Abstract
The human-specific Salmonella enterica serovar Typhi (S. Typhi) causes typhoid fever, a systemic disease with no known reservoir. Curli fimbriae are major components of biofilm produced by Salmonella and are encoded by the csg gene cluster (csgBAC and csgDEFG). The role of curli in S. Typhi is unknown, although detection of anti-curli antibodies suggests they are produced during host infection. In this study, we investigated curli gene expression in S. Typhi. We demonstrated that the CsgD regulatory protein binds weakly to the csgB promoter. Yet, replacing S. Typhi csgD with the csgD allele from S. Typhimurium did not modify the curli negative phenotype on Congo Red medium suggesting that differential regulation of curli gene expression in S. Typhi is not dependent on modification of the CsgD regulator. The entire csg gene cluster from S. Typhimurium was also cloned into S. Typhi, but again, despite introduction of a fully functional csg gene cluster from S. Typhimurium, curli were still not detected in S. Typhi. Thus, in addition to intrinsic genomic differences in the csg gene cluster that have resulted in production of a modified CsgD protein, S. Typhi has likely undergone other changes independent of the csg gene cluster that have led to distinctive regulation of csg genes compared to other Salmonella serovars.
Similar content being viewed by others
Introduction
Salmonella enterica serovar Typhi (S. Typhi) is a human-specific pathogen that causes a systemic disease known as typhoid fever. Typhoid fever affects an estimated 11–21 million people yearly and causes 128,000–161,000 deaths1. Typhoid fever has become a global public health concern as many endemic areas are facing the emergence of multiresistant strains2,3. Among recovered patients, 2–5% become chronic carriers4, that may develop due to colonization and biofilm formation in the gallbladder5,6,7. These chronic carriers currently represent the only known reservoir, and mechanisms controlling biofilm formation by S. Typhi are not well characterized. On the other hand, Salmonella enterica serovar Typhimurium (S. Typhimurium) is a non-typhoidal Salmonella and one of the main causes of human foodborne gastroenteritis. S. Typhimurium can colonize a large variety of hosts ranging from mammals to birds8,9. The host range and diseases caused by these two serovars are distinct although S. Typhimurium and S. Typhi share 89% of their genomes10. Among the genomic differences, S. Typhi contains more than 200 pseudogenes and 600 Typhi-specific genes compared to S. Typhimurium10,11,12,13,14,15. While the specific components and mechanisms underlying biofilm formation by S. Typhi are mostly unknown, S. Typhimurium biofilm includes cellulose, the BapA protein and curli fimbriae16,17,18,19,20. Curli are amyloid fibrils produced by certain bacteria such as E. coli and S. Typhimurium. Curli can mediate initiation of biofilm formation18,20. However, the role of curli in S. Typhi is still poorly understood. Curli are encoded by genes present in the two divergent operons, csgBAC and csgDEFG. CsgB is the nucleator mediating polymerisation of CsgA major structural subunits21,22. CsgC may help prevent intracellular polymerization of CsgA23. CsgG is a porin-like protein involved in secretion of extracellular curli components (CsgB, CsgA and CsgF) and has a chaperone-like function that promotes intracellular stabilization of CsgA and CsgB20,21,24,25. CsgF is an assembly protein which can localize CsgB on the bacterial surface26. CsgE is an accessory protein with a chaperone-like function that modulates which components can be secreted by the porin-like CsgG protein23. Finally, CsgD is a positive regulator belonging to the UhpA/FixJ/LuxR family, and is a DNA-binding protein. It is the main activating regulator of csgBAC27. In addition to regulation of curli-encoding genes, CsgD also plays a role in regulation of the O-antigen capsule, c-di-GMP, RpoS stress response and other biofilm components including regulation of BapA and cellulose synthesis in S. Typhimurium and E. coli17,21,28,29. Curli production is also controlled by several regulators that respond to different environmental stimuli such as temperature, oxygen, and osmolarity21,30. Under laboratory conditions, it was shown that the expression of curli by S. Typhimurium was highest after growth at 30 °C in lysogeny broth (LB) without salt or yeast extract, also called tryptone medium31. On Tryptone agar medium with Congo red dye, colonies are red, dry, and rough (rdar), corresponding to colonies that are producing both curli and cellulose. However, under the same growth conditions, S. Typhi colonies are smooth and white (saw), suggesting a lack of cellulose and/or curli expression19,31.
Thus, based on these results and on the fact that S. Typhi and S. Typhimurium have different pathogenic lifestyles, we hypothesized that in S. Typhi curli fimbriae are differently regulated compared to S. Typhimurium. The level of expression from the divergent promoters encoding each of the curli operons csgBAC and csgDEFG were compared between S. Typhi and S. Typhimurium under various growth conditions including different temperatures and oxygen levels. The role of CsgD on csgB expression was also investigated by determining DNA-binding to the csgB promoter region. S. Typhi csgD and curli gene cluster were replaced by the S. Typhimurium allele to investigate the impact of genetic polymorphisms on curli regulation.
Results
csgB expression profile
Although curli expression is well characterized in S. Typhimurium, the optimal laboratory expression conditions in S. Typhi remain unclear. Thus, curli gene expression in S. Typhi (STy) was evaluated in different media using a lacZ reporter gene (pcsgBSTy-lacZ) and was compared with expression in standard LB (Fig. 1). csgB expression varied and was significantly lower on solid medium, under SPI-1 inducing conditions, and with 0.3 M NaCl, whereas the highest expression was in tryptone medium. These results suggested that S. Typhi csgB expression was regulated by differences in nutrient and oxygen levels. Tryptone medium was selected to further investigate csg expression.
S. Typhi csgB expression in different growth media. csgB expression of WT S. Typhi was evaluated by β-galactosidase assays using pcsgBSTy-lacZ fusion vector. Expression is presented as a ratio comparing various media to expression levels after growth in LB. All cultures were incubated at 37 °C overnight. The experiments were repeated at least 3 times with 2 technical replicates. The negative control, WT S. Typhi transformed with pRS415 empty vector did not present significant expression level in all studied conditions. *p < 0.001.
Impacts of temperature and oxygen levels on csgB expression were investigated using the S. Typhi (STy) pcsgBSTy-lacZ reporter (Fig. 2A) and the S. Typhimurium (STm) pcsgBSTm-lacZ reporter (Fig. 2B). In S. Typhi, csgB was always significantly more expressed at 37 °C, regardless of the oxygen level (Fig. 2A, red). While, the highest expression was under aerobic growth, csgB expression dropped under microaerobic and anaerobic environments, at both 30 °C and 37 °C, without changing expression pattern. In S. Typhimurium, the highest level of csgB was observed at 30 °C under aerobic conditions and was reduced at least sixfold at 37 °C. While csgB expression was reduced with a lower level of oxygen at 30 °C, the expression at 37 °C remained similar in all tested oxygen levels (Fig. 2B, blue). Overall, temperature and oxygen seem to impact csgB expression differently between serovars.
Effect of temperature and oxygen on csgB expression in S. Typhi and S. Typhimurium. csgB expression was evaluated by β-galactosidase assays in aerobic, microaerobic and anaerobic conditions, at 30 °C and 37 °C. (a) csgB expression in S. Typhi: pcsgBSTy-lacZ is in solid red and pcsgBSTm-lacZ is in red and light blue stripes. (b) csgB expression in S. Typhimurium: pcsgBSTm-lacZ is in solid blue and pcsgBSTy-lacZ is in blue and light red stripes. The experiments were performed at least 3 times with two technical replicates. The negative controls, WT S. Typhi and WT S. Typhimurium transformed with pRS415 empty vector did not present significant expression level in all studied conditions. *p < 0.01.
Knowing that there are polymorphisms within the genomes of these serovars10,15,32, the influence of the genetic background on csgB expression was investigated using the S. Typhimurium promoter (pcsgBSTm-lacZ) in S. Typhi (Fig. 2A, stripes) and the S. Typhi promoter (pcsgBSTy-lacZ) in S. Typhimurium (Fig. 2B, stripes). In all studied conditions, csgB expression in S. Typhi was 2 to 2.5-fold higher when expressing pcsgBSTm-lacZ. Interestingly, the expression patterns were still conserved, with the highest expression at 37 °C and no major oxygen effects on the expression pattern. When S. Typhimurium expressed the S. Typhi csgB promoter (pcsgBSTy-lacZ), no significant changes were observed compared with the S. Typhimurium csgB promoter. Thus, it was suspected that the genetic background might play a role in csgB regulatory differences between S. Typhi and S. Typhimurium.
csgD expression profiles
CsgD is the main activator of the csgBAC operon27. Since the csgB expression profile differs between S. Typhi and S. Typhimurium, we evaluated the csgD expression profile under the same conditions. Surprisingly, in S. Typhi, the highest expression of pcsgDSTy-lacZ was at 30 °C under aerobic conditions, and significantly decreased when oxygen was less present (Fig. 3A, red). This pattern was different from what was observed for csgB expression (Fig. 2A, red) and was more similar to S. Typhimurium csgB expression (Fig. 2B, blue). In S. Typhimurium, the highest level of csgD was observed at 30 °C under aerobic conditions at a similar level to S. Typhi (Fig. 3A, red). Expression decreased with a lowest level in the absence of oxygen, at both 30 °C and 37 °C (Fig. 3B, blue).
Effect of temperature and oxygen on csgD expression in S. Typhi and S. Typhimurium. csgD expression was evaluated by β-galactosidase assay in aerobic, microaerobic and anaerobic conditions at 30 °C and 37 °C. (a) csgD expression in S. Typhi: pcsgDSTy-lacZ is in solid red and pcsgDSTm-lacZ is in red and light blue stripes. (b) csgD expression in S. Typhimurium: pcsgDSTm-lacZ is in blue and pcsgDSTy-lacZ is in blue/light red stripes. The experiments were repeated at least 3 times with 2 technical replicates each. The negative controls, WT S. Typhi and WT S. Typhimurium transformed with pRS415 empty vector did not present significant expression level in all studied conditions. *p < 0.01.
Then, the influence of the genetic background was also investigated, by using the csgD promoter from S. Typhimurium (pcsgDSTm-lacZ) in S. Typhi (Fig. 3A, stripes) and the promoter from S. Typhi (pcsgDSTy-lacZ) in S. Typhimurium (Fig. 3B, stripes). When the pcsgDSTm-lacZ expression was evaluated in S. Typhi, a significant decrease of expression was observed in aerobic conditions at 30 °C, while a significant increase was observed in microaerobic conditions and no change in anaerobic conditions. No difference was observed at 37 °C, regardless of the oxygen level. When pcsgDSTy-lacZ was expressed in S. Typhimurium, the expression profiles were similar and conserved the same tendency, which was a decrease in expression with lower levels of oxygen, even if slightly higher in some conditions (Fig. 3B, stripes).
CsgD binding capacity
The csgD expression profiles were similar for both S. Typhi and S. Typhimurium strains whereas csgB expression, which is known to be regulated by the CsgD regulator, differed considerably. We therefore investigated the binding capacity of CsgD proteins to the csgB promotor regions of each of these serovars. The DNA sequence of the csgB promotor regions in both S. Typhi and S. Typhimurium strains is identical.
Interestingly, CsgD of S. Typhi lacks the last 8 amino acids of the S. Typhimurium CsgD protein due to a single-nucleotide polymorphism (SNP) that introduces a premature stop codon15,32. It is unknown whether the truncated CsgD protein present in S. Typhi is functional32. Moreover, these missing amino acids are located in the DNA binding region of the regulator33. To evaluate the capacity of S. Typhi CsgD to bind to csgB promoter region, an EMSA (electrophoretic motility shift assay) was performed using purified S. Typhi 6XHis-CsgD (6XHis-CsgDSTy) with a DNA probe (PcsgB) (Fig. 4). S. Typhimurium 6XHis-CsgD (6XHis-CsgDSTm) was used as a control. 6XHis-CsgDSTy binding to the csgB promoter region was reduced when compared to CsgD of S. Typhimurium. For 6XHis-CsgDSTy, a partial shift only occurred when 2 μg of protein were present. By contrast, 6XHis-CsgDSTm binding to the csgB promoter region began with 0.5 μg of protein and the band shift was complete with 1 μg of protein. These results demonstrate that the CsgD protein of S. Typhi has a lower DNA-binding capacity to the csgB promotor region when compared to CsgDSTm.
Capacity of CsgD proteins to bind the csgB promoter. (a) S. Typhi purified 6XHis-CsgDSTy and (b) S. Typhimurium purified 6XHis-CsgDSTm serving as a control. DNA probe PcsgB consists of the csgB promoter region fused with fluorescein FAM. The promoter sequence is identical between S. Typhi and S. Typhimurium. (1) Control well using 6XHis-CsgDSTm and the DNA probe. (2) Control well with DNA probe only. (3) Control well with purified proteins only at 4 μg. (4–10) Increasing concentrations of 6XHis-CsgD from 0.1 to 8 µg with DNA probe.
Effect of genomic substitution of csgD alleles in Salmonella Typhi and Typhimurium serovars
Knowing that CsgDSTy demonstrated a reduced DNA-binding capacity, we investigated the outcome of replacing the endogenous csgD gene in S. Typhi and S. Typhimurium with a heterogenous allele from the other serovar by allelic exchange in the genome. We then evaluated the effect of introduction of the different csgD alleles on csgB expression using the pcsgBSTy-lacZ or pcsgBSTm-lacZ fusion vectors (Fig. 5). As the greatest difference in csgB expression between S. Typhi and S. Typhimurium was observed during growth in aerobic conditions, this growth condition was selected for these experiments. In S. Typhi, introduction of the CsgDSTm allele increased csgB expression by fourfold at 30 °C and by 1.5-fold at 37 °C. However, this increase in expression still did not reach the same level as observed in WT S. Typhimurium. Interestingly, when grown at 30 °C, csgB expression decreased drastically by 14-fold in S. Typhimurium when the native csgD gene was replaced by csgDSTy. By contrast, when grown at 37 °C, substitution of the native CsgDSTm with CsgDSTy in S. Typhimurium did not change csgB expression levels. These results suggest that the CsgD from S. Typhi is less able to activate csgB gene expression especially at 30 °C. However, in S. Typhi it is likely that the decreased activity of CsgDSTy alone is not the only factor contributing to differences in curli gene expression.
Effect of substituting csgD alleles in S. Typhi and S. Typhimurium on csgB expression. Expression of csgB in Miller units at 30 °C and 37 °C was evaluated by β-galactosidase Assay. Expression of csgB of S. Typhi in S. Typhi WT (STy) and in S. Typhi expressing S. Typhimurium csgD (CsgDSTm) are presented in red. Expression of S. Typhimurium csgB in S. Typhimurium WT (STm) and in S. Typhimurium expressing S. Typhi csgD (CsgDSTy) are presented in blue. The experiment was repeated at least 3 times with 2 biological replicates each. The negative controls, WT S. Typhi and WT S. Typhimurium transformed with pRS415 empty vector did not present significant expression level in all studied conditions. *p < 0.0001.
As the csgB expression levels were modified after substitution of the different csgD alleles, CsgD production was evaluated by Western Blot in these strains at 30 °C (Fig. 6A) and 37 °C (Fig. 6B). Surprisingly, despite the increase in csgB expression observed in S. Typhi containing csgD from S. Typhimurium (Fig. 5), no CsgDSTm was detected in S. Typhi at either 30 °C or 37 °C. On Congo red agar, a slight phenotypic change was observed with the appearance of a red ring within the colony at 30 °C but this was absent at 37 °C. For S. Typhimurium strains, only the WT and the control strain ∆csgDSTm::csgDSTm (CsgDSTm reintroduced to complement the ∆csgDSTm mutant) at 30 °C showed production of a CsgD-specific band. Introduction of csgDSTy in S. Typhimurium caused loss of the typical rdar phenotype for a saw phenotype and no CsgD was detected by Western Blot (Fig. 6).
Impact of substituting csgD alleles on colonial morphology and production of CsgD protein. Detection of CsgD by Western Blot was performed on S. Typhi expressing csgD of S. Typhimurium (CsgDSTm) and S. Typhimurium expressing csgD of S. Typhi (CsgDSTy) and compared with wild-type protein expression—S. Typhi (STy) and S. Typhimurium (STm). Complementation controls were also tested—S. Typhi mutant ∆csgDSTy complemented with csgDSTy (∆csgDSTy::csgDSTy) and S. Typhimurium mutant ∆csgDSTm complemented with csgDSTm (∆csgDSTm::csgDSTm). All strains were grown in tryptone medium at (a) 30 °C and (b) 37 °C. Upper panels present detection of GroEL (~ 60 kDa) and lower panels present detection of CsgD (~ 25 kDa) by Western blot. Original blots are presented in Supplementary Fig. S3. Colonial morphology after 72 h was also evaluated by using Congo Red Plate Assay.
The previous set of experiments indicated that in S. Typhi, neither of the csgD alleles were able to produce a detectable protein, at either 30 °C or 37 °C. Further, when the csgDSTy allele was introduced into the S. Typhimurium strain background, no CsgD protein could be detected. These results suggest that CsgD production in S. Typhi may be silenced or nonfunctional. Further, in S. Typhimurium, lack of detection of any CsgDSTy protein could be due either instability of the truncated protein or differences in the csgDEFG promoter region in S. Typhimurium. In these strains, although the heterogenous copies of csgD alleles were introduced, the endogenous promoter regions were maintained.
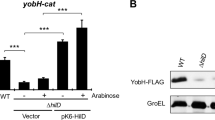
Thus, to verify if the inability of detecting CsgDSTm in S. Typhi was due to possible protein degradation or expression inhibition, we complemented the csgD mutants (∆csgDSTy and ∆csgDSTm) with csgD from each serovar expressed from a low-copy plasmid (pWSK-csgDSTy and pWSK-csgDSTm). Production of CsgD at 30 °C and 37 °C was evaluated by Western blot (Fig. 7). The control ∆csgDSTm complemented with pWSK-csgDSTm has a band for CsgD detection and the rdar phenotype was restored, but again, no CsgD was detected when CsgD of S. Typhi (pWSK-csgDSTy) was cloned into S. Typhimurium and the rdar phenotype was not restored. For S. Typhi, results were different then with the genomic substitutions of csgD alleles. For csgDSTm expressed from the vector promoter, CsgD was detected by Western Blot in S. Typhi (∆csgDSTy pWSK-csgDSTm) at both temperatures, but at lower level after growth at 37 °C. However, on Congo red agar, colonial morphology remained white and smooth with a red ring as in Fig. 6A.
Introduction of csgDSTy or csgDSTm alleles cloned on a low-copy vector to ΔcsgD mutants. Expression of CsgD was evaluated by Western blot of complemented strains grown in tryptone medium at 30 °C and 37 °C. (a) GroEL detection (~ 60 kDa). (b) CsgD detection (~ 25 kDa). Original blots are presented in Supplementary Fig. S3. (c) Congo Red Plate Assay of complemented and substituted strains of S. Typhi (∆csgDSTy) or S. Typhimurium (∆csgDSTm) with either pWSK-csgDSTy or pWSK-csgDSTm. (d) Congo Red Plate Assay on ∆csgD strains without plasmid as controls. Colonial morphology was evaluated after 72 h of incubation.
Introduction of a complete functional csg gene cluster to S. Typhi and S. Typhimurium strains
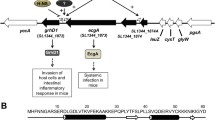
Since there were important differences in the csgD gene between S. Typhi and S. Typhimurium, we investigated whether other genes in the csg clusters contained polymorphisms that could affect production of curli. We compared S. Typhi and S. Typhimurium csg gene clusters and found multiple SNPs (silent, missense and nonsense SNPs) in the S. Typhi csg gene cluster (Fig. 8A and Supplementary Fig. S1). Due to the presence of multiple SNPs in different csg genes in S. Typhi, we introduced the entire csg gene cluster (csgABCDEFG) of S. Typhimurium into a csg mutant ∆csgSTy (∆csgBACDEFG) of S. Typhi using a low-copy pWSK plasmid (pWSK-csgSTm) and evaluated the phenotype by Congo Red Plate Assay (Fig. 8B). pWSK-csgSTm was also electroporated into a ∆csgSTm mutant of S. Typhimurium as a control. In the S. Typhimurium ∆csg strain, introduction of the csg gene clusters resulted in a regain of the rdar phenotype, confirming that production of curli can be complemented by this plasmid. However, when this same pWSK-csgSTm plasmid was cloned into the S. Typhi ∆csg mutant, no changes in colonial morphology were observed and S. Typhi colonies retained a saw phenotype. These results indicate that in addition to polymorphisms in the csg gene cluster that may alter csg gene expression and curli production, S. Typhi also likely contains other polymorphisms external to csg that reduce or inhibit csg gene expression or curli production under the growth conditions used in this study. To ensure that the lack of colonial morphology changes was not due to the presence of the Vi capsule- as extracellular components such as polysaccharide might interfere with Congo Red binding34, we also investigated colonial morphology in a Vi capsule-negative ∆tviB mutant of S. Typhi with pWSK-csgSTm. Regardless, loss of the Vi capsule did not alter the saw colony phenotype. Lastly, we investigated whether this lack of rdar phenotype was strain specific by introducing pWSK-csgSTm in different S. Typhi strains (Ty2, SarB63 and SarB64) (Fig. 8C). Again, no significant morphology change was observed suggesting a potential serovarspecific curli down regulation.
Complementation of curli operons. (a) Presence of single-nucleotide polymorphisms (SNPs) in the S. Typhi csg gene cluster. Missense SNPs are presented in green, nonsense SNPs in red and silent SNPs are in yellow. The nucleotides and amino acid sequences are presented in Fig. S1. (b) Congo Red Plate Assay at 30 °C or 37 °C of S. Typhi (STy) and S. Typhimurium (STm) curli ∆csg (∆csgBACDEFG) mutant and ∆csg complemented with a complete csg gene cluster from S. Typhimurium present on vector pWSK-csgSTm (pWSK-csgBACDEFGSTm). A ∆tviB mutant and ∆tviB::pWSK-csgSTm containing a functional Csg system from S. Typhimurium were used as controls to determine if the Vi capsule could interfere with Congo Red binding and colonial morphology. (c) Congo Red Plate Assay at 30 °C or 37 °C of other S. Typhi strains: SarB63 and SarB64.
Discussion
Curli are known to be an important component of biofilm formation by non-typhoidal Salmonella enterica serovars such as S. Typhimurium18,20. S. Typhi persistence in asymptomatic carrier hosts was demonstrated to involve biofilm formation in the gallbladder5,6,7. Expression and regulation of curli in S. Typhi are poorly understood. It was suggested that curli were non-functional in S. Typhi, as a white and smooth phenotype (saw) was observed on Congo red agar using standard growth conditions for the curli positive rdar (red, dry and rough) morphotype21,31,32. However, antibodies against curli components have been found in patients with typhoid fever35, suggesting a potential for curli production during typhoid fever infections.
By comparing media mimicking conditions encountered at different infection stages, we have determined that the highest level of expression of csgB in laboratory conditions was in tryptone medium, a low salt and low nutrient medium (Fig. 1). This medium is also known to promote curli production in S. Typhimurium31. Other growth conditions such as temperature and oxygen levels encountered during infection were also investigated and their effect on curli gene expression in S. Typhi was compared to a S. Typhimurium strain. The highest expression of csgB in S. Typhi was always at 37 °C (Fig. 2). This is in line with a potential for csg gene expression by S. Typhi in the human host. Expression of csgB in S. Typhimurium was sixfold higher at 30 °C than at 37 °C. This was only observed under aerobic conditions, while reducing oxygen levels decreased the effect of temperature. Temperature and oxygen levels seem to have an interdependent effect on S. Typhimurium. However, oxygen levels did not affect expression in S. Typhi. These results confirm that csg gene expression in S. Typhi does not follow the same regulation as S. Typhimurium. Thus, we investigated the role of the genetic background on curli expression by measuring expression of the S. Typhimurium csgB promoter including the intergenic region in S. Typhi (Fig. 2). About 2–2.5-fold higher expression was observed in all studied conditions and growth at 37 °C promoted the highest level of expression. However, strikingly, expression levels were distinct compared to those in S. Typhimurium. Based on these results, it was expected that in S. Typhimurium the csgB promoter of S. Typhi should have a decreased expression profile. Surprisingly, no difference in expression was observed in all the studied conditions. This further implies that regulation of curli in S. Typhi is different.
Many regulators are implicated in curli expression. Each one has various roles responding to environmental stress and culture conditions and temperature20,21. CsgD is the main regulator that activates curli expression. Thus, we evaluated its expression profile to compare it with csgB expression (Fig. 3)27. The most striking result was that both serovars presented similar expression profiles at 30 °C under aerobic conditions. Knowing that csgB expression was significantly different (Fig. 2), these results suggested that the potential expression differences between these serovars may be due to post-transcriptional regulation that implicates the CsgD regulator. It was previously reported that csgD expression was highest in microaerobic conditions in S. Typhimurium21,30. While our results propose that expression levels of csgB and csgD were higher in aerobic conditions. However, the growth conditions were different between these studies.
Since the expression level of csgD was similar between S. Typhi and S. Typhimurium but expression of csgB was lower in S. Typhi, we investigated the impact of CsgD regulation on csgB expression. CsgD is a member of the UhpA/FixJ/LuxR family with a DNA binding site in the C-terminal portion27,33. S. Typhi CsgD (CsgDSTy) lacks the last 8 amino acids compared to the CsgDSTm protein due to a nonsense polymorphism32. Thus, we investigated if CsgDSTy binding to the csgB promoter is altered compared to CsgDSTm (Fig. 4). We found that CsgDSTy was only able to partially bind the csgB promoter region even at high concentration while CsgDSTm was able to effectively bind at lower protein concentrations. Our results demonstrated that CsgDSTy had reduced functionality and binding-capacity to the csgB promoter region. This might explain the significantly lower expression of csgB in S. Typhi compared to S. Typhimurium (Fig. 2). To confirm if this reduced binding profile alters curli regulation and expression, csgDSTy was integrated into the S. Typhimurium genome to substitute for the native csgDSTm open reading frame without altering the native promoter region. In this strain, csgB expression was drastically reduced (Fig. 5) and the rdar phenotype of S. Typhimurium on Congo Red agar was lost (Fig. 6). Interestingly, the CsgDSTy protein could not be detected by Western Blot despite growth conditions in S. Typhimurium that favour CsgD expression. The possibility of issues with antibody specificity were eliminated since the monoclonal anti-CsgD antibody also effectively detected the 6XHis-CsgDSTy protein (Supplementary Fig. S2). The absence of production of CsgDSTy suggests that the protein is degraded or not functional in S. Typhimurium. CsgDSTy in S. Typhi was also never detected under any conditions, and will be discussed below.
When the functional csgDSTm gene was introduced into S. Typhi, csgB expression did increase by 1.5- to 4-fold depending on the growth conditions. However, expression was still significantly lower than that observed in WT S. Typhimurium (Fig. 5). This suggests that the CsgDSTy was only one of a number of other factors leading to differences in csg gene expression between serovars. Other genomic polymorphisms outside of the csg gene cluster are likely to prevent high level expression of curli in S. Typhi under the growth conditions used in the study. Surprisingly, despite the increase of gene expression, no CsgDSTm protein was detected by Western Blot (Fig. 6). Since the csgD allele substitution experiments only replaced the open reading frames encoding the CsgD regulatory proteins, it is possible that differences in csgDEFG promoter regions could also affect expression of the csg gene clusters. We therefore evaluated the effect of csgDSTm expressed from its native promoter region in a low-copy plasmid to complement ∆csgD mutant to bypass this regulation. In this way, we were able to detect CsgDSTm in S. Typhi (Fig. 7). This result refutes the likelihood that in S. Typhi the CsgDSTm protein may be unstable or is degraded and confirms that in S. Typhi expression of csgD from the endogenous CsgDSTy promoter is inhibited under the growth conditions used in the laboratory. Despite the production of CsgDSTm from the low-copy plasmid in S. Typhi, this strain still maintained a saw phenotype on Congo Red Agar, indicating that other polymorphisms or modifications in the S. Typhi genome may further affect production of curli.
The nonsense SNP in the CsgDSTy sequence reduced DNA-binding efficiency of this regulatory protein, suggesting an underlying reason why curli gene expression may be altered in S. Typhi. However, as the rdar morphotype was not recovered after introduction of a functional CsgDSTm regulatory protein, it is possible that polymorphisms in other genes involved in curli expression or synthesis could also alter curli function in S. Typhi. In fact, analysis of the S. Typhi genome in the csg gene cluster identified several additional SNPs and most of the genes encoding curli assembly components present SNPs, with the majority being silent, although two are missense and only CsgD has a nonsense SNP (Fig. 8). To determine if a combination of modifications within the csg gene cluster in S. Typhi was potentially responsible for differences in curli gene expression or production in S. Typhi, we cloned the entire functional gene cluster from S. Typhimurium into a low-copy vector and introduced this plasmid into S. Typhi and S. Typhimurium ∆csg (∆csgBACDEFG) mutants. This pWSK-csgSTm plasmid was confirmed to be functional as complementation of the S. Typhimurium ∆csg mutant which had a saw colonial morphotype regained the rdar phenotype, that is associated with production of curli (Fig. 8). By contrast, the pWSK-csgSTm plasmid when introduced into the S. Typhi ∆csg mutant, did not alter the saw phenotype at either 30 °C or 37 °C. The lack of a rdar phenotype after introduction of the csgSTm gene cluster was also not due to presence of the Vi capsule which might interfere with the Congo Red Plate Assay. Congo Red dye could react differently with various cellular surface components34,36. Loss of the Vi capsule in a strain containing the plasmid encoding the csgSTm gene cluster remained white and smooth (Fig. 8B), as well as other Typhi strains like Ty2, SarB63 and SarB64 expressing csgSTm gene (Fig. 8C). Thus, this lack of morphology change seems to be serovar-specific.
Taken together, these results suggest that in S. Typhi, altered regulation or inhibition of curli production is multitiered and involves a missense SNP in CsgD that truncates the protein and reduces its DNA-binding capacity. In addition, there are likely additional as yet undefined modifications or polymorphisms beyond the csg gene cluster that lead to inhibition of curli production. Since the functional csg gene cluster from S. Typhimurium cloned into S. Typhi did not confer production of curli or the rdar colonial morphotype even under conditions favourable for curli production at 30 °C. Conditions that may promote expression of curli fimbriae in S. Typhi are still unknown as well as overall mechanisms underlying differences in its multitiered regulatory control.
Curli are known to promote adherence to intestinal epithelial cells and activate the immune system via toll-like receptor TLR-237,38. TLR-2 also plays a role in PI3K mediated tight-junction barrier response which is a host innate immune mechanism to prevent bacterial dissemination. Hence, S. Typhimurium causes a local intestinal inflammation39. Contrary to non-typhoidal Salmonella infection, S. Typhi does not cause inflammation in the gut and it is able to bypass some innate immune defenses during infection40,41. Thus, in the intestine inhibition of curli expression may be critical for S. Typhi to prevent TLR-2 activation leading to increased intestinal epithelial barrier integrity. However, curli might nevertheless play a role at other stages of S. Typhi infection under specific conditions yet to be determined such as for biofilm production to promote persistence and the chronic carrier state. We are currently investigating potential alternative regulation mechanisms for curli expression in S. Typhi that may be distinct from regulation in most other Salmonella enterica serovars.
Conclusions
Curli fimbriae encoded by the csg gene cluster have been characterized extensively in E. coli and other Salmonella. However, little is known specifically about curli gene expression in Salmonella Typhi. Here, we demonstrate that S. Typhi curli gene expression does not respond to the same environmental cues as the csg genes in S. Typhimurium. Importantly, it was demonstrated that the regulatory protein CsgD, which is the major regulator of curli production and biofilm formation in S. Typhimurium, has a reduced DNA-binding capacity to the csgB promoter, which controls expression of the curli subunit encoding gene. Further, introduction of a functional csg gene cluster cloned from S. Typhimurium was unable to promote production of curli in S. Typhi under conditions that are normally favourable for its production in Salmonella and E. coli. Taken together, results confirm that the regulatory mechanisms underlying curli gene regulation in S. Typhi are multitiered and include genomic polymorphism within the csgD gene and other as yet undefined modifications elsewhere in the genome. Results also suggest that if curli are produced by S. Typhi, expression is subject to regulatory cues that are distinct from those regulating curli in Salmonella Typhimurium and E. coli.
Materials and methods
Conditions and media used for bacterial growth
All bacterial strains and plasmids used are listed in Supplementary Table S1. All strains were routinely grown in lysogeny broth (LB) or LB agar at 37 °C overnight (18 h). When required, antibiotics or supplements were added at the following concentrations: 100 μg/mL for ampicillin, 34 μg/mL for chloramphenicol, 50 μg/mL for diaminopimelic acid (DAP) and 0.5 mM for IPTG.
Cloning of curli promoters and β-galactosidase assays
Promotor regions of csgBAC (PcsgB) and csgDEFG (PcsgD) from S. Typhi (STy) ISP1820 and S. Typhimurium (STm) SL1344 were amplified by PCR using csg_prom_F_short_EcoRI/csg_prom_R_BamHI for PcsgB and csgD_Prom_F_EcoRI/csgD_Prom_R_BamHI for promoter PcsgD (Supplementary Table S2). The promoters were inserted in vector pRS415 to generate lacZ fusions using restriction enzymes (Anza) EcoRI and BamHI, and T4 ligase (NEB). The resulting plasmid was transformed into WT ISP1820 and SL1344 by electroporation as described in O’Callaghan et al.42. Clones were confirmed by PCR and sequenced using respective promoter forward primer and the lacZ_alpha_R primer from the pRS415 vector. Empty vector (pRS415) was also electroporated in wild-type strains to serve as negative control for β-galactosidase assay.
After growth under various culture conditions, promoter expression was evaluated by β-galactosidase assay as described by Miller43. For csgB expression, various media were used to mimic different environments encountered during the infection process. For host invasion, SPI-1 was induced in LB with 0.3 M NaCl without agitation for low oxygenation44,45. To mimic intracellular conditions encountered in macrophages, SPI-2 was induced in low phosphate medium (LPM)46. For high osmolarity conditions, salt (0.3 M NaCl) was added to LB and incubated with agitation. M63 minimal medium, was used as a defined medium47. Effect of bile was tested using LB with 3% ox-bile (Sigma), as S. Typhi produces biofilm in gallbladders of asymptomatic chronic hosts7,20. LB without any salt or yeast extract also called tryptone medium, was also tested as it is the optimal medium for S. Typhimurium curli expression under laboratory conditions31. For determination of csgB and csgD expression levels, bacteria were grown at different temperatures. 37 °C was studied as S. Typhi is a human specific pathogen. Experiments were also performed at 30 °C, as it is the optimal temperature for curli expression in S. Typhimurium31. For oxygen impact on expression, bacteria were incubated with agitation for aerobic condition, without agitation for microaerobic conditions and using a GasPak system (MGC) for anaerobic growth. Experiments were done at least three times in duplicate. In all the studied conditions, no significant expression was observed for the negative control.
Purification of 6XHis-CsgD protein and electrophoretic motility shift assay (EMSA)
High levels of ISP1820 and SL1344 CsgD were produced using the pET system (Novagen). The csgD gene of both serovars was cloned in expression vector pET14b by PCR using primers CsgD-F-NdeI/CsgD-R-BamHI and transformed in E. coli host strain BL21(DE3). To harvest 6XHis-CsgD, a preculture was performed overnight, then diluted 1:50 in 1 L of fresh LB. The culture was incubated until OD600 reached 0.5–0.6 followed by addition of 0.5 mM IPTG and induction at 25 °C overnight. Bacteria were then treated and lysed following as described in the pET System Manual (Novagen) to isolate the soluble fraction. 6XHis-CsgD protein was then purified by gravity affinity chromatography (Bio-RAD) using Ni-NTA resin (Thermo-Fisher).
To perform the EMSA, a fluorescent probe of the PcsgB promoter region was synthesized by PCR amplification using a fluorescein fused primer curli-inter-F-FAM and curli-inter-R. The selected csgB promoter region sequences of S. Typhi ISP1820 and S. Typhimurium SL1344 are identical. Different concentrations of previously purified 6×His-CsgD with DNA probes were used. A 20 μL volume of each mixture was prepared in EMSA buffer (50 mM NaCl, 20 mM Tris pH 7.4 and 0.02% sodium azide). Every reaction was incubated for 40 min at 30 °C to allow protein binding before adding 5 μL of loading buffer (50 mM Tris pH7.4, 50 mM EDTA, 0.1% Bromophenol Blue and 12.5% sucrose). Migration of the reactions was performed using 5% native polyacrylamide gel containing 0.5×TAE buffer (20 mM Tris Base, 10 mM acetic acid, 0.5 mM EDTA) and 2% glycerol during 30 min at 150 V. Imaging of fluorescent probes was performed using a ChemiDoc imaging system (Bio-RAD) and Image Lab Software.
Cloning, deletion and allelic exchange of csgD
Mutagenesis of csgD was performed by allelic exchange with suicide vector pMEG375 as described in Forest et al.48 using two pairs of primers, CsgD_F1/CsgD_R2 and CsgD_F3/CsgD_R4, targeting the upstream and downstream region of the gene. csgD markerless and nonpolar deletion was done in S. Typhi ISP1820 and S. Typhimurium SL1344.
For gene complementation by low-copy plasmid pWSK29, csgD from S. Typhi or S. Typhimurium was amplified by PCR using primer pair csgD_Prom_F_BamHI and csgD_R_XbaI. The generated insert was digested with BamHI and XbaI (Anza) and ligated with T4 ligase (NEB). The same procedure was used for cloning of the csg gene cluster with primer pair Csg_F and Csg_R. Plasmids were transformed into the following mutants and Salmonella strains: ∆csgD, ∆csg (∆csgBACDEFG), ∆tviB, wild type Ty2, SarB63 and SarB64.
Introduction of the heterogenous csgD gene from the other serovar was achieved by cloning the csg genes from S. Typhi ISP1820 or S. Typhimurium SL1344 by PCR using primer pair csgD_swap F/csgD_swap_R. The generated amplicons were inserted into suicide vector pMEG375 following the same procedure as the described for mutagenesis. Allelic exchange was performed using the ΔcsgD mutant of S. Typhi ISP1820 and S. Typhimurium SL1344 as recipients. Each serovar received the heterogenous csgD from the other serovar. Complementation with the endogenous csgD gene was also performed as a control. All vectors were confirmed by PCR and sequencing.
Congo red plate assay
Strains were grown overnight in LB with agitation and ampicillin when required. A volume of 3.5 μL of culture was plated on Congo Red Agar (1% tryptone, 1% agar, 40 μL/mL of Congo Red Dye, 0.8 μL/mL of Coomassie Blue) and ampicillin when required. Plates were incubated at 30 °C or 37 °C for 72 h. Pictures of colonies were taken using an Olympus DP22 digital camera and its software with Olympus SZ61 stereomicroscope.
SDS-PAGE and western blot
Bacteria were grown in 50 mL of tryptone medium at 30 °C or 37 °C overnight with agitation. When needed, ampicillin was added to cultures. Cells were harvested and lysed as described above in the 6XHis-CsgD purification section. Protein concentration from lysates was determined by Bradford Assay. Roughly 0.04–0.05 mg of total proteins were loaded per well. SDS-PAGE was performed with 5% stacking and 12% resolving mini gels. Proteins were transferred on PVDF membranes for 1 h at 100 V in a Mini Trans-Blot Cell. A volume of 1 μg/mL of monoclonal mouse antibodies against CsgD were used to detect the protein of interest32. Goat antibodies against mouse IgG coupled with HRP were used as secondary antibodies at a dilution of 1:10,000 (GenScript). GroEL was detected using rabbit anti-GroEL at a dilution of 1:80,000 (GenScript) with rabbit anti-IgG coupled with HRP as secondary antibodies at 1:10,000 dilution (GenScript). Chemiluminescence was detected using SuperSignal West Pico PLUS (Thermo Fisher) and visualized with a Amersham Imager 600.
Statistical analysis
All statistical analysis was performed using two-tailed unpaired parametric Student’s t-test and the mean ± SEM is represented using the software GraphPad Prism 8.0.1.
Data availability
The data generated during the current study are available from the corresponding author on reasonable request.
References
World Health Organization, Typhoid Fever. https://www.who.int/news-room/q-a-detail/typhoid-fever (2020).
Rowe, B., Ward, L. R. & Threlfall, E. J. Multidrug-resistant Salmonella typhi: A worldwide epidemic. Clin. Infect. Dis. 24(Suppl 1), S106–S109. https://doi.org/10.1093/clinids/24.supplement_1.s106 (1997).
Marchello, C. S., Birkhold, M. & Crump, J. A. Complications and mortality of typhoid fever: A global systematic review and meta-analysis. J. Infect. 81, 902–910. https://doi.org/10.1016/j.jinf.2020.10.030 (2020).
Levine, M. M., Black, R. E., Lanata, C., Chilean Typhoid Committee. Precise estimation of the numbers of chronic carriers of Salmonella typhi in Santiago, Chile, an endemic area. J. Infect. Dis. 146, 724–726 (1982).
Gunn, J. S. et al. Salmonella chronic carriage: Epidemiology, diagnosis, and gallbladder persistence. Trends Microbiol. 22, 648–655. https://doi.org/10.1016/j.tim.2014.06.007 (2014).
Prouty, A. M., Schwesinger, W. H. & Gunn, J. S. Biofilm formation and interaction with the surfaces of gallstones by Salmonella spp. Infect. Immun. 70, 2640–2649. https://doi.org/10.1128/iai.70.5.2640-2649.2002 (2002).
Di Domenico, E. G., Cavallo, I., Pontone, M., Toma, L. & Ensoli, F. Biofilm producing Salmonella Typhi: Chronic colonization and development of gallbladder cancer. Int. J. Mol. Sci. https://doi.org/10.3390/ijms18091887 (2017).
Rabsch, W. et al. Salmonella enterica serotype Typhimurium and its host-adapted variants. Infect. Immun. 70, 2249–2255. https://doi.org/10.1128/iai.70.5.2249-2255.2002 (2002).
Rabsch, W., Tschäpe, H. & Bäumler, A. J. Non-typhoidal salmonellosis: Emerging problems. Microbes Infect. 3, 237–247. https://doi.org/10.1016/s1286-4579(01)01375-2 (2001).
McClelland, M. et al. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413, 852–856. https://doi.org/10.1038/35101614 (2001).
Sabbagh, S. C., Forest, C. G., Lepage, C., Leclerc, J.-M. & Daigle, F. So similar, yet so different: Uncovering distinctive features in the genomes of Salmonella enterica serovars Typhimurium and Typhi. FEMS Microbiol. Lett. 305, 1–13. https://doi.org/10.1111/j.1574-6968.2010.01904.x (2010).
Liu, S. L. & Sanderson, K. E. Rearrangements in the genome of the bacterium Salmonella typhi. Proc. Natl. Acad. Sci. U.S.A. 92, 1018–1022. https://doi.org/10.1073/pnas.92.4.1018 (1995).
Liu, S.-L. & Sanderson, K. E. Highly plastic chromosomal organization in Salmonella typhi. Proc. Natl. Acad. Sci. 93, 10303–10308 (1996).
Alokam, S., Liu, S.-L., Said, K. & Sanderson, K. E. Inversions over the terminus region in Salmonella and Escherichia coli: IS 200 s as the sites of homologous recombination inverting the chromosome of Salmonella enterica serovar Typhi. J. Bacteriol. 184, 6190–6197 (2002).
Parkhill, J. et al. Complete genome sequence of a multiple drug resistant Salmonella enterica serovar Typhi CT18. Nature 413, 848–852. https://doi.org/10.1038/35101607 (2001).
Latasa, C. et al. BapA, a large secreted protein required for biofilm formation and host colonization of Salmonella enterica serovar enteritidis. Mol. Microbiol. 58, 1322–1339. https://doi.org/10.1111/j.1365-2958.2005.04907.x (2005).
Zogaj, X., Nimtz, M., Rohde, M., Bokranz, W. & Römling, U. The multicellular morphotypes of Salmonella typhimurium and Escherichia coli produce cellulose as the second component of the extracellular matrix. Mol. Microbiol. 39, 1452–1463. https://doi.org/10.1046/j.1365-2958.2001.02337.x (2001).
Jonas, K. et al. Roles of curli, cellulose and BapA in Salmonella biofilm morphology studied by atomic force microscopy. BMC Microbiol. 7, 70. https://doi.org/10.1186/1471-2180-7-70 (2007).
Römling, U., Sierralta, W. D., Eriksson, K. & Normark, S. Multicellular and aggregative behaviour of Salmonella typhimurium strains is controlled by mutations in the agfD promoter. Mol. Microbiol. 28, 249–264. https://doi.org/10.1046/j.1365-2958.1998.00791.x (1998).
Barnhart, M. M. & Chapman, M. R. Curli biogenesis and function. Annu. Rev. Microbiol. 60, 131–147. https://doi.org/10.1146/annurev.micro.60.080805.142106 (2006).
Römling, U., Bian, Z., Hammar, M., Sierralta, W. D. & Normark, S. Curli fibers are highly conserved between Salmonella typhimurium and Escherichia coli with respect to operon structure and regulation. J. Bacteriol. 180, 722–731 (1998).
Hammar, M., Bian, Z. & Normark, S. Nucleator-dependent intercellular assembly of adhesive curli organelles in Escherichia coli. Proc. Natl. Acad. Sci. 93, 6562. https://doi.org/10.1073/pnas.93.13.6562 (1996).
Gibson, D. L., White, A. P., Rajotte, C. M. & Kay, W. W. AgfC and AgfE facilitate extracellular thin aggregative fimbriae synthesis in Salmonella enteritidis. Microbiology (Reading, England) 153, 1131–1140. https://doi.org/10.1099/mic.0.2006/000935-0 (2007).
Loferer, H., Hammar, M. & Normark, S. Availability of the fibre subunit CsgA and the nucleator protein CsgB during assembly of fibronectin-binding curli is limited by the intracellular concentration of the novel lipoprotein CsgG. Mol. Microbiol. 26, 11–23. https://doi.org/10.1046/j.1365-2958.1997.5231883.x (1997).
Robinson, L. S., Ashman, E. M., Hultgren, S. J. & Chapman, M. R. Secretion of curli fibre subunits is mediated by the outer membrane-localized CsgG protein. Mol. Microbiol. 59, 870–881. https://doi.org/10.1111/j.1365-2958.2005.04997.x (2006).
Nenninger, A. A., Robinson, L. S. & Hultgren, S. J. Localized and efficient curli nucleation requires the chaperone-like amyloid assembly protein CsgF. Proc. Natl. Acad. Sci. U.S.A. 106, 900–905. https://doi.org/10.1073/pnas.0812143106 (2009).
Hammar, M., Arnqvist, A., Bian, Z., Olsén, A. & Normark, S. Expression of two csg operons is required for production of fibronectin- and congo red-binding curli polymers in Escherichia coli K-12. Mol. Microbiol. 18, 661–670 (1995).
Gibson, D. L. et al. Salmonella produces an O-antigen capsule regulated by AgfD and important for environmental persistence. J. Bacteriol. 188, 7722–7730. https://doi.org/10.1128/jb.00809-06 (2006).
Römling, U., Rohde, M., Olsén, A., Normark, S. & Reinköster, J. AgfD, the checkpoint of multicellular and aggregative behaviour in Salmonella typhimurium regulates at least two independent pathways. Mol. Microbiol. 36, 10–23. https://doi.org/10.1046/j.1365-2958.2000.01822.x (2000).
Gerstel, U. & Römling, U. Oxygen tension and nutrient starvation are major signals that regulate agfD promoter activity and expression of the multicellular morphotype in Salmonella typhimurium. Environ. Microbiol. 3, 638–648. https://doi.org/10.1046/j.1462-2920.2001.00235.x (2001).
Collinson, S. K., Emödy, L., Müller, K. H., Trust, T. J. & Kay, W. W. Purification and characterization of thin, aggregative fimbriae from Salmonella enteritidis. J. Bacteriol. 173, 4773–4781. https://doi.org/10.1128/jb.173.15.4773-4781.1991 (1991).
MacKenzie, K. D. et al. Parallel evolution leading to impaired biofilm formation in invasive Salmonella strains. PLoS Genet. 15, e1008233. https://doi.org/10.1371/journal.pgen.1008233 (2019).
Ogasawara, H., Yamamoto, K. & Ishihama, A. Role of the biofilm master regulator CsgD in cross-regulation between biofilm formation and flagellar synthesis. J. Bacteriol. 193, 2587–2597. https://doi.org/10.1128/jb.01468-10 (2011).
Teather, R. M. & Wood, P. J. Use of Congo red-polysaccharide interactions in enumeration and characterization of cellulolytic bacteria from the bovine rumen. Appl. Environ. Microbiol. 43, 777–780. https://doi.org/10.1128/aem.43.4.777-780.1982 (1982).
Harris, J. B. et al. Identification of in vivo-induced bacterial protein antigens during human infection with Salmonella enterica serovar Typhi. Infect. Immun. 74, 5161–5168. https://doi.org/10.1128/iai.00488-06 (2006).
Yakupova, E. I., Bobyleva, L. G., Vikhlyantsev, I. M. & Bobylev, A. G. Congo Red and amyloids: History and relationship. Biosci. Rep. https://doi.org/10.1042/bsr20181415 (2019).
Tükel, C. et al. Responses to amyloids of microbial and host origin are mediated through toll-like receptor 2. Cell Host Microbe 6, 45–53. https://doi.org/10.1016/j.chom.2009.05.020 (2009).
Tükel, C. et al. CsgA is a pathogen-associated molecular pattern of Salmonella enterica serotype Typhimurium that is recognized by Toll-like receptor 2. Mol. Microbiol. 58, 289–304. https://doi.org/10.1111/j.1365-2958.2005.04825.x (2005).
Oppong, G. O. et al. Epithelial cells augment barrier function via activation of the Toll-like receptor 2/phosphatidylinositol 3-kinase pathway upon recognition of Salmonella enterica serovar Typhimurium curli fibrils in the gut. Infect. Immun. 81, 478–486. https://doi.org/10.1128/iai.00453-12 (2013).
Wangdi, T., Winter, S. E. & Bäumler, A. J. Typhoid fever: “you can’t hit what you can’t see”. Gut Microbes 3, 88–92. https://doi.org/10.4161/gmic.18602 (2012).
Dougan, G. & Baker, S. Salmonella enterica serovar Typhi and the pathogenesis of typhoid fever. Annu. Rev. Microbiol. 68, 317–336. https://doi.org/10.1146/annurev-micro-091313-103739 (2014).
O’Callaghan, D. & Charbit, A. High efficiency transformation of Salmonella typhimurium and Salmonella typhi by electroporation. Mol. Gen. Genet. MGG 223, 156–158. https://doi.org/10.1007/bf00315809 (1990).
Miller, J. H. Assay of β-galactosidase. Experiments in Molecular Genetics (1972).
Lee, C. A., Jones, B. D. & Falkow, S. Identification of a Salmonella typhimurium invasion locus by selection for hyperinvasive mutants. Proc. Natl. Acad. Sci. U.S.A. 89, 1847–1851. https://doi.org/10.1073/pnas.89.5.1847 (1992).
Weinstein, D. L., O’Neill, B. L., Hone, D. M. & Metcalf, E. S. Differential early interactions between Salmonella enterica serovar Typhi and two other pathogenic Salmonella serovars with intestinal epithelial cells. Infect. Immun. 66, 2310–2318. https://doi.org/10.1128/iai.66.5.2310-2318.1998 (1998).
Coombes, B. K., Brown, N. F., Valdez, Y., Brumell, J. H. & Finlay, B. B. Expression and secretion of Salmonella pathogenicity island-2 virulence genes in response to acidification exhibit differential requirements of a functional type III secretion apparatus and SsaL. J. Biol. Chem. 279, 49804–49815. https://doi.org/10.1074/jbc.M404299200 (2004).
Leclerc, J. M., Dozois, C. M. & Daigle, F. Role of the Salmonella enterica serovar Typhi Fur regulator and small RNAs RfrA and RfrB in iron homeostasis and interaction with host cells. Microbiology (Reading) 159, 591–602. https://doi.org/10.1099/mic.0.064329-0 (2013).
Forest, C. et al. Contribution of the stg fimbrial operon of Salmonella enterica serovar Typhi during interaction with human cells. Infect. Immun. 75, 5264–5271. https://doi.org/10.1128/iai.00674-07 (2007).
Acknowledgements
We thank Dr. Karine Dufresne (Western University) for her participation in the initiation of this project. We thank Dr. Aaron P. White (University of Saskatchewan) for providing the monoclonal anti-CsgD antibodies. This work was supported by a grant RGPIN-2020-05233 from the Natural Sciences and Engineering Research Council of Canada and by funding from Swine and Poultry Infectious Diseases Research Center (CRIPA) funded by Fonds de recherche du Québec- Nature et technologies (FRQNT).
Author information
Authors and Affiliations
Contributions
All authors conceptualized the project. C.O. contributed to the methodology, investigation, validation and data curation. C.O. and F.D. contributed to the formal analysis and original draft preparation. F.D. supervised and administrated the project and contributed for the resources. All authors reviewed and edited the manuscript. All authors have read and approved the submitted manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Ou, C., Dozois, C.M. & Daigle, F. Differential regulatory control of curli (csg) gene expression in Salmonella enterica serovar Typhi requires more than a functional CsgD regulator. Sci Rep 13, 14905 (2023). https://doi.org/10.1038/s41598-023-42027-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-42027-y
- Springer Nature Limited