A., Rosso, J. J., Souza, M. F. B., Astarloa, J. M. D. & Rodrigues, L. R. R. Integrative taxonomy reveals disjunct distribution and first record of Hoplias misionera (Characiformes: Erythrinidae) in the Amazon River basin: morphological (2021).
Brasil. Portaria MMA no. 148, de 7 de junho de 2022. Atualização da Lista Nacional de Espécies Ameaçadas de Extinção. Diário Oficial da União. p.74. 8 jun. 2022. https://www.icmbio.gov.br/cepsul/destaques-e-eventos/704-atualizacao-da-lista-oficial-das-especies-ameacadas-de-extincao.html (2022).
IUCN. International Union for Conservation of Nature. Red List of Threatened Species. Available online at: https://www.iucnredlist.org (accessed 10 april, 2023).
Guimarães-Costa, A. J. et al. Fish diversity of the largest deltaic formation in the Americas-a description of the fish fauna of the Parnaíba Delta using DNA Barcoding. Sci. Rep. 9, 1–8 (2019).
Lutz, I. et al. Quality analysis of genomic DNA and authentication of fisheries products based on distinct methods of DNA extraction. PLoS One. 18(2), e0282369 (2023).
Freire, J. L., Silva, B. B. & Souza, A. Aspectos econômicos e higiênico-sanitários da comercialização do pescado no município de Bragança (PA). Biota Amazônica. 1, 17–28 (2011).
DNA barcoding and cytogenetic considerations. Neotrop. Ichthyol. 19, 1–20 (2021).
Rosso, J. J. et al. Integrative taxonomy reveals a new species of the Hoplias malabaricus species complex (Teleostei: Erythrinidae). Ichthyol. Explor. Freshw. 1, 1–18 (2018).
Jacobina, U. P. et al. DNA barcode sheds light on systematics and evolution of neotropical freshwater trahiras. Genetica 146, 505–515 (2018).
Marceniuk, A. P. et al. Incipient speciation, driven by distinct environmental conditions, in the marine catfishes of the genus Aspistor (Siluriformes, Ariidae), from the Atlantic coast of South America. J. Zool. Syst. Evol. Res. 57, 400–417 (2019).
Menezes, N. A., Nirchio, M., Oliveira, C. D. & Siccharamirez, R. Taxonomic review of the species of Mugil (Teleostei: Perciformes: Mugilidae) from the Atlantic South Caribbean and South America, with integration of morphological, cytogenetic and molecular data. Zootaxa. 3918, 1–38 (2015).
Durand, J. D., Hubert, N., Shen, K. N. & Borsa, P. DNA barcoding grey mullets. Rev. Fish Biol. Fish. 27, 233–243 (2017).
Sousa, R. P. D. et al. Range distribution and contributions to taxonomy of Elops smithi (Elopiformes: Elopidae). An. Acad. Bras. Cienc. 91, e20181240 (2019).
Sousa, R. P. C. et al. Exploring the diversity of elopidae (Teleostei; Elopiformes) using DNA barcoding analysis. Diversity. 14, 1008 (2022).
Carvalho, C. O. et al. Integrative taxonomy of the species complex Haemulon steindachneri (Eupercaria; Haemulidae) with a description of a new species from the western Atlantic. Zoology. 141, 125782 (2020).
Brasil. MPA. Ministério da Pesca e Aquicultura. Boletim estatístico de pesca e aquicultura 2011. https://www.icmbio.gov.br/cepsul/images/stories/biblioteca/download/estatistica/est_2111_bol__bra.pdf (2013).
Brasil. Instituto Brasileiro do Meio Ambiente e Recursos Naturais Renováveis. Portaria no 121, de 20 de setembro de 2002. Proíbe, nas águas jurisdicionais brasileiras, por um período de cinco anos, a captura de Epinephelus itajara. Diário Oficial da União: seção 1, Brasília, DF, n. 184, 23 set. 2002 https://www.icmbio.gov.br/cepsul/images/stories/legislacao/Portaria/2002/p_ibama_121_2002_moratoria5anospescadomero_alterada_p_ibama_42_2007.pdf (2002).
Figueiredo, J. L. & Menezes N. A. Manual de peixes marinhos do sudeste do Brasil: Teleostei. Universidade de São Paulo. (1980).
Ward, R. D., Zemlak, T. S., Innes, B. H., Last, P. R. & Hebert, P. D. DNA barcoding Australia’s fish species. Philos. Trans. R. Soc. Lond. B: Biol. Sci. 360, 1847–1857 (2005).
Paithankar, K. R. & Prasad, K. S. Precipitation of DNA by polyethylene glycol and ethanol. Nucleic Acids Res. 19, 1346 (1991).
Sanger, F., Nicklen, S. & Coulson, A. R. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. U.S.A. 74, 5463–5467 (1977).
Hall, T. A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41, 95–98 (1999).
Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F. & Higgins, D. G. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25, 4876–4882 (1997).
Rozas, J. et al. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 34, 3299–3302 (2017).
Altschul, S. F. et al. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 (1997).
Ratnasingham, S., & Hebert, P. D. BOLD: The barcode of life data system (http://www.barcodinglife.org). Mol. Ecol. 7, 355–364 (2007).
Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16, 111–120 (1980).
Tamura, K., Stecher, G. & Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38, 3022–3027 (2021).
Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution. 39, 783–791 (1985).
Miller, M. A., Pfeiffer, W., & Schwartz, T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In 2010 gateway computing environments workshop (GCE). Ieee. pp. 1–8. (2010).
Darriba, D., Taboada, G. L., Doallo, R. & Posada, D. jModelTest 2: more models, new heuristics and parallel computing. Nat. Methods. 9, 772–772 (2012).
Drummond, A. J. & Rambaut, A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7, 214 (2007).
Drummond, A. J., Suchard, M. A., Xie, D. & Rambaut, A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 29, 1969–1973 (2012).
Rambaut, A., Drummond, A. J., Xie, D., Baele, G. & Suchard, M. A. Posterior summarization in Bayesian phylogenetics using tracer 1.7. Syst. Biol. 67, 901–904 (2018).
Rambaut, A. FigTree, a graphical viewer of phylogenetic trees (Version 1.4. 4). Institute of evolutionary biology, University of Edinburgh. (2018).
Guimarães-Costa, A. J et al. T. DNA Barcoding for the assessment of the taxonomy and conservation status of the fish bycatch of the northern Brazilian shrimp trawl fishery. Front. Mar. Sci. 7, 566021 (2020).
Acknowledgements
This work was financed by CNPq, through the Universal Call (process: 439113/2018-0) and by the Coordination for the Improvement of Higher Education Personnel (CAPES), through the granting of a doctoral scholarship for the doctoral candidate Paula Santana. Article processing fees were granted by the Qualified Publication Support Program (PAPQ)/Federal University of Pará (UFPA).
Author information
Authors and Affiliations
Contributions
P.S: literature review; sample collection; sample processing, image database, tissue database; laboratory experiments; sequence alignment; genetic analyses; analyses in public databases; interpretation and writing of results. T.M.: sample collection; sample processing; image database; tissue database; image editing. J.M.: laboratory experiments; sequence alignment; genetic analyzes. I.L.: sample collection; sample processing; image database; tissue database; image editing. R.S.: sample collection; genetic analyzes. D.M.: sample collection; sample processing. R.M.: sample collection; sample processing. I.V.: sample collection; sample processing. M.V.: financing; article correction and analysis. I.S.: financing; article correction and analysis. G.E.G.: project design; financing; literature review; laboratory experiments; sequence alignment; genetic analyses; analyses in public databases; interpretation and writing of results; article correction and analysis.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Santana, P., Martins, T., Lutz, Í. et al. DNA barcode reveals occurrence of threatened species and hidden diversity on Teleost fish trade in the Coastal Amazon. Sci Rep 13, 19749 (2023). https://doi.org/10.1038/s41598-023-47063-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-47063-2
- Springer Nature Limited