Abstract
This study aimed to characterize the molecular features and virulence profiles of carbapenem-resistant Acinetobacter baumannii (CRAB) isolates. Clinical CRAB isolates were obtained from blood cultures of adult patients with CRAB bacteremia, collected between July 2015 and July 2021 at a Korean hospital. Real-time polymerase chain reaction was used to detect 13 virulence genes, genotyping was conducted via multilocus sequence typing (MLST), and a Tenebrio molitor infection model was selected for survival analysis. Herein, 170 patients, from whom CRAB isolates were collected, showed the in-hospital mortality rate of 57.6%. All 170 clinical CRAB isolates harbored blaOXA-23 and blaOXA-51. MLST genotyping identified 11 CRAB sequence types (STs), of which ST191 was predominant (25.7%). Virulence genes were distributed as follows: basD, 58.9%; espA, 15.9%; bap, 92.4%; and ompA, 77.1%. In the T. molitor model, ST195 showed a significantly higher mortality rate (73.3% vs. 66.7%, p = 0.015) than the other groups. Our findings provide insights into the microbiological features of CRAB blood isolates associated with high mortality. We suggest a potential framework for using a T. molitor infection model to characterize CRAB virulence. Further research is warranted to elucidate the mechanisms by which virulence improves clinical outcomes.
Similar content being viewed by others
Introduction
Acinetobacter baumannii is a significant nosocomial pathogen with attributable mortality rates ranging from 8.4 to 36.5%1. Bacteremia and ventilator-associated pneumonia are major nosocomial infections caused by this pathogen. Owing to its multidrug resistance and virulence, A. baumannii is associated with unfavorable treatment outcomes. Carbapenem-resistant A. baumannii (CRAB) is ranked as a high-priority antibiotic-resistant pathogen and is designated as an urgent threat by the Centers for Disease Control and Prevention in the United States2. According to the Korean Antimicrobial Resistance Monitoring System and Korean Nosocomial Infections Surveillance System data, the resistance rate of A. baumannii to imipenem in the Republic of Korea (ROK) will increase to 85% in 2015 and 93.5% in 2022, thereby representing a major public health threat3,4.
Identifying the molecular features and mechanisms of virulence is crucial for containing transmission and reducing mortality associated with an increasing number of CRAB infections. In the ROK, a drastic increase in Acinetobacter isolates with blaOXA-23 genes has been observed since the mid-2000s5. Moreover, ST191 A. baumannii harboring blaOXA-23 has been held responsible for the high rate of carbapenem resistance recorded in the ROK between 2009 and 20126,7. In addition to the complex mechanism of antibiotic resistance, CRAB carries several virulence genes that may result in clinical deterioration in patients with other co-morbidities; though, little is known about the distribution and clinical implications of the virulence genes in this bacterium. Previous studies suggested that several virulence genes are prevalent in A. baumannii isolates and may be associated with antibiotic resistance8,9,10,11. Recent evidence suggests that a hypervirulent strain of CRAB has evolved (theory of evolution), and is associated with high mortality and clonal transmission within hospitals12,13,14. However, few studies have focused on the distribution of virulence genes via multilocus sequence typing (MLST) of CRAB blood isolates. The aim of this study was to investigate the molecular characteristics of CRAB blood isolates and evaluate the survival rates of Tenebrio molitor that had been infected with CRAB strains exhibiting different molecular features.
Methods
Hospital setting and study design
This prospective cohort study was performed in a 1048-bed, university-affiliated hospital in ROK. Clinical CRAB isolates were obtained from blood cultures of adult patients (≥ 18 years of age) diagnosed with CRAB bacteremia between July 2015 and July 2021. Patients with polymicrobial bacteremia were excluded from the study. Patients had multiple episodes of CRAB bacteremia, and only the first episode was included. Clinical information associated with CRAB bacteremia was collected from the electronic medical records of each patient, including age, sex, source of CRAB bacteremia, and mortality. This study was approved by the Institutional Review Board of the Korea University Anam Hospital [No. 2022AN0292], and the requirement for written informed consent was waived. This study was conducted in accordance with the ethical guidelines and regulations outlined in the Declaration of Helsinki.
Bacterial strains and antibiotic susceptibility test
All isolates were collected on day 1 of bacteremia. Non-repetitive CRAB isolates were identified using a MicroScan WalkAway-96 Plus system (Beckman Coulter, Inc., Fullerton, CA, USA) using a routine laboratory diagnostic process. Two reference bacterial strains, namely Escherichia coli ATCC® 25922 and Pseudomonas aeruginosa ATCC® 27853, were used for the internal quality control of the antibiotic susceptibility test. Minimal inhibitory concentrations (MICs) were determined using the agar dilution method and the breakpoints of the antibiotic susceptibility tests were interpreted according to the 2017 Clinical Laboratory Standards Institute guidelines15. Carbapenem resistance was defined as resistance to imipenem, at MICs of ≥ 8 μg/mL15.
Virulence and antimicrobial resistant genes
Multiplex real-time polymerase chain reaction (PCR) assays were performed to detect virulence and antimicrobial resistance genes in CRAB isolates16,17,18. Thirteen virulence genes (basD, espA, ompA, bap, bfmR, pbpG, fhaB, cpaA, ata, recA, lipA, abeD, and chop) and two carbapenemase genes (blaOXA-23 and blaOXA-51) were detected using specific primers and positive controls, which were previously confirmed as positive by sequencing (Supplementary Table S1). Virulence genes were associated with siderophore synthesis (basD), bacterial adhesion (espA, bap, ata, chop), biofilm formation (ompA, pbpG), glycoconjugates (bfmR), host cell death (fhaB, abeD), toxin production (cpaA, lipA), and stress response (recA)19,20,18,21. The PCR cycling conditions were as follows: denaturation at 95 ℃ for 5 min, annealing between 50 ℃ and 61 ℃ for 30–45 s, and extension at 72 ℃ for 1 min (for 34 cycles). The presence of amplified PCR products was determined by electrophoretic separation on 2% agarose gels and visualized using a transilluminator.
Genotypes according to MLST
For MLST, seven housekeeping genes (gltA, gyrB, gdhB, recA, cpn60, gpi, and rpoD) of A. baumannii were amplified and sequenced for A. baumannii, as described previously22, using specific primers (Supplementary Table S2). MLST was performed using the Oxford scheme, and the sequence types (STs) were assigned using tools available in the A. baumannii MLST database (https://pubmlst.org/organisms/acinetobacter-baumannii).
Virulence assay
A virulence assay was performed using T. molitor larvae (mealworms) as described previously23. Larvae were maintained in a plastic box containing wheat bran. CRAB isolates were placed in a brain–heart infusion broth and incubated overnight at 37 °C. Next, subcultures were grown on blood agar plates that were incubated for 24 h at 37 °C. The cultured CRAB were then suspended in insect saline (130 mM NaCl, 5 mM KCl, and 1 mM CaCl2) at a concentration of 10 CFU/mL. Using an insulin syringe, 10 µL of the CRAB suspension (at 106 CFU/µl) was injected into a T. molitor larva; in the control group, 10 µL of sterilized insect saline solution was injected. However, a positive control group has not been established. Post injection, all larvae were raised in Petri dishes for 72 h at 25 °C. Survival rates were calculated according to melanization (using three sets of five larvae per strain).
Statistical analyses
Categorical variables were summarized as frequencies and analyzed using Pearson’s chi-square or Fisher’s exact test. Continuous variables were expressed as medians with interquartile ranges and analyzed using the Student’s t test. The difference in the mortality rates of T. molitor larvae infected with CRAB isolates containing different STs was evaluated using a one-way analysis of variance. Statistical significance was set at p < 0.05. SPSS v23.0 for Windows software (SPSS Inc., Chicago, IL., USA) was used to conduct statistical analyses.
Results
Patients with CRAB bacteremia
During the study period, 170 patients, from whom CRAB isolates were collected, were enrolled. The mean age of the patients was 66 years (inter quartile range, 56–77 years), and 60.0% were male. The most common source of CRAB bacteremia was catheter-related bloodstream infections (50.6%), followed by primary bloodstream infections (23.5%), intra-abdominal infections (16.5%), pneumonia (8.2%), and skin and soft tissue infections (0.6%). Seventy-three patients (43.5%) experienced septic shock. The in-hospital and 30-day mortality rates were 57.6% and 21.2%, respectively.
Antimicrobial susceptibility of CRAB isolates
A total of 170 CRAB blood isolates were collected; all were resistant to three or more antimicrobial agents and were therefore considered multidrug-resistant. Antibiotic resistance patterns of the isolates are listed in Table 1. The highest resistance rates were observed for ciprofloxacin and imipenem (100%), followed by those for piperacillin (99.4%), cefepime (99.4%), ampicillin/sulbactam (97.6%), gentamicin (90%), and minocycline (10%). blaOXA-23 and blaOXA-51 were identified in all strains (Table 2).
MLST analysis of CRAB isolates
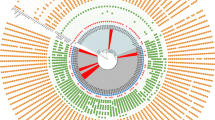
MLST analysis revealed 11 different STs in 160 (94.1%) CRAB isolates: ST191, ST195, ST208, ST357, ST369, ST451, ST469, ST491, ST784, ST1599, and ST1653 (Supplementary Table S3). Of these, ST191 was the most abundant, occurring in 49 (28.8%) isolates. ST191 was mostly isolated between 2015 and 2018, whereas ST195 was mainly found in samples collected between 2019 and 2021. ST451 (n = 27) was detected throughout the study. ST491 (n = 4), ST1599 (n = 9), ST469 (n = 2), and ST357 (n = 1) were detected after 2017 (Fig. 1).
Distribution of virulence factors in CRAB isolates
The distribution of virulence genes was as follows: 58.9% basD (siderophore synthesis); 15.9% espA, 92.4% bap, 86.5% ata, and 7.1% chop (bacterial adhesion); 77.1% ompA and 93.5% pbpG (biofilm formation); 92.9% bfmR (glycoconjugates); 70.6% fhaB and 99.4% abeD (host cell death); 0.6% cpaA and 99.4% lipA (toxin production); and 100% recA (stress response) (Table 2). We found that one isolate carried 12 virulence genes, 11 isolates carried 11 virulence genes, 49 isolates carried 10 virulence genes, 52 isolates carried nine virulence genes, 37 isolates carried eight virulence genes, 14 isolates carried seven virulence genes, five isolates carried six virulence genes, and one isolate carried five virulence genes. Occasionally, the frequency of a virulence gene differed according to the ST (Table 2).
Virulence assay
The overall mortality rate of CRAB-inoculated T. molitor was 70.4%. The mortality rates in our T. molitor model showed the following differences depending on the virulence genes carried by CRAB isolates: 71.9% (1079/1500) basD (siderophore synthesis); 72.6% (294/405) espA, 68.8% (1620/2355) bap, 68.1% (1502/2205) ata, and 71.7% (129/180) chop (bacterial adhesion); 69.9% (1374/1965) ompA and 68.1% (1625/2385) pbpG (biofilm formation); 68.3% (1619/2370) bfmR (glycoconjugates); 67.9% (1222/1800) fhaB and 68.5% (1737/2535) abeD (host cell death); 80.0% (12/15) cpaA and 68.5% (1737/2535) lipA (toxin production); and 68.5% (1747/2550) recA (stress response). The mortality rates of T. molitor larvae infected with CRAB isolates were statistically significantly different among the different STs (Table 3): ST1599 vs. ST191 (p = 0.004), ST1599 vs. ST195 (p = 0.002), ST1599 vs. ST208 (p = 0.042), and ST1599 vs. ST369 (p = 0.01).
Differences in microbiological characteristics of CRAB among major STs
CRAB isolates with ST191 displayed a significantly lower rate of resistance to minocycline than the non-ST191 group (2.0% vs. 13.3%, p = 0.027), whereas ST451 isolates showed a higher rate of resistance to minocycline than the non-ST451 group (33.3% vs. 5.6%, p < 0.001). The ST195 group showed a lower tigecycline resistance rate than the non-ST195 group (4.2% vs. 22.1%, p = 0.042). Additionally, the ST191 group showed a higher prevalence than the non-ST191 group in virulence genes such as fhaB (host cell death; 98% vs. 59.5%, p < 0.001) and ata (bacterial adhesion; 98.0% vs. 81.8%, p = 0.005). In contrast, the ST195 group was more predominant than the non-ST195 group in terms of basD (siderophore synthesis: 92.9% vs. 52.1%, p < 0.001) and ompA (biofilm formation: 100% vs. 72.5%, p = 0.002) genes. Notably, the ST195 group showed a significantly higher T. molitor mortality rate than the non-ST195 group (73.3 vs. 66.7%, p = 0.015), whereas the mortality rate was lower in the ST451 group than in the non-ST451 group (60.0% vs. 73.3%, p = 0.007) (Table 4).
Discussion
We demonstrated an evolutionary change in CRAB from blood isolates collected from a hospital, as evidenced by the emergence of new bacterial STs. During the study period, ST191, harboring the blaOXA-23 carbapenemase gene, was the most prevalent genotype, and a marked increase in genomic variation was observed after 2019. To the best of our knowledge, this study is the first to evaluate the virulence of CRAB isolates and the associated mortality in a T. molitor larval infection model.
CRAB isolates displayed an extensive drug-resistant phenotype, being highly resistant to all clinically available antimicrobial agents, which is consistent with previous findings in the ROK9,24. Our results also revealed that all CRAB isolates harbored blaOXA-23, which previous studies have identified as the most prevalent carbapenemase gene carried by A. baumannii in Asian countries25,26,27,28. Our findings suggest that the genetic evolution enabling blaOXA-23 expression in CRAB contributes to this bacterium, establishing it as a highly successful nosocomial pathogen in clinical settings.
Furthermore, ST191 was the predominant CRAB isolate until 2018, whereafter ST195 became more prevalent. In 2018, ST195 was the most prevalent ST in China29. The same shift in the predominant ST from ST191 to ST195 was also reported in Hong Kong following the emergence of CRAB ST195 harboring the blaOXA-23 gene30. Our study showed that the ST195 harboring the blaOXA-23 and blaOXA-51 genes was associated with higher mortality in a T. molitor infection model, compared to those of the non-ST195. The shift in the predominant sequence type of CRAB strains from ST191 to ST195 may be an adaptation through the acquisition of higher virulence. In contrast, our study demonstrated that ST451 first emerged in 2016 and was steadily discovered during the study period. Similarly, a previous study in the ROK reported that ST451 was the most prevalent ST between 2016 and 2018 in the context of clonal evolution related to antimicrobial resistance9. Indeed, all the prevalent STs recorded (ST191, ST195, and ST451—are well-known multidrug-resistant clones, causing a decrease in the likelihood of appropriate antibiotic selection11. There appears to be a clonal spread of several epidemic lineages that predominate over the rest and play a pivotal role in nosocomial transmission.
Several virulence factors may influence disease progression to critical illness and are related to functions, such as transmission, binding to host structures, cellular damage, and invasion. Consistent with previous studies, we found that stress response-associated recA was present in all CRAB isolates10. The remaining virulence genes showed differential distributions according to ST. A correlation between virulence genes and multidrug-resistant phenotypic differences in CRAB isolates has been previously suggested9,10,11. Selection pressure may drive the formation of virulence factors and the acquisition of multidrug resistance during the process of adaptation, facilitating the survival of these isolates in a clinical setting.
Previous reports suggested a positive relationship between the expression of virulence genes associated with biofilm formation and antibiotic resistance in A. baumannii isolates31,32,33. Biofilm formation is responsible for various types of medical device-related infections. Notably, CRAB strains, as biofilm-formers, are also advantageous for survival and dissemination in the hospital environment and the acquisition of antimicrobial resistance33. Although our study did not confirm the biofilm-forming capacity of the CRAB isolates, we assessed the frequency of biofilm-associated gene occurrence in CRAB harboring blaOXA-23, recording 15.9, 92.4, 86.5, 7.1, 77.1, and 93.5% for espA, bap, ata, chop, ompA, and pbpG, respectively. In particular, ompA and bap are essential for bacterial adhesion to human epithelial cells, development of biofilms, and antimicrobial resistance34,35. The virulence genes identified in our study may lead to treatment challenges owing to their high pathogenicity and antimicrobial resistance36. In Iran and Korea, A. baumannii isolates harboring ompA (81% and 69%) and bap (92% and 100%), respectively, have been similarly identified32,37,38,39. Surveillance and control measures against CRAB isolates containing biofilm-associated genes are essential to prevent their emergence and transmission.
Furthermore, we evaluated the virulence of the CRAB blood isolates using T. molitor larvae as an infection model. Mealworms are widely used to assess the pathogenic virulence of bacteria such as Listeria monocytogenes, Staphylococcus aureus, and Aeromonas hydrophila40,41,42. Galleria mellonella is another reliable model for the evaluation of CRAB strain pathogenicity43. Although none of the insect species can replace mammalian models, T. molitor larvae offer an attractive alternative for investigating host–pathogen interactions involving CRAB isolates because of their cost-effectiveness and ease of handling for housing and breeding44. In addition, its short lifecycle allows for large-scale experiments over a short period. In addition to sharing all of the merits already listed for G. mellonella, T. molitor larvae are larger.
Interestingly, our study showed that the mortality rates of T. molitor larvae infected with CRAB isolates differed according to the ST. The relative rarity of some STs, resulting in a correspondingly small sample size of related blood isolates, prevented us from drawing definite conclusions. Nevertheless, ST195 larvae exhibited a significantly higher mortality rate than the non-ST195 group, whereas ST451 mealworms exhibited a lower mortality rate than the non-ST451 group. ST191 also displayed a trend of a higher mortality rate among larvae than the non-ST191 group, although this difference was not statistically significant. A previous study reported that patients infected with ST191, ST195 or ST208 may develop severe infections accompanied by organ damage45. Furthermore, ST191 and ST195 strains are considered highly virulent in serum complement killing assays and G. mellonella models45,46. In a clinical study with similar results, Cox regression multivariate analysis found that ST191 was a risk factor associated with 30-day mortality in patients with A. baumannii bloodstream infections, whereas ST451 served as a protective factor11. The distribution of STs varies according to geographical location14,47. In Taiwan, ST218 harboring blaOXA-72 gene (which was not detected in our study) was also reported as a hypervirulent strain, raising the possibility of intra-hospital transmission and mortality14.
Our study demonstrated that ST191 isolates exhibited a significantly lower resistance rate to minocycline than non-ST191 isolates, whereas ST451 showed the opposite. Considering that the most dominant ST is shifting from ST191 to ST451, it is evident that CRAB isolates are becoming more multidrug-resistant and virulent. Consequently, delays in appropriate antibiotic administration and rapid disease progression may have synergistic effects, contributing to treatment failure. Antibiotic susceptibility, genetic type, and virulence genes of CRAB blood isolates that constitute the microbiological characteristics may be related to each other.
Our study has some limitations. First, all CRAB blood isolates were collected from a single hospital in the ROK, and generalizing the results to other regions or healthcare facilities may be problematic. Further studies with larger sample sizes and in different clinical settings are necessary to fully understand the microbiological characteristics of CRAB. Second, we analyzed only a limited number of virulence types and antibiotic resistance genes, disregarding other factors that could potentially contribute to CRAB infection pathogenicity. In addition, the presence of virulence and antibiotic resistance genes may not necessarily indicate their expression. However, clear evidence is needed to confirm whether their expression is related to pathogenicity. Finally, our virulence assay relied on an insect model, which may not accurately reflect the virulence of human CRAB isolates. Compared to non-ST191, the relatively high mortality rate of ST191 identified in T. molitor infection models should be verified in patients with CRAB bacteremia. In future, various clinical variables and microbiological characteristics should be simultaneously considered as confounding variables to identify significant risk factors related to treatment outcomes.
Conclusion
Our findings showed that ST191 and ST195, which are associated with high antimicrobial resistance and virulence in the ROK, are prevalent in clinical settings. Insights into the dynamic changes in antibiotic resistance mechanisms and virulence factors among CRAB isolates are necessary for the effective control and treatment of CRAB infections. We propose the use of the T. molitor infection model as an alternative approach for evaluating the pathogenicity of CRAB isolates. Research with larger sample sizes obtained from a variety of settings is essential to further explore the mechanism of virulence of CRAB isolates to reduce the mortality associated with CRAB bacteremia.
Data availability
The datasets generated in the current study are available in the NCBI GenBank database under accession numbers OR498912–OR499081, [https://www.ncbi.nlm.nih.gov/nucleotide/]. Further information is provided by the corresponding author upon request.
References
Falagas, M. E. & Rafailidis, P. I. Attributable mortality of Acinetobacter baumannii: No longer a controversial issue. Crit Care 11, 1–3. https://doi.org/10.1186/cc5911 (2007).
CDC. Antibiotic resistance threats in the United States. Atlanta, GA: U.S. Department of Health and Human Services, CDC. https://www.cdc.gov/drugresistance/biggest-threats.html (2019).
Kim, D. et al. Increasing resistance to extended-spectrum cephalosporins, fluoroquinolone, and carbapenem in gram-negative bacilli and the emergence of carbapenem non-susceptibility in Klebsiella pneumoniae: Analysis of Korean Antimicrobial Resistance Monitoring System (KARMS) data from 2013 to 2015. Ann. Lab. Med. 37, 231–239. https://doi.org/10.3343/alm.2017.37.3.231 (2017).
Korean nosocomial infections surveillance system. Intensive care unit module report: data summary July 2021 through June 2022. http://konis.cafe24.com/xe/reports_icu_y.
Lee, Y. et al. Increasing prevalence of blaOXA-23-carrying Acinetobacter baumannii and the emergence of blaOXA-182-carrying Acinetobacter nosocomialis in Korea. Diagn. Microbiol. Infect. Dis. 77, 160–163. https://doi.org/10.1016/j.diagmicrobio.2013.06.009 (2013).
Gurung, M. et al. Emergence and spread of carbapenem-resistant Acinetobacter baumannii sequence type 191 in a Korean hospital. Infect. Genet. Evol. 19, 219–222. https://doi.org/10.1016/j.meegid.2013.07.016 (2013).
Selasi, G. N. et al. Genetic basis of antimicrobial resistance and clonal dynamics of carbapenem-resistant Acinetobacter baumannii sequence type 191 in a Korean hospital. Infect. Genet. Evol. 36, 1–7. https://doi.org/10.1016/j.meegid.2015.09.001 (2013).
Ayoub Moubareck, C. & Hammoudi Halat, D. Insights into Acinetobacter baumannii: A review of microbiological, virulence, and resistance traits in a threatening nosocomial pathogen. Antibiotics 9, 119. https://doi.org/10.3390/antibiotics9030119 (2013).
Jun, S. H. et al. Clonal change of carbapenem-resistant Acinetobacter baumannii isolates in a Korean hospital. Infect. Genet. Evol. 93, 104935. https://doi.org/10.1016/j.meegid.2021.104935 (2021).
Bahador, A. et al. Association of virulence gene expression with colistin-resistance in Acinetobacter baumannii: Analysis of genotype, antimicrobial susceptibility, and biofilm formation. Ann. Clin. Microbiol. Antimicrob. 17, 24. https://doi.org/10.1186/s12941-018-0277-6 (2018).
Yoon, E. J. et al. Counter clinical prognoses of patients with bloodstream infections between causative Acinetobacter baumannii clones ST191 and ST451 belonging to the international clonal lineage II. Front. Public Health 7, 233. https://doi.org/10.3389/fpubh.2019.00233 (2019).
Jones, C. L. et al. Fatal outbreak of an emerging clone of extensively drug-resistant Acinetobacter baumannii with enhanced virulence. Clin. Infect. Dis. 61, 145–154. https://doi.org/10.1093/cid/civ225 (2015).
Ou, H. Y. et al. Complete genome sequence of hypervirulent and outbreak-associated Acinetobacter baumannii strain LAC-4: Epidemiology, resistance genetic determinants and potential virulence factors. Sci. Rep. 5, 1–13. https://doi.org/10.1038/srep08643 (2015).
Liu, C. P., Lu, H. P. & Luor, T. Clonal relationship and the association of the ST218 strain harboring blaOXA-72 gene to mortality in carbapenem-resistant Acinetobacter baumannii bacteremia. J. Microbiol. Immunol. Infect. 52, 297–303. https://doi.org/10.1016/j.jmii.2018.10.005 (2019).
Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing 27th edn. (Clinical and Laboratory Standards Institute, 2017).
Chen, T. L. et al. Comparison of one-tube multiplex PCR, automated ribotyping and intergenic spacer (ITS) sequencing for rapid identification of Acinetobacter baumannii. Clin. Microbiol. Infect. 1, 801–806. https://doi.org/10.1111/j.1469-0691.2007.01744.x (2007).
Woodford, N. et al. Multiplex PCR for genes encoding prevalent OXA carbapenemases in Acinetobacter spp. Int. J. Antimicrob. Agents 27, 351–353. https://doi.org/10.1016/j.ijantimicag.2006.01.004 (2006).
Mozafari, H., Mirkalantari, S., Kalani, B. S. & Amirmozafari, N. Prevalence determination of virulence related and biofilm formation genes in Acinetobacter baumannii isolates from clinical respiratory samples in Imam Khomeini Hospital, Tehran, Iran in 2018. Iran. J. Med. Microbiol. 15, 266–280. https://doi.org/10.30699/ijmm.15.3.266 (2021).
Shirazi, A. S. et al. Different virulence capabilities and ompA expressions in ST2 and ST513 of multidrug-resistant Acinetobacter baumannii. Curr. Microbiol. 76, 723–731. https://doi.org/10.1007/s00284-019-01686-9 (2019).
Tanaka, D. et al. Acinetobacter sp. Ud-4 efficiently degrades both edible and mineral oils: Isolation and characterization. Curr. Microbiol. 60, 203–9. https://doi.org/10.1007/s00284-009-9525-5 (2010).
Lin, F. et al. Molecular characterization of reduced susceptibility to biocides in clinical isolates of Acinetobacter baumannii. Front. Microbiol. 8, 1836. https://doi.org/10.3389/fmicb.2017.01836 (2017).
Bartual, S. G. et al. Development of a multilocus sequence typing scheme for characterization of clinical isolates of Acinetobacter baumannii. J. Clin. Microbiol. 43, 4382–4390. https://doi.org/10.1128/JCM.43.9.4382-4390.2005 (2005).
Yeom, D. H., Kim, S. K., Lee, M. N. & Lee, J. H. Pleiotropic effects of acyltransferases on various virulence-related phenotypes of Pseudomonas aeruginosa. Genes Cells 18, 682–693. https://doi.org/10.1111/gtc.12076 (2013).
Son, H. J. et al. Clinical and microbiological analysis of risk factors for mortality in patients with carbapenem-resistant Acinetobacter baumannii bacteremia. Open Forum Infect. Dis. 7, ofaa378. https://doi.org/10.1093/ofid/ofaa378 (2020).
Andriamanantena, T. S. et al. Dissemination of multidrug resistant Acinetobacter baumannii in various hospitals of Antananarivo Madagascar. Ann. Clin. Microbiol. Antimicrob. 9, 17. https://doi.org/10.1186/1476-0711-9-17 (2010).
Salehi, B. et al. Emergence and characterization of nosocomial multidrug-resistant and extensively drug-resistant Acinetobacter baumannii isolates in Tehran, Iran. J. Infect. Chemother. 24, 515–523. https://doi.org/10.1016/j.jiac.2018.02.009 (2018).
Jain, M. et al. Phenotypic and molecular characterization of Acinetobacter baumannii isolates causing lower respiratory infections among ICU patients. Microb. Pathog. 128, 75–81. https://doi.org/10.1016/j.micpath.2018.12.023 (2019).
Kim, M. H. et al. Using comparative genomics to understand molecular features of carbapenem-resistant Acinetobacter baumannii from South Korea causing invasive infections and their clinical implications. PLoS One 15, e0229416. https://doi.org/10.1371/journal.pone.0229416 (2020).
Ning, N. Z. et al. Molecular epidemiology of bla OXA-23 -producing carbapenem-resistant Acinetobacter baumannii in a single institution over a 65-month period in north China. BMC Infect. Dis. 17, 14. https://doi.org/10.1186/s12879-016-2110-1 (2017).
Leung, E. C., Leung, P. H. & Lai, R. W. Emergence of carbapenem-resistant Acinetobacter baumannii ST195 harboring blaOXA-23 isolated from bacteremia in Hong Kong. Microb. Drug Resist. 25, 1199–1203. https://doi.org/10.1089/mdr.2018.0433 (2019).
Khoshnood, S., Savari, M., Abbasi Montazeri, E. & Farajzadeh Sheikh, A. Survey on genetic diversity, biofilm formation, and detection of colistin resistance genes in clinical isolates of Acinetobacter baumannii. Infect. Drug Resist. 13, 1547–1558. https://doi.org/10.2147/IDR.S253440 (2020).
Zeighami, H., Valadkhani, F., Shapouri, R., Samadi, E. & Haghi, F. Virulence characteristics of multidrug resistant biofilm forming Acinetobacter baumannii isolated from intensive care unit patients. BMC Infect. Dis. 19, 629. https://doi.org/10.1186/s12879-019-4272-0 (2019).
Yang, C. H., Su, P. W., Moi, S. H. & Chuang, L. Y. Biofilm formation in Acinetobacter Baumannii: Genotype-phenotype correlation. Molecules 2, 1849. https://doi.org/10.3390/molecules24101849 (2019).
Thummeepak, R., Kongthai, P., Leungtongkam, U. & Sitthisak, S. Distribution of virulence genes involved in biofilm formation in multi-drug resistant Acinetobacter baumannii clinical isolates. Int. Microbiol. 19, 121–129. https://doi.org/10.2436/20.1501.01.270 (2016).
Brossard, K. A. & Campagnari, A. A. The Acinetobacter baumannii biofilm-associated protein plays a role in adherence to human epithelial cells. Infect. Immun. 80, 228–233. https://doi.org/10.1128/iai.05913-11 (2012).
Liu, C. et al. Distribution of virulence-associated genes and antimicrobial susceptibility in clinical Acinetobacter baumannii isolates. Oncotarget 9, 21663–21673. https://doi.org/10.18632/oncotarget.24651 (2018).
Sung, J. Y. Molecular characterization and antimicrobial susceptibility of biofilm-forming Acinetobacter baumannii clinical isolates from Daejeon, Korea. Korean J. Clin. Lab. Sci. 50, 100–109. https://doi.org/10.15324/kjcls.2018.50.2.100 (2018).
Fallah, A., Rezaee, M. A., Hasani, A., Barhaghi, M. H. S. & Kafil, H. S. Frequency of bap and cpaA virulence genes in drug resistant clinical isolates of Acinetobacter baumannii and their role in biofilm formation. Iran. J. Basic Med. Sci. 20, 849–855. https://doi.org/10.22038/IJBMS.2017.9105 (2017).
Sung, J. Y., Koo, S. H., Kim, S. & Kwon, G. C. Persistence of multidrug-resistant Acinetobacter baumannii isolates harboring blaOXA-23 and bap for 5 years. J. Microbiol. Biotechnol. 26, 1481–1489. https://doi.org/10.4014/jmb.1604.04049 (2016).
Tindwa, H. et al. Cloning, characterization and effect of TmPGRP-LE gene silencing on survival of Tenebrio molitor against Listeria monocytogenes infection. Int. J. Mol. Sci. 14, 22462–22482. https://doi.org/10.3390/ijms141122462 (2013).
Dorling, J., Moraes, C. & Rolff, J. Recognition, survival and persistence of Staphylococcus aureus in the model host Tenebrio molitor. Dev. Comp. Immunol. 48, 284–290. https://doi.org/10.1016/j.dci.2014.08.010 (2015).
Noonin, C. et al. Melanization and pathogenicity in the insect, Tenebrio molitor, and the crustacean, Pacifastacus leniusculus, by Aeromonas hydrophila AH-3. PLoS One 5, e15728. https://doi.org/10.1371/journal.pone.0015728 (2010).
Tao, Y., Duma, L. & Rossez, Y. Galleria mellonella as a good model to study Acinetobacter baumannii pathogenesis. Pathogens 10, 1483. https://doi.org/10.3390/pathogens10111483 (2021).
Petronio Petronio, G. et al. Emerging evidence on Tenebrio molitor immunity: A focus on gene expression involved in microbial infection for host-pathogen interaction studies. Microorganisms 10, 1983. https://doi.org/10.3390/microorganisms10101983 (2022).
Niu, T. et al. Prevalent dominant Acinetobacter baumannii ST191/195/208 strains in bloodstream infections have high drug resistance and mortality. Infect. Drug Resist. 16, 2417–27. https://doi.org/10.2147/IDR.S403604 (2023).
Hu, L. et al. Capsule Thickness, not biofilm formation, gives rise to mucoid Acinetobacter baumannii phenotypes that are more prevalent in long-term infections: A study of clinical isolates from a hospital in China. Infect. Drug Resist. 13, 99–109. https://doi.org/10.2147/IDR.S230178 (2020).
Huang, G. et al. Multilocus sequence typing analysis of carbapenem-resistant Acinetobacter baumannii in a Chinese burns institute. Front. Microbiol. 7, 1717. https://doi.org/10.3389/fmicb.2016.01717 (2016).
Funding
This study was supported by grants from the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health and Welfare, Republic of Korea (Grant Number: HI23C1297). Funding sources had no role in the study design, data collection, data analysis, decision to publish, or manuscript preparation.
Author information
Authors and Affiliations
Contributions
Y.K.Y. conceived, designed, and conducted the study. J.W.S., J.Y.K., S.B.K. and J.W.S. contributed to data collection and analysis. S.M.P. and Y.K.J. performed the experiments. Y.K.Y., J.W.S. and S.M.P. drafted the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Park, S.M., Suh, J.W., Ju, Y.K. et al. Molecular and virulence characteristics of carbapenem-resistant Acinetobacter baumannii isolates: a prospective cohort study. Sci Rep 13, 19536 (2023). https://doi.org/10.1038/s41598-023-46985-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-46985-1
- Springer Nature Limited
This article is cited by
-
Co-production of metallo-β-lactamase and OXA-type β-lactamases in carbapenem-resistant Acinetobacter baumannii clinical isolates in North East India
World Journal of Microbiology and Biotechnology (2024)