Abstract
Pseudomonas aeruginosa is resistant to a wide range of extended spectrum-lactamases (ESBLs) antibiotics because it produces several kinds of ESBLs. The goal of the current investigation was to identify the bacteria that produce extended spectrum -lactamases and the genes that encode three different ESBLs, such as blaOXA-10, blaPER-1 and blaSHV genes in Pseudomonas aeruginosa isolated from burn patients. In this investigation, 71 Pseudomonas aeruginosa isolates were isolated from burn wounds in Burn and Plastic Surgery Hospital, Duhok City between July 2021 to June 2022. For the purpose of finding the blaOXA-10, blaPER-1, and blaSHV ESBL expressing genes, Polymerase Chain Reaction (PCR) was used. Among 71 Pseudomonas aeruginosa isolates, 26.36% (29/71) were isolated from males and 38.18% (42/71) from females, and 76.06% (54/71) of the isolates were multidrug resistant. They exhibited higher resistance against Piperacillin with resistance rates of 98.59%. Among the ESBL-producing isolates tested, blaOXA-10 was found in 59.26% (32), blaPER-1 was found in 44.44% (24), and blaSHV was found in 11.11% (6). All isolates must undergo antimicrobial susceptibility testing because only a few numbers of the available antibiotics are effective for the treatment of this bacterium. This will prevent the development of resistance in burn units and aids in the management of the treatment plan.
Similar content being viewed by others
Introduction
Burns can be induced by a variety of factors, including as heat, radiation, electricity, friction, or chemical contact. Burn-related fatalities are estimated to be 180,000 every year, due to the fact that low- and middle-income developing countries account for the majority of these deaths1. Burn wounds are an ideal environment for opportunistic organisms of both endogenous and foreign origin to flourish in infected sites. Infections can develop in burn patients for a variety of reasons, including exposed body surfaces, immunocompromised states, invasive operations performed in the hospital, and extended hospital stays. Invasive infection is influenced by both patient-related parameters like age, total body surface area (TBSA), and burn wound depth as well as microbiological organism-related ones like organisms’ variety and number, enzyme/toxin production, and motility2. For patients with severe burns covering more than 40% of their (TBSA), sepsis due to burn wound infection or other infection complications and/or inhalation injury accounts for 75% of all deaths3.
Pseudomonas aeruginosa is a non-fermentative, aerobic, Gram-negative rod that is widely present in the environment. It is one of the most significant opportunistic pathogens that has been linked to both community- and hospital-acquired illnesses, including nosocomial infections, otitis media, burns, and respiratory tract infections4. Pseudomonas aeruginosa infections are occurring more often, and isolates that are multidrug-resistant (MDR) are becoming more common in patients who are hospitalized5,6. This bacterium has the capacity to infect almost all tissues, leading to a significant increase in morbidity and mortality7. Pseudomonas aeruginosa is physiologically resistant to many antibiotics and disinfectants, and it is resistant to extended-spectrum antibiotics. Because of the synthesis of numerous kinds of extended spectrum-lactamases, cephalosporins such as ceftazidime, ceftriaxone, and cefotaxime (ESBLs). The prevalence of ESBLs in P. aeruginosa has risen8,9,10. The Ambler classification that found in ESBLs Gram-negative bacteria divide them into four groups: A, B, C, and D. These ESBLs play a significant role in the development of ß-lactam antibiotic resistance11. Mobile genetic elements (MGEs) such as plasmids, transposons, insertion sequences, integrons, and bacteriophages contribute to the dissemination of various ESBL-encoding genes. MGEs can move themselves and/or genes from one location to another within the cell or be transferred from cell to cell horizontally by conjugation, transformation or, in the case of bacteriophages, by transduction12.
Pseudomonas aeruginosa possesses a number of ESBLs, including those of the Ambler class A, which contain a variety of enzymes, such as Pseudomonas extended resistance bla (PER-1). In 1993, a Turkish patient being treated at a French hospital provided the first proof of PER-1, which only P. aeruginosa produces13. Displaying resistance to Cephalosporins and inhibition to Clavulanate, this enzyme hydrolyzed most Penicillins well and Cephalosporins including Cefalotin, Cefoperazone, Cefuroxime, Ceftriaxone, and Ceftazidime. PER-1 did not hydrolyse Oxacillin, Cephamycins or Imipenem. PER enzymes are most commonly found in isolates from Turkey and Mediterranean countries14.
Pseudomonas aeruginosa is the documented source of oxacillinase (OXA type), a class D -lactamase that hydrolyzes Oxacillin, but it has also been found in numerous other gram-negative bacteria, including Enterobacteriaceae. In general, OXA-type enzymes are a diverse category that exhibits variation in amino acid sequences and substrate profiles. However, it has been shown that a number of OXA-type variations hydrolyze cephalosporins, cephems, and/or monobactams. According to a recent review, there are 27 oxacillinase enzymes described as extended-spectrum. These enzymes’ substrates include third- and/or fourth-generation Cephalosporins in addition to Penicillins and early Cephalosporins. Most extended-spectrum oxacillinases derive from OXA-10 and OXA-2. The OXA-10 derivatives include OXA-11, OXA-13, OXA-14, OXA-16, OXA-17, OXA-19 and OXA-2815. In general, -lactamase inhibitors have no effect on OXA-type enzymes16. The occurrence of SHV-type ESBLs has been recorded in a number of European nations, including Austria, France, Italy, and Greece, as well as the United States and Australia. Perhaps more so than any other form of ESBL, they will be found in clinical isolates17.
The goal of the current investigation was to identify the patterns of antibiotic susceptibility against various antibiotics because P. aeruginosa is becoming increasingly resistant to multiple ESBLs. In addition, genes encoding blaOXA-10, blaPER-1, and blaSHV were analyzed in clinical isolates of P. aeruginosa from infected hospitalized burn patients to assess their prevalence.
Results
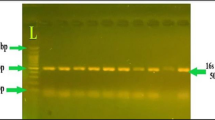
In this current study, a total of 110 burn patient samples of both genders and different ages were collected from July 2021 to May 2022. From 110 samples, P. aeruginosa was the predominant pathogenic bacteria, and 71 (64.55%) of them were confirmed phenotypically and molecularly by the species-specific gene (16S rDNA). Phenotypically, P. aeruginosa was identified on MacConkey, Cetrimide, Nutrient, and Blood agars as well as Gram stain and biochemical tests (Oxidase, Citrate, Catalase, TSI). In addition, all isolated P. aeruginosa were genotypically confirmed by the species-specific gene (16S rDNA).
Among 71 P. aeruginosa, 26.36% (29/71) were isolated from males and 38.18% (42/71) from females. There was a statistically non-significant (P > 0.05) difference between both genders. Most isolates were collected from ages between 21 and 30 years (13.64%). Statistical analysis revealed that there were significant (P < 0.04) differences across age groups. The demographic information for 71 positive patients from whom P. aeruginosa was isolated are displayed in Table 1. The most frequent types of burn wounds were those caused by flame, accounting for 38.18% (42/110) of the patients; electric burns affected 2.73% (3/110) of the patients. Statistical analysis showed highly significant (P < 0.01) differences in the causes of burn. Patients with second-degree burns had a higher risk of contracting infection (45.45%) than third- and mix-degree burns (6.36% and 12.73%) respectively. Statistical analysis revealed the presence of highly significant (P < 0.01) differences between degrees of burns. Patients with TBSA burns between 20 and 40 percent (48.18%) had a higher risk of infection than those with burns between 20 and 40 percent (7.27% and 9.09%), but these differences were statistically non-significant (P > 0.05) (Table 2).
Antimicrobial susceptibility test revealed that 76.06% (54/71) P. aeruginosa isolates were multidrug-resistant (MDR), showing resistance to β-lactams, aminoglycosides, and/or fluoroquinolones (at least three different types of antimicrobial medicines).
All of the isolated P. aeruginosa in the present study were sensitive to Colistin (100%). While, they displayed strong levels of resistance against Meropenem, Ceftazidime, Cefepime, and Piperacillin at rates of 74.65%, 84.50%, 85.92%, and 98.59% respectively. However, they experienced moderate resistance to Imipenem, Amikacin, Ciprofloxacin, Levofloxacin, and Gentamicin at rates of 60.56%, 63.38%, 64.79%, 67.61%, and 69.01%, respectively (Table 3).
Using PCR amplification, the prevalence of the antibiotic resistance genes blaOXA-10, blaPER-1, and blaSHV in the ESBL-producing P. aeruginosa isolates were examined and confirmed by agarose gels. Among the ESBL genes, blaOXA-10, and blaPER-1 were more abundant at 59.26% (32), and 44.44% (24), respectively, and the blaSHV had the lowest abundance of 11.11% (6). The main point of interest is that 14 of the isolated P. aeruginosa strains had both blaOXA-10, and blaPER-1 genes, and 6 strains had blaOXA-10, and blaSHV. Also, blaOXA-10, blaPER-1, and blaSHV genes had 5 strains. Furthermore, four strains possess blaPER-1, and blaSHV. (Table 4).
The isolates carrying the bla OXA-10 gene were completely resistant to Piperacillin, Cefepime, and Ceftazidime, and showed high to moderate resistance to other tested antibiotics, while isolates with bla PER-1 gene, were 100% resistant to Piperacillin, Ceftazidime, and Meropenem, and showed high to moderate resistance to other tested antibiotics, finally isolates with bla SHV gene were 100% almost resistant to all antibiotics. We can show all isolates with bla OXA-10, bla PER-1, and bla SHV genes were 100% sensitive to Colistin (Table 5).
Discussion
Recently, ESBL-producing P. aeruginosa has become a significant source of infections in healthcare settings, particularly in immunocompromised individuals and burn victims18. Because the surface of moist wounds provides a habitat that is suited for their existence, it thrives in the clinical setting, proving that this environment is perfect for their colonization. Because of the high levels of resistance to the most often given antibiotics in hospital settings, the treatment of infections brought on by these multidrug-resistant organisms is becoming more difficult19.
Our study revealed the prevalence of P. aeruginosa isolates among burn patients at the Duhok Burn Hospital, at a rate of 64.55% (71/110), a similar finding was made in an Algerian study, where the rate was recorded at 62%20. On the other hand, another study conducted in Iraq recorded a high prevalence of P. aeruginosa isolates at 97.6%21. however, results from other studies from other nations showed lower prevalence rates, including Morocco (15.1%)22, and Egypt (19.8%)23. This difference might be attributed to antibiotics abuse, different hospital strategies for the management of infections, hygiene, and geographic climatic.
The current study showed the highest rate of 38.18% from flame burn followed by scald burn (15.45%). Regarding gender and age, females showed the highest rate of burn wounds than males (38.18% vs 26.36%), with the ages of 21–30 showing the highest rate (13.64%). These results are somewhat similar to the study conducted in Basra/Iraq24 they showed that females had a higher proportion of burns than males (57.5% vs. 19.16%), with flame burns accounting for 76.6%, followed by scald burns (19.1%). Additionally, studies from Iraq and Iran have reported somewhat lower rates; 57% in the Iraqi city of Suleimani25, 54.84%, and 56% in Iran1,26 respectively. Due to the fact that most flame injuries occur at home and that most women in our society perform daily household tasks like heating and cooking in areas of the kitchen where there is a greater danger of flame burns, females were significantly more likely than males to sustain flame burns.
While a study conducted in Suleimani/Iraq showed the highest infection in males (56.2%) with scald (Hot water) being the highest (72.5%) group involved, followed by flame (22.8%)27. According to this research's findings, the highest rate 45.45% were second-degree burn, followed by 12.73% of mixed burn infections and only 6.36% had third-degree burns. Similarly, Al-Aali et al.3 revealed that 72.1% of burn patients had second-degree burns. According to the present study observations, patients who had TBSA burns were 20–40% more likely to contract an infection (48.18%), this finding is consistent with the research conducted by Rashid et al.27 in Suleimani/Iraq found infection rates were higher in patients with TBSA burns of 20–40% compared to those with burns of 40%. The higher the total body surface area damaged by the thermal assault the higher the potential for the bacteria to colonize and proliferate increasing the wound thickness and depth making way to the bloodstream involvement28.
Regardless to the etiology of the burns, female patients over the age of ten years were the most prevalent patient demographics linked to a higher prevalence of P. aeruginosa infection. In the present investigation, 76.06% of the patients had MDR P. aeruginosa infection which is higher than that reported by other studies in Iran (16.5–41%), Iraq (12.4%)29,30,31 , Brazil (71.4%) and Egypt (70%)32,33. On the other hand, another study in Iran found a much higher rate (89.24%) of patients infected with MDR P. aeruginosa26. Worldwide increasing of MDR P. aeruginosa could be attributed to improper use of antibiotics in hospitals and communities in addition to the accumulation of a variety of resistance mechanisms34.
In the current investigation, 100% of the isolates demonstrated sensitivity to Colistin. In contrast, a substantially lower percentage of Colistin-sensitive individuals (53.1%) were reported by Jalil et al.24 in their investigation in Iraq. Another study in Iraq also reported resistance to colistin at a rate of 7.4%21. However, susceptibility to Colistin remains vastly high against P. aeruginosa approaching 100% in most countries in the area of the Middle East and North Africa35.
In this study, 60.56% of P. aeruginosa isolates were resistant to Imipenem, which is higher than that reported by a study conducted in Iran at a rate of 41.3%17 and Iraq at a rate of 47%36, and lower than that reported by studies conducted in Iran at a rate (83.9%)26, in Iraq 68.40%21. Multifactorial agents, such as increased carbapenemase production, oprD mutation, AmpC, and efflux pumps overexpression, a shift in drug target sites, and other mechanisms, are contributing to P. aeruginosa increasing resistance to carbapenems37.
The current study reported the highest resistance of P. aeruginosa to Piperacillin (98.59%). This finding is higher than previous studies conducted in Iran 74.8%38 and in South Africa, 94%39. These findings raise a major concern that requires Health Authorities to urgently work on finding rapid and accurate diagnostic procedures and regulating the dispensing of antibiotics, in addition to tightening microbiological control systems in hospitals.
ESBLs are one of the most common sources of ß-lactam antibiotic resistance in Gram-negative bacteria. These enzymes are plasmid-encoded ß-lactamases that have been discovered in Klebsiella pneumoniae and Escherichia coli, as well as clinical isolates of Enterobacteriaceae and Pseudomonas19. In the current study, the genes linked to blaOXA-10, blaPER-1, and blaSHV were found in 59.26% (32), 44.44% (24), and 11.11% (6) of the ESBL-producing P. aeruginosa isolates. The level of resistance was even higher in isolates carrying the OXA-10 gene. The results were consistent with a study conducted in Iran by Farshadzadeh40, which used the same primers for the OXA-10 and PER-1 genes and produced a band of the same molecular weight. Another study conducted in Shiraz, Iran reported a rate of 76.2% blaOXA-10, and 40% of resistant isolates contain blaPER-1-related genes13. The prevalence of the blaSHV gene in the current study is consistent with the rate of 10.7% reported by Peymani et al.17 in Iran.
Different environments may have a different distribution of ESBL-resistant bacteria, as evidenced by the varying prevalence of ESBL-encoding genes. In addition, the prevalence of blaPER-1 in Ahvaz, Iran, was 62.75%, which was higher than the prevalence of blaPER-1 in Italy (34.61%)41, Hungary (1.3%), and Belgium (2%), respectively42, but less than the incidence of blaPER-1 in Turkey (86.75%)43. The results of our study showed a high prevalence of the ESBL-producing P. aeruginosa isolates in burn patients, which is an alarming sign and should be taken into consideration because increasing of the antimicrobial-resistant bacteria isolated from burn patients is an important issue.
Methods
Sample collection
In the current study, one hundred ten clinical samples were collected from patients attending Burn and Plastic Surgery Hospital in Duhok City, Iraq over a period from July 2021 to May 2022. These samples were collected from hospitalized patients of both genders and different ages (1 month–90 years). The clinical samples were collected from infected burn wounds at the same time the dressings were changed using cotton disposable swabs and transported into the sterile medium in plastic bottles.
Isolation and Identification of Pseudomonas aeruginosa
After taking the samples with sterile cotton swabs and placed in a tube containing 5 ml of brain heart infusion broth, the samples were incubated at 37 °C for 24 h. Then the samples were cultured by a sterile loop using the streak method on, MacConkey agar. After the incubation for 24 h at 37 °C, the subculture of the pure colonies of suspected P. aeruginosa were done every 48 h on Blood agar, Cetrimide agar, and Nutrient agar plates. Initial diagnosis of isolates was made on the basis of Gram’s staining of culture, hemolysis on blood agar, pigment production, and the smell in cultures. Based on the morphological and biochemical characteristics of these pure colonies were identified according to a study by44.
Antimicrobial susceptibility test
All isolated P. aeruginosa were subjected to antibiotic susceptibility test using Kirby–Bauer disk diffusion method by spreading the inoculated sterile swab on Muller–Hinton agar and incubated overnight aerobically at 37 °C according to45. Ten antibiotics were used, supplied by (Bioanalyses, Turkey). The tested antimicrobial agents included: Piperacillin (PI; 100 mg), Ceftazidime (CAZ; 30 mg), Cefepime (CPM; 30 mg), Imipenem (IPM; 10 mg), Meropenem (MEM; 10 mg), Amikacin (AK; 30 mg), Gentamicin (CN; 10 mg), Levofloxacin (LEV; 5 mg), Ciprofloxacin (CIP; 5 mg), Colistin (CL; 10 mg). The diameter of the inhibition zone around antibiotic disks was measured according to the Clinical and Laboratory Standards Institute46.
Bacterial DNA extraction from P. aeruginosa
Genomic DNA was extracted from bacterial isolates using High yield DNA Purification Kit according to the manufacturer’s instructions (Genomic DNA mini kit, Favorgen, Taiwan). Bacterial DNA quality was achieved by (NanoDrop™ One UV–Vis Spectrophotometer, Thermo Fisher Scientific, Waltham, MA, USA) and then stored in a freezer at – 20 °C, ready to be used for PCR. Different primers were used for amplification of these genes as described in Table 6. Purified DNA was used for PCR amplification of 4 genes (Table 7). A species-specific primer for P. aeruginosa, 16S rDNA gene. After confirming P. aeruginosa, 3 primers were used to detect ESBL genotypes, blaOXA-10, blaPER-1, and blaSHV β-lactamase genes.
The amplification of DNA for each gene was carried out in PCR tubes containing 10 µl of PCR master mix (ADDBIO.INC, South Korea), 3 µL of DNA, 5 µL of distilled deionized water, and the forward and reverse primers, totaling 1 µL of each primer (10 pmol/µL), to make a final volume of 20 µL. The amplification conditions of each gene are described in Table 7.
The DNA lengths of each fragment produced with these 4 primers were confirmed by running 5 µL of each PCR product on 1.5% agarose gels in 1 × TAE buffer and added 8 µL of Safe Gel stain Dye (Guangzhou Dongsheng Biotech Co., Ltd., Guangzhou, China) and the electrophoresis was performed at 85 V for 45 min. The agarose gel was visualized under UV radiation (Cleaver Scientific Ltd., Rugby, UK). The images of DNA bands were captured, and the estimated amplicon size were compared with the 1500–100 bp DNA ladder (Guangzhou Dongsheng Biotech Co., Ltd., Guangzhou, China)51.
Ethics declarations
The study protocol was received and approved by the Duhok Directorate General of Health, Directorate of Planning, Scientific Research Division, Institutional Ethics Committee (approval No. 13072021-7-10).
Approval for human experiments
-
1.
The study was approved by the Duhok Directorate General of Health, Directorate of Planning, Scientific Research Division, Institutional Ethics Committee (approval No. 13072021-7-10).
-
2.
I confirm that all experiments were performed in accordance with relevant named guidelines and regulations.
Informed consent statement
The request for verbal consent is part of the original approval (with reference number 13072021-7-10 on 21 May 2023) and the researcher was not asked for written informed consent.
Statistical analysis
Statistical analysis was performed using GraphPad Prism version 9.3.1 (471). The chi-square test and Odd (ratio 95% CI) test were performed to determine statistical significance at P-value of less than 0.05.
Conclusion
In conclusion, the current study's high proportion of ESBL-producing P. aeruginosa highlights a serious health issue that requires further attention. All isolates must undergo an antimicrobial susceptibility test prior to therapy because there are only a small number of medications that are effective against this bacterium. The execution of this test reduces the indiscriminate use of antibiotics and resistance development in burn units and aids in the management of treatment plan.
Data availability
All data supporting the present work are available from the corresponding author upon reasonable request.
References
Khosravi, A. D. et al. Investigation of the prevalence of genes conferring resistance to carbapenems in Pseudomonas aeruginosa isolates from burn patients. Infect. Drug Resist. 12, 1153–1159. https://doi.org/10.2147/IDR.S197752 (2019).
Chaudhary, N. A. et al. Epidemiology, bacteriological profile, and antibiotic sensitivity pattern of burn wounds in the burn unit of a tertiary care hospital. Cureus 11(6), e4794. https://doi.org/10.7759/cureus.4794 (2019).
Al-Aali, K. Y. Microbial profile of burn wound infections in burn patients, Taif, Saudi Arabia. Arch. Clin. Microbiol. 7(2), 1–9 (2016).
Pachori, P., Gothalwal, R. & Gandhi, P. Emergence of antibiotic resistance Pseudomonas aeruginosa in intensive care unit; a critical review. Genes. Dis. 6(2), 109–119. https://doi.org/10.1016/j.gendis.2019.04.001 (2019).
Pang, Z. et al. Antibiotic resistance in Pseudomonas aeruginosa: Mechanisms and alternative therapeutic strategies. Biotechnol. Adv. 37(1), 177–192. https://doi.org/10.1016/j.biotechadv.2018.11.013 (2019).
Abdelrahman, D. N. et al. β-lactamases (bla TEM, bla SHV, bla CTXM-1, bla VEB, bla OXA-1) and class C β-lactamases gene frequency in Pseudomonas aeruginosa isolated from various clinical specimens in Khartoum State, Sudan: A cross-sectional study. F1000Research 9, 774. https://doi.org/10.12688/F1000RESEARCH.24818.2 (2021).
Najem, S. A. Bacteriological study on multidrug resistance genes in Pseudomonas aeruginosa isolated from different clinical samples in Al-Najaf Province. MS.C.-University of Kufa, Al-Najaf/Iraqi (2022).
Malekzadegan, Y. et al. Antimicrobial resistance pattern and frequency of multiple-drug resistant Enterobacter spp. at a tertiary care hospital in Southwest of Iran. J. Krishna Inst. Med. Sci. 6(2), 33–39 (2017).
Hosseinzadeh, Z. et al. Emerge of bla NDM-1 and bla OXA-48-like harboring carbapenem-resistant Klebsiella pneumoniae isolates from hospitalized patients in southwestern Iran. J. Chin. Med. Assoc. 81(6), 536–540. https://doi.org/10.1016/j.jcma.2017.08.015 (2018).
Soltani, B. et al. Molecular characteristics of multiple and extensive drug-resistant Acinetobacter baumannii isolates obtained from hospitalized patients in Southwestern Iran. Infez. Med. 26(1), 67–76 (2018).
Tavakoly, T. et al. The prevalence of CMY-2, OXA-48 and KPC-2 genes in clinical isolates of Klebsiella spp.. Cell Mol. Biol. 64(3), 40–44. https://doi.org/10.14715/cmb/2018.64.3.7 (2018).
Castanheira, M., Simner, P. J. & Bradford, P. A. Extended-spectrum β-lactamases: An update on their characteristics, epidemiology and detection. JAC Antimicrob. Resist. 3(3), dlab092 (2021).
Emami, A. et al. Detection of blaPER-1 & blaOxa10 among imipenem resistant isolates of Pseudomonas aeruginosa isolated from burn patients hospitalized in Shiraz Burn Hospital. Iran J. Microbiol. 7(1), 7–11 (2015).
Alikhani, M. Y. et al. Antimicrobial resistance patterns and prevalence of blaPER-1 and blaVEB-1 genes among ESBL-producing Pseudomonas aeruginosa isolates in West of Iran. Jundishapur. J. Microbiol. 7(1), e8888. https://doi.org/10.5812/jjm.8888 (2014).
Evans, B. A. & Amyes, S. G. OXA β-lactamases. Clin. Microbiol. Rev. 27(2), 241–263 (2014).
Al-Kaisse, A. A., Al-Thwani, A. N. & Al-Segar, R. Q. PCR detection of some ESBLs (bla) genes in Pseudomonas aeruginosa isolated from burn’s units in Bagdad hospitals. J. Biotechnol. Res. Center 9(2), 74–80. https://doi.org/10.24126/jobrc.2015.9.2.439 (2015).
Peymani, A. et al. Distribution of blaTEM, blaSHV, and blaCTX-M genes among ESBL-producing P. aeruginosa isolated from Qazvin and Tehran hospitals, Iran. J. Prev. Med. Hyg. 58(2), 155–160 (2017).
Litwin, A. et al. Pseudomonas aeruginosa device associated—healthcare associated infections and its multidrug resistance at intensive care unit of University Hospital: Polish, 8.5-year, prospective, single-center study. BMC. Infect. Dis. 21(1), 180. https://doi.org/10.1186/s12879-021-05883-5 (2021).
Amirkamali, S. et al. Distribution of the bla OXA, bla VEB-1, and bla GES-1 genes and resistance patterns of ESBL-producing Pseudomonas aeruginosa isolated from hospitals in Tehran and Qazvin, Iran. Rev. Soc. Bras. Med. Trop. 50, 315–320 (2017).
Meradji, S. et al. Epidemiology and virulence of VIM-4 metallo-beta-lactamase-producing Pseudomonas aeruginosa isolated from burn patients in eastern Algeria. Burns 42(4), 906–918 (2016).
Alkhulaifi, Z. M. & Mohammed, K. A. The Prevalence of Cephalosporins resistance in Pseudomonas aeruginosa isolated from clinical specimens in Basra, Iraq. UTJsci. 10(1), 896 (2023).
Essayagh, M. et al. Épidémiologie de l’infection des plaies des brûlés de Rabat, Maroc: Expérience de trois ans. Méd. Santé Trop. 24(2), 157–164 (2014).
Mahmoud, A. B. et al. Prevalence of multidrug-resistant Pseudomonas aeruginosa in patients with nosocomial infections at a university hospital in Egypt, with special reference to typing methods. J. Virol. Microbiol. 13, 165–259 (2013).
Jalil, M. B., Abdul-Hussien, Z. R. & Al-Hmudi, H. A. Isolation and identification of multi drug resistant biofilm producer Pseudomonas aeruginosa from patients with burn wound infection in Basra province/Iraq. IJDR 7(11), 17258–17262 (2017).
Othman, N. et al. Pseudomonas aeruginosa infection in burn patients in Sulaimaniyah, Iraq: Risk factors and antibiotic resistance rates. J. Infect. Dev. Ctries 8(11), 1498–1502. https://doi.org/10.3855/jidc.4707 (2014).
Khoshnood, S. et al. Distribution of extended-spectrum β-lactamase genes in antibiotic resistant strains of Pseudomonas aeruginosa obtained from burn patients in Ahvaz, Iran. J. Acute. Dis. 8(2), 53–57. https://doi.org/10.4103/2221-6189.254426 (2019).
Rashid, K. J., Babakir-Mina, M. & Abdilkarim, D. A. Characteristics of burn injury and factors in relation to infection among pediatric patients. MOJ Gerontol. Ger. 1(3), 57–66. https://doi.org/10.15406/MOJGG.2017.01.00013 (2017).
Sewunet, T. et al. Bacterial profile and antimicrobial susceptibility pattern of isolates among burn patients at Yekatit 12 hospital burn center, Addis Ababa, Ethiopia. Ethiop. J. Health Sci. 23(3), 209–216 (2013).
Mirzaei, B. et al. Prevalence of multi-drug resistant (MDR) and extensively drug-resistant (XDR) phenotypes of Pseudomonas aeruginosa and Acinetobacter baumannii isolated in clinical samples from Northeast of Iran. BMC Res. Notes. 13, 380 (2020).
Ahmadian, L. et al. Distribution and molecular characterization of resistance gene cassettes containing class 1 integrons in multi-drug resistant (MDR) clinical isolates of Pseudomonas aeruginosa. Infect. Drug Resist. 13, 2773–2781 (2020).
Al-Khudhairy, M. K. & Al-Shammari, M. M. M. Prevalence of metallo-β-lactamase–producing Pseudomonas aeruginosa isolated from diabetic foot infections in Iraq. New Microbes New Infect. 35, 100661 (2020).
De-Almeida-Silva, K. C. F. et al. Molecular characterization of multidrug-resistant (MDR) Pseudomonas aeruginosa isolated in a burn center. Burns 43, 137–143 (2017).
Kishk, R. M. et al. Efflux MexAB-mediated resistance in P. aeruginosa isolated from patients with healthcare associated infections. Pathogens. 9, 471 (2020).
Alkhulaifi, Z. M. & Mohammed, K. A. Prevalence and molecular analysis of antibiotic resistance of Pseudomonas aeruginosa isolated from clinical and environmental specimens in Basra, Iraq. Iran J. Microbiol. 15(1), 45 (2023).
Al-Orphaly, M. et al. Epidemiology of multidrug-resistant Pseudomonas aeruginosa in the Middle East and North Africa Region. mSphere 6(3), e00202-e221 (2021).
Qader, M. K., Solmaz, H. & Merza, N. S. Molecular typing and virulence analysis of Pseudomonas aerugınosa isolated from burn infections recovered from Duhok and Erbil Hospitals/Iraq. UKH J. Sci. Eng. 4(2), 1. https://doi.org/10.25079/ukhjse.v4n2y2020.pp1-10 (2020).
Doi, Y. Treatment options for carbapenem-resistant Gram-negative bacterial infections. Clin. Infect. Dis. 69(Suppl 7), S565–S575. https://doi.org/10.1093/cid/ciz830 (2019).
Haghighifar, E., Dolatabadi, R. K. & Norouzi, F. Prevalence of blaVEB and blaTEM genes, antimicrobial resistance pattern and biofilm formation in clinical isolates of Pseudomonas aeruginosa from burn patients in Isfahan, Iran. Gene Rep. 23, 101157 (2021).
Adjei, C. B. et al. Molecular characterisation of multidrug-resistant Pseudomonas aeruginosa from a private hospital in Durban, South Africa. South. Afr. J. Infect. Dis. 33(2), 38–41 (2018).
Farshadzadeh, Z. et al. Spread of extended-spectrum β-lactamase genes of bla OXA-10, bla PER-1 and bla CTX-M in Pseudomonas aeruginosa strains isolated from burn patients. Burns 40(8), 1575–1580. https://doi.org/10.1016/j.burns.2014.02.008 (2014).
Endimiani, A. et al. Pseudomonas aeruginosa bloodstream infections: Risk factors and treatment outcome related to expression of the PER-1 extended-spectrum beta-lactamase. BMC Infect. Dis. 6, 52. https://doi.org/10.1186/1471-2334-6-52 (2006).
Ranellou, K. et al. Detection of Pseudomonas aeruginosa isolates of the international clonal complex 11 carrying the bla PER-1 extended-spectrum β-lactamase gene in Greece. J. Antimicrob. Chemother. 67(2), 357–361. https://doi.org/10.1093/jac/dkr471 (2012).
Aktaş, Z. et al. PER-1- and OXA-10-like beta-lactamases in ceftazidime-resistant Pseudomonas aeruginosa isolates from intensive care unit patients in Istanbul, Turkey. Clin. Microbiol. Infect. 11(3), 193–198. https://doi.org/10.1111/j.1469-0691.2004.01067.x (2005).
Leboffe, M. J. & Pierce, B. E. A Photographic Atlas for the Microbiology Laboratory 4th edn. (UK, Morton Pub. Co, 2011).
Hudzicki, J. Kirby-Bauer disk diffusion susceptibility test protocol. Am. Soc. Microbiol. 15, 55–63 (2009).
CLSI (Clinical and Laboratory Standards Institute). In Performance Standards for Antimicrobial Susceptibility Testing, Twentieth Informational Supplement, CLSI Document M100- Ed32 February 2022 Replaces M100-Ed31 (2022).
Spilker, T. et al. PCR-based assay for differentiation of P. aerugjnosa from other Pseudomonas species recovered from cystic fibrosis patients. J. Clin. Microbiol. 42(5), 2074–2079 (2004).
Pai, H. et al. Carbapenem resistance mechanisms in Pseudomonas aeruginosa clinical isolates. Antimicrob. Agents Chemother. 45(2), 480–484 (2001).
Celenza, G. et al. Spread of bla CTX-M-type and bla PER-2 β-lactamase genes in clinical isolates from Bolivian hospitals. J. Antimicrob. Chemother. 57(5), 975–978. https://doi.org/10.1093/jac/dkl055 (2006).
Kim, Y. T., Kim, T. U. & Baik, H. S. Characterization of extended-spectrum beta-lactamase genotype TEM, SHV, and CTX-M producing Klebsiella pneumoniae isolated from clinical specimens in Korea. J. Microbiol. Biotechnol. 16(6), 889–895 (2006).
Maniatis, T., Fritsch, E. F. & Sambrook, J. Molecular cloning a Laboratory manual gold spring harber laboratory. N. Y. Biotechnol. 5, 257–261 (1982).
Acknowledgements
The authors would like to thank the burn patients who participated in the study. We also thank the Burn and Plastic Surgery Hospital workers for supplying the samples and fundamental data needed for our inquiry.
Author information
Authors and Affiliations
Contributions
Conceptualization, P.R.F.; data curation, K.H.M. and M.W.M.S.; formal analysis, P.R.F.; methodology, P.R.F.; project administration, P.R.F., K.H.M. and M.W.M.S.; visualization, M.W.M.S.; writing—original draft, P.R.F.; writing—review & editing, P.R.F., K.H.M. and M.W.M.S.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Polse, R.F., Khalid, H.M. & Mero, W.M.S. Distribution of blaOXA-10, blaPER-1, and blaSHV genes in ESBL-producing Pseudomonas aeruginosa strains isolated from burn patients. Sci Rep 13, 18402 (2023). https://doi.org/10.1038/s41598-023-45417-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-45417-4
- Springer Nature Limited