Abstract
Ultraviolet (UV) irradiation offers an effective and convenient method for the disinfection of pathogenic microorganisms. However, UV irradiation causes protein and/or DNA damage; therefore, further insight into the performance of different UV wavelengths and their applications is needed to reduce risks to the human body. In this paper, we determined the efficacy of UV inactivation of the SARS-CoV-2 omicron BA.2 and BA.5 variants in a liquid suspension at various UV wavelengths by the 50% tissue culture infection dose (TCID50) method and quantitative polymerase chain reaction (qPCR) assay. The inactivation efficacy of 220 nm light, which is considered safe for the human body, was approximately the same as that of health hazardous 260 nm light for both BA.2 and BA.5. Based on the inactivation rate constants determined by the TCID50 and qPCR methods versus the UV wavelength, the action spectra were determined, and BA.2 and BA.5 showed almost the same spectra. This result suggests that both variants have the same UV inactivation characteristics.
Similar content being viewed by others
Introduction
With the global outbreak of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and the emergence of their new variants, there is a great demand for developing and demonstrating efficient disinfection technologies to protect against various pathogenic viruses and bacteria1,2,3. In this case, vaccines provide effective protection against the infection, the efficacy and supply speed of these vaccines against future emerging SARS-CoV-2 variants are not clear at the present stage4. Therefore, it is important to prepare additional strategies to mitigate public health risks during the pre-vaccine development period against emerging pathogens.
Disinfection by ultraviolet (UV) irradiation is attracting special interest to reduce SARS-CoV-2 transmission because UV irradiation offers an effective and convenient method for the inactivation of pathogenic microorganisms, including SARS-CoV-25,6,7,8,9,10. In particular, the wavelength range from 200 to 235 nm, often referred to as far-UVC, has attracted increasing attention as a novel disinfection wavelength. Far-UVC light shows a strong germicidal effect on pathogenic viruses and bacteria11,12,13,14,15 and has been shown to be harmless to mammalian cells due to the strong absorption effect of the stratum corneum layer16,17,18,19,20. However, its safety profile in mammalian cells has been much less thoroughly documented, and there are numerous reports suggesting that far-UVC light is not as safe as irradiation far beyond threshold levels21,22,23,24,25,26 since it significantly damages epidermal cells, leading to the formation of erythema and cyclopyrimidine dimers21,22,23,24,26.
Furthermore, the inactivation dose reported to achieve a certain log-reduction varies widely from approximately 1 to 20 mJ/cm25,10,27,28,29,30,31,32,33,34. Such inconsistencies might be caused by varied experimental conditions and setups employed. For example, many light sources, such as UV-LEDs, KrCl-excimer lamps, and metal vapor discharge lamps, have been used to inactivate SARS-CoV-25,14,27,28,29,30,31,32,33,34; however, it is difficult to compare the magnitude of dose and the inactivation efficacy for these different UV wavelength regions due to the differences in both the strains of SARS-CoV-2 and experimental conditions such as the spectrum of the light sources. Therefore, there is a substantial need for systematic experiments with varying UV wavelengths and without variance in other experimental conditions.
In this paper, we describe the inactivation efficacy of the SARS-CoV-2 omicron BA.2 and BA.5 variants in a viral suspension as a function of UV wavelength with 10 nm bandwidth based on the construction of a UV wavelength tunable irradiation source. We employed the standard 50% tissue culture infection dose (TCID50) method and a quantitative polymerase chain reaction assay (qPCR) to detect UV damage to the viral genome. We have found a strong correlation between the TCID50 and qPCR. Based on the inactivation rate constants determined by the TCID50 and qPCR methods versus UV wavelength, the action spectra of both BA.2 and BA.5 were determined, and these two variants showed almost the same spectra. This result suggests that both variants have the same UV inactivation characteristics and that the action spectra of SARS-CoV-2 were quantitatively explained by the absorption spectra of both RNA and protein, where the protein layer shields the RNA from the UV light.
Materials and methods
Cells
VeroE6/TMPRSS2 cells (African green monkey kidney-derived cells expressing human TMPRSS2) were obtained from the Japanese Collection of Research Bioresources (JCRB) Cell Bank (#JCRB1819). The cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM, low-glucose, Sigma–Aldrich, #D6046) supplemented with 10% fetal bovine serum (Gibco, #10270-106), penicillin/streptomycin (Sigma–Aldrich, #P0781), and 1 mg/mL G418 (Wako, #070-06803) at 37 °C with 5% CO2. The concentration of cells was approximately 1.4 × 105 cells/cm2.
Virus preparation, stocks and infectivity assays
Two types of SARS-CoV-2 variants, omicron BA.2 (hCoV-19/Japan/TKYS02037/2022) and omicron BA.5 (hCoV-19/Japan/TKYS14631/2022), were obtained from Tokyo Metropolitan Institute of Public Health. These viruses were propagated in VeroE6/TMPRSS2 cells cultured in medium A (DMEM containing penicillin/streptomycin and 1 mg/mL G418) for infection and incubated for 3 days at 37 °C with 5% CO2. After infection, the virus-containing supernatant was collected and the cellular debris was removed by centrifugation at 3000 rpm (= 1700 g) for 5 min. The virus stocks were then aliquoted and stored at − 80 °C until use. We measured viral infectivity with the standard TCID50 method to determine the viral titer of the collected viral samples. TCID50/mL values were calculated 4 days after the infection using the Behrens–Karber method35. The viral titer of BA.2 was 4.9 × 105 TCID50/mL and that of BA.5 was 2.1 × 105 TCID50/mL.
Quantitative reverse transcription polymerase chain reaction (RT‒qPCR)
SARS-CoV-2 RNA was extracted from the collected viral samples of each well using TRIzol Reagent following the manufacturer’s protocol. RT‒qPCR for SARS-CoV-2 was performed using a PrimeScript RT Reagent Kit with gDNA Eraser (Takara Bio Inc., #RR047A) according to the manufacturer’s protocol. qPCR was performed using TB Green Premix Ex Taq (Takara Bio Inc., #RR420A) according to the manufacturer’s protocol. The following primers were used: qCoV2 forward, 5´-GCCTCTTCTCGTTCCTCATCAC-3´; qCoV2 reverse, 5´-AGCAGCATCACCGCCATTG-3´; Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) forward, 5´-ACACCCACTCCTCCACCTTT-3´; and GAPDH reverse, 5´-TAGCCAAATTCGTTGTCATACC-3´. Thermal cycling was carried out as follows: initial denaturation at 95 °C for 30 s, 40 cycles of denaturation at 95 °C for 5 s, and a final annealing/extension at 60 °C for 30 s. For BA.2, the value of the threshold cycle (Ct) without UV irradiation was Ct = 13 and that with UV irradiation (260 nm, 18 mJ/cm2) was Ct = 16. For BA.5, Ct = 13 without UV irradiation and Ct = 17 with UV irradiation (260 nm, 18 mJ/cm2). In both cases, we used GAPDH as a reference gene and its Ct value was 27. As a dye for staining DNA, we used SYBR@ FAM for the fluorescence detection. We set the fluorescent intensity as level 10 (Thermal Cycler Dice Real Time System Software, Thermo Fisher Scientific Inc. Massachusetts, USA) to determine all Ct values. All experiments with SARS-CoV-2 were performed in a biosafety level 3 (BSL3) containment facility at Kumamoto University.
Plating and counting method for inactivated virus
We applied from 200 to 260 nm-UV irradiation to inactivate virus suspensions. For each wavelength, we varied dose from 0 to 18 mJ/cm2. A total of 600 μL of viral suspension (200 μL of virus stock mixed with 400 μL of PBS) was irradiated for each wavelength and dose. VeroE6/TMPRSS2 cells were plated in both 96-well plates for TCID50 assays and 24-well plates for qPCR one day prior to infection. Just before the infection experiments, the medium on VeroE6/TMPRSS2 cells was aspirated, and 50 μL of the medium A was added to each well. We used the TCID50 method to determine viral infectivity. Inactivated virus or control virus suspensions were plated into the first column, and then the threefold-diluted suspensions were successively plated into the adjacent columns. This dilution plating was performed for all 96 wells. The plated 96 wells were incubated for one hour at 37 °C. After incubation, the viral supernatants were aspirated, and 100 μL of medium B (DMEM supplemented with 10% FBS, penicillin/streptomycin, and 1 mg/mL G418) for culture was added to each well. The plate was incubated for four days at 37 °C in a 5% CO2. Cytopathic effects (CPEs) were scored under a bright field microscope (10 ×) as cytoplasm vacuolization, cell rounding and sloughing. TCID50/mL values were calculated by the Behrens–Karber method35. Viral genome integrity was analyzed using reverse-transcription quantitative-polymerase chain reaction (RT‒qPCR). We used the same inactivated virus suspensions as those used in the TCID50 assay without dilution. A 100 μL viral suspension was plated on VeroE6/TMPRSS2 cells in a 24-well multiwell dish. This plate was incubated for one hour at 37 °C in a 5% CO2. After incubation, the viral supernatants were aspirated, and then 500 μL of the medium B was added to each well. This plate was incubated for one day at 37 °C in a 5% CO2. SARS-CoV-2 RNA was extracted from the collected viral samples of each plate using TRIzol Reagent (Thermo Fisher, #15,596,018). All experimental results are reported as the means across 3 replicates.
Wavelength-tunable UV light source
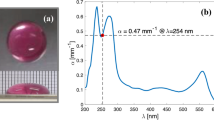
Figure 1a shows the wavelength-tunable UV light source used to compare the efficacy of the far-UV light region (200–230 nm) and the deep UV (DUV) light region (230–260 nm). A Laser-Driven Light Source (LDLS EQ-99X, Energetiq Technology, Inc. Wilmington, USA), which emits radiation of 170–2100 nm, was used as a broadband emission source. The emission was selected by using a UV bandpass filter from 200 to 260 nm (200 nm, 220 nm, 240 nm, and 260 nm) with a 10 nm bandwidth (Edmond Optics Japan, Tokyo, Japan). The spectrum of UV radiation that the virus was exposed to, which is shown in Fig. 1b, was measured by a spectrometer through an optical fiber. The viral suspension (600 μL) was added to a single well (10 mm in diameter) of a microplate (48 wells), and during UV irradiation, the viral suspension was agitated by a microplate shaker (TM-1FN, AS ONE Corp. Osaka, Japan). The irradiance of UV radiation that the virus was exposed to was measured by setting a UV-extended Si photodiode with an aperture of 10 mm (S120VC, Thorlabs Inc. New Jersey, USA) at the surface of the viral suspension.
(a) Optical setup of the wavelength-tunable UV inactivation system, (b) transmission spectrum, and (c) absorbance spectra of DMEM diluted with PBS. A laser-driven light source, which emits radiation of 170–2100 nm, was used as a broadband emission source, and the emission was selected by using a UV bandpass filter from 200 to 260 nm with a 10 nm bandwidth. We used a DMEM:PBS = 1:2 solution (blue line) because the absorbance between 200 and 260 nm is approximately the same magnitude.
Absorbance correction of viral suspensions
Generally, the medium A is used to maintain viral viability and infectivity, and this medium contains proteins and amino acids which strongly absorb UV light36,37,38. To extract the viral particles from the medium A, an ultracentrifugation followed by buffer exchange can be used. However, various problems such as the shed of the S protein during the ultracentrifugation are pointed out39,40,41. Therefore, to measure correct viability and infectivity versus UV irradiation, the absorbance of the viral solution was adjusted by PBS dilution42. The absorbance spectra of DMEM diluted with PBS were measured using a UV–visible spectrometer through an optical fiber (BIM-6002A, Brolight Technology Corporation, Hangzhou, China). Here, all the absorbance spectra were calibrated by a spectral calibrated 150 W-xenon standard light source with an emission wavelength from 185 to 2000 nm (L7810-03, Hamamatsu Photonics Corporation, Hamamatsu, Japan), and a fused silica cell with an optical path length of 1 cm (T-3-ES-10, Tosoh Quartz Corporation, Tokyo, Japan) was used for the absorbance measurements. As shown in Fig. 1c, DMEM exhibited absorption peaks at approximately 230 nm and 280 nm, which were due to proteins or amino acids in DMEM. The absorbance by PBS (not shown), which contains NaCl, KCl, and sodium phosphate, was much lower than 0.1 cm−1 at all wavelengths (200–300 nm). To avoid the influence of absorption by DMEM components, the viral suspension could be diluted with PBS. However, in this case, the virus titer would also decrease. Therefore, we used a DMEM:PBS = 1:2 solution (blue line), for which the absorbance at 200 nm, 220 nm, 240 nm, and 260 nm did not show significantly different magnitudes (α200 nm = 0.12 cm−1, α220 nm = 0.32 cm−1, α240 nm = 0.30 cm−1, and α260 nm = 0.35 cm−1). The viral suspension used here was 0.6 cm in height (L). The irradiance that the virus was exposed to differed up to 30% between the top and bottom layers; e.g. 30 µW/cm2 for the top layer and 20 µW/cm2 for the bottom layer at the wavelength of 260 nm. Therefore, to determine the effective irradiance (Ie) correctly, we subtracted the reflection loss (R) at the air/suspension interface and averaged the absorption effect in the height direction as
where I0 is the irradiance measured at the top layer. The reflection loss was determined by using the Fresnel equation43, and R is approximately 0.02 to 0.03, as we assume that the refractive index of the suspension has a value similar to that of water44,45. Based on the irradiance determined by Eq. (1), the dose was varied from 0 to 18 mJ/cm2 (0 mJ/cm2, 3 mJ/cm2, 6 mJ/cm2, 9 mJ/cm2 and 18 mJ/cm2) by changing the UV irradiation duration.
Ethical approval statement
All protocols were reviewed and approved by the Ethics Committee for Faculty of Life Science, Kumamoto University (approval number 49).
Results
Dose response of SARS-CoV-2 at various wavelengths measured by infectivity and RNA amplification
Inactivation of SARS-CoV-2 omicron BA.2 and BA.5 using the wavelength tunable UV light source are presented in Fig. 2(a; 200 nm), (b; 220 nm), (c; 240 nm), and (d; 260 nm) as a function of UV dose. According to comparison of Fig. 2b,d, omicron BA.2 and BA.5 show approximately the same reduction in viral infectivity (solid circles) and in RNA amplification (solid squares) for both 220 nm (BA.2: dark green, BA.5: light green) and 260 nm (BA.2: dark red, BA.5: light red) UV irradiation. This result indicates that the BA.2 and BA.5 variants have almost the same UV irradiation inactivation properties. The fact that the inactivation rates obtained with 220 nm light show approximately the same value as that obtained with 260 nm light highlights the significance of disinfection by far-UVC light because far-UVC light is attracting special attention as a safe germicidal light for the human body16,17,18,19,20.
Inactivation of SARS-CoV-2 BA.2 and BA.5 using the wavelength-tunable UV light source. Inactivation at (a) 200 nm (BA.2; blue), (b) 220 nm (BA.2; dark green, BA.5; light green), (c) 240 nm (BA.2; orange), and (d) 260 nm (BA.2; dark red, BA.5; light red) as a function of UV dose. Solid circles show the viral infectivity obtained by TCID50 assay, and solid squares show the reduction in RNA amplification determined by qPCR, where the relative ratio to those of unexposed controls was used. Inactivation rate constants at each wavelength were determined by linear regression lines (solid line: TCID50, broken line: qPCR). These linear inactivation rate constants for each wavelength are summarized in Table 1.
Figure 2 shows a correlation between the reduction in viral infectivity (solid circles) and the reduction in RNA amplification (solid squares) for these wavelengths of UV irradiation. The highest inactivation rate constant (Γ, cm2/mJ) was obtained at 260 nm; for cell culture infectivity, the rate of BA.2 was 0.40 (p < 0.05), and that of BA.5 was 0.38 (p < 0.05); for the qPCR assay analyzing 111 bp fragments, the rate of BA.2 was 0.25 (p < 0.05), and the rate of BA.5 was 0.23 (p < 0.05). These linear inactivation rate constants (cm2/mJ) for each wavelength are summarized in Table 1. As shown in Fig. 2 and Table 1, the obtained rates are different between TCID50 and qPCR. However, there is a correlation between these rate constants. Both the difference and the correlation between TCID50- and qPCR-rates are considered to originate from the fact that the TCID50 measures the number of infectious virus, while the qPCR measures both non-infectious and infectious viruses.
Determination of the spectral sensitivities of SARS-CoV-2
Figure 3a shows the spectral sensitivity (action spectra) of SARS-CoV-2 omicron BA.2 inactivation (red circles) and genome damage (red squares) and of omicron BA.5 inactivation (orange circles) and genome damage (orange squares) determined by calculating the inactivation rate constants relative to their peak values at 260 nm. Both the spectral sensitivities obtained by TCID50 assays and those obtained by qPCR assays coincide when multiplying the inactivation rate constants obtained by qPCR by 1.6, which shows the correlation between these methods. Notably, the obtained spectral sensitivity is almost identical to that obtained by Schuit et al.7.
(a) Spectral sensitivity (action spectra) of SARS-CoV-2 BA.2 and BA.5 inactivation (BA.2: red circles, BA.5: orange circles) and genome damage (BA.2: red squares, BA.5: orange squares), obtained by calculating the inactivation rate constants relative to their peak values at 260 nm. Both the spectral sensitivities obtained by TCID50 assay and by qPCR coincide with each other after multiplying the inactivation rate constants obtained by qPCR by 1.6. As a comparison, the spectral sensitivity of E. coli determined by CFU experiments (blue rhombus) is also shown. The solid blue line and solid red line are theoretically fitted action spectra for SARS-CoV-2 (red line) and E. coli (blue line) determined by weighting the absorption coefficient of the protein layer (broken brown line) to that of DNA or RNA (broken green line), where the action spectra for SARS-CoV-2 is fitted by αSARS (λ) = αDNA (λ)–1.1 × αPROTEIN (λ) (red line) and αE. Coli (λ) = αDNA (λ)–0.85 × αPROTEIN (λ) (blue line), respectively. (b) UV irradiance (mW/cm2) shield model for a protein layer to explain the difference in the spectral sensitivity between SARS-CoV-2 and E. coli below 240 nm.
As a comparison, the spectral sensitivity of Escherichia coli (E. coli) determined by a colony-forming unit (CFU) experiment (blue rhombus) is also shown in this figure46. The figure highlights approximately the same value for SARS-CoV-2 BA.2 and BA.5 inactivation, SARS-CoV-2 BA.2 and BA.5 genome damage, and E. coli inactivation, namely, above 240 nm. These values are aligned with the absorbance spectrum of RNA, which is shown by the green broken line47,48. However, the inactivation rate constants as well as the genome damage show significant differences between SARS-CoV-2 variants and E. coli below 240 nm.
Discussion
If we consider that both RNA and protein absorbance play a role in inactivation, this difference below the 240 nm region can be quantitatively understood by considering the thickness of the protein layer covering DNA or RNA, as shown in Fig. 3b. That is, E. coli DNA is covered by a thick protein layer, whereas SARS-CoV-2 RNA is covered by a thin protein layer. This protein layer strongly absorbs UV light below 240 nm (shield effect); thus, the UV irradiance (mW/cm2) of E. coli DNA is significantly reduced compared to that to SARS-CoV-2 RNA. The solid blue line and solid red line shown in Fig. 3a are theoretically fitted action spectra for SARS-CoV-2 (red line) and E. coli (blue line) determined by weighting the absorption coefficient of the protein layer (broken brown line) to that of DNA or RNA (broken green line)47,48, where the action spectra for SARS-CoV-2 is fitted by αSARS (λ) = αDNA (λ)–1.1 × αPROTEIN (λ) and αE. coli (λ) = αDNA (λ)–0.85 × αPROTEIN (λ), respectively. Notably, the above theoretical analysis is based on the fact that the excitation of peptide bonds plays a minor role in both RNA modification and bacterial inactivation because protein consists of a much larger number of molecules than DNA and protein can be repaired using genetic information when necessary49,50,51.
As shown in Table 1, we determined the linear inactivation rate constants (cm2/mJ) of BA.2 and BA.5 for each UV wavelength. These values were obtained using a viral suspension and were significantly different from the values obtained for coronavirus in aerosols12. For example, there is a large difference in the inactivation rate constant at 220 nm obtained here (suspension: 0.28 cm2/mJ) and that reported by Buonanno et al. (aerosol: 4–6 cm2/mJ)12. It is likely that some physical and/or biochemical mechanisms are responsible for this large difference. We note here that this large difference between aerosol and liquid suspensions is widely recognized for many viruses, such as SARS-CoV10,52, murine hepatitis virus (MHV) coronavirus53, adenovirus serotype 2 (VR-846)53, influenza virus H1N113,54, and bacteriophage MS253. This comparison shows the definite enhancement of efficacy in aerosols compared to that in liquid suspensions regardless of the size of the virus (90–100 nm or 30–40 nm), the type of nucleic acid (DNA or RNA), and the viral structure (naked or enveloped). The difference is quantitatively understood based on the optical Mie scattering theory55,56. Our calculation shows that the inactivation rate constant in the aerosol state is enhanced by a factor of 10 compared to that in the liquid suspension. The quantitative evaluation of the enhancement factor as a function of the droplet size is provided in Supplemental information (Fig. S1). The Mie scattering effect is therefore a possible candidate to explain this significant enhancement of the UV irradiance inside an aerosol droplet.
The inactivation rate constants obtained at 220 nm is smaller than those obtained at 260 nm. However, if we consider the threshold level that can be irradiated to the human body57,58, far-UVC radiation (220 nm) can be effective compared to Deep-UVC radiation (260 nm). For example, the total amount of UV radiation that can be irradiated per day is 25 mJ/cm2 for 220 nm, and 3 mJ/cm2 for 260 nm57,58. Multiplying these values by the rate constants obtained here yields a 3-log inactivation efficacy at 220 nm, whereas only 30% inactivation efficacy can be obtained by 260 nm irradiation. Therefore, considering the safety level to the human body, far-UVC can efficiently inactivate SARS-CoV-2 compared to generally used UVC wavelength region (250–270 nm).
Conclusions
To our knowledge, this is the first investigation of the effect of the UV susceptibility of SARS-CoV-2 omicron BA.2 and BA.5. We determined the inactivation rate constant by TCID50 and qPCR methods as a function of UV irradiation wavelength. The spectral sensitivity of SARS-CoV-2 omicron variants was derived from these inactivation rate constants. Difference in the inactivation rate constants obtained by TCID50 and qPCR is an issue to be resolved. The fact that the inactivation efficacy of 220 nm light is approximately the same as that of 260 nm light shows a promising aspect that far-UVC light can be used to prevent airborne virus transmission in a simple and safe manner.
Data availability
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Cimolai, N. Disinfection and decontamination in the context of SARS-CoV-2-specific data. J. Med. Virol. 94, 4654–4668 (2022).
Van Doremalen, N. et al. Aerosol and surface stability of SARS-CoV-2 as compared with SARS-CoV-1. N. Engl. J. Med. 382, 1564–1567 (2020).
Cimolai, N. Environmental and decontamination issues for human coronaviruses and their potential surrogates. J. Med. Virol. 92, 2498–2510 (2020).
CDC. Variants of the viruses. https://www.cdc.gov/coronavirus/2019-ncov/variants/index.html. Accessed on February 2023.
Raeiszadeh, M. & Adeli, B. A critical review on ultraviolet disinfection systems against COVID-19 outbreak: Applicability, validation, and safety considerations. ACS Photonics 7, 2941–2951 (2020).
Gerchman, Y., Mamane, H., Friedman, N. & Mandelboim, M. UV-LED disinfection of coronavirus: Wavelength effect. J. Photochem. Photobiol. B. 212, 112044 (2020).
Schuit, M. A. et al. SARS-CoV-2 inactivation by ultraviolet radiation and visible light is dependent on wavelength and sample matrix. J. Photochem. Photobiol. B. 233, 112503 (2022).
Hadi, J., Dunowska, M., Wu, S. & Brightwell, G. Control measures for SARS-CoV-2: A review on light-based inactivation of single-stranded RNA viruses. Pathogens 9, 737 (2020).
Sellera, F. P., Sabino, C. P., Cabral, F. V. & Ribeiro, M. S. A systematic scoping review of ultraviolet C (UVC) light systems for SARS-CoV-2 inactivation. J. Photochem. Photobiol. 8, 100068 (2021).
Heßling, M., Hönes, K., Vatter, P. & Lingenfelder, C. Ultraviolet irradiation doses for coronavirus inactivation—Review and analysis of coronavirus photoinactivation studies. GMS Hyg. Infect. Control 15, Doc08 (2020).
Welch, D. et al. Far-UVC light: A new tool to control the spread of airborne-mediated microbial diseases. Sci. Rep. 8, 2752 (2018).
Buonanno, M., Welch, D., Shuryak, I. & Brenner, D. J. Far-UVC light (222 nm) efficiently and safely inactivates airborne human coronaviruses. Sci. Rep. 10, 10285 (2020).
Narita, K. et al. Ultraviolet C light with wavelength of 222 nm inactivates a wide spectrum of microbial pathogens. J. Hosp. Infect. 105, 459–467 (2020).
Ma, B., Gundy, P. M., Gerba, C. P., Sobsey, M. D. & Linden, K. G. UV Inactivation of SARS-CoV-2 across the UVC Spectrum: KrCl* Excimer, Mercury-Vapor, and Light-Emitting-Diode (LED) Sources. Appl. Environ. Microbiol. 87, e01532-e1621 (2021).
Welch, D. et al. Inactivation rates for airborne human coronavirus by low doses of 222 nm Far-UVC radiation. Viruses 14, 684 (2022).
Blatchley III, E.R. et al. Far UV-C radiation: An emerging tool for pandemic control. Crit. Rev. Environ. Sci. Technol. 0, 1–21 (2022).
Buonanno, M. et al. Germicidal efficacy and mammalian skin safety of 222-nm UV light. Radiat. Res. 187, 483–491 (2017).
Narita, K. et al. Effect of ultraviolet C emitted from KrCl excimer lamp with or without bandpass filter to mouse epidermis. PLoS ONE 17, e0267957 (2022).
Yamano, N. et al. Long-term Effects of 222-nm ultraviolet radiation C Sterilizing Lamps on Mice Susceptible to Ultraviolet Radiation. Photochem. Photobiol. 96, 853–862 (2020).
Yamano, N. et al. Evaluation of acute reactions on mouse skin irradiated with 222 and 235 nm UV-C. Photochem. Photobiol. 97, 770–777 (2021).
Freeman, S. et al. Systematic evaluating and modeling of SARS-CoV-2 UVC disinfection. Sci. Rep. 12, 5869 (2022).
Ong, Q., Wee, W., Dela Cruz, J., Teo, J. W. & Han, W. 222-nanometer far-UVC exposure results in DNA damage and transcriptional changes to mammalian cells. Int. J. Mol. Sci. 23, 9112 (2022).
Otake, M., Okamoto Yoshiyama, K., Yamaguchi, H. & Hidema, J. 222 nm ultraviolet radiation C causes more severe damage to guard cells and epidermal cells of Arabidopsis plants than does 254 nm ultraviolet radiation. Photochem. Photobiol. Sci. 20, 1675–1683 (2021).
Eadie, E., Barnard, I. M. R., Ibbotson, S. H. & Wood, K. Extreme exposure to filtered far-UVC: A case study†. Photochem. Photobiol. 97, 527–531 (2021).
Eadie, E. et al. Computer modeling indicates dramatically less DNA damage from far-UVC krypton chloride lamps (222 nm) than from sunlight exposure. Photochem. Photobiol. 97, 1150–1154 (2021).
Woods, J. A. et al. The effect of 222-nm UVC on healthy volunteer skin. Photodermatol. Photoimmunol. Photomed. 31, 159–166 (2015).
Inagaki, H., Saito, A., Sugiyama, H., Okabayashi, T. & Fujimoto, S. Rapid inactivation of SARS-CoV-2 with deep-UV LED irradiation. Emerg. Microbes Infect. 9, 1744–1747 (2020).
Heilingloh, C. S. et al. Susceptibility of SARS-CoV-2 to UV irradiation. Am. J. Infect. Control 48, 1273–1275 (2022).
Storm, N. et al. Rapid and complete inactivation of SARS-CoV-2 by ultraviolet-C irradiation. Sci. Rep. 10, 22421 (2020).
Chiappa, F. et al. The efficacy of ultraviolet light-emitting technology against coronaviruses: A systematic review. J. Hosp. Infect. 114, 63–78 (2021).
Trivellin, N. et al. UV-based technologies for SARS-CoV2 inactivation: Status and perspectives. Electronics 10, 1703 (2021).
Ma, B. et al. Inactivation of coronaviruses and phage Phi6 from irradiation across UVC wavelengths. Environ. Sci. Technol. Lett. 8, 425–430 (2021).
Robinson, R. T., Mahfooz, N., Rosas-Mejia, O., Liu, Y. & Hull, N. M. UV222 disinfection of SARS-CoV-2 in solution. Sci. Rep. 12, 14545 (2022).
Widera, M. et al. Evaluation of stability and inactivation methods of SARS-CoV-2 in context of laboratory settings. Med. Microbiol. Immunol. 210, 235–244 (2021).
Kärber, G. Beitrag zur kollektiven behandlung pharmakologischer reihenversuche. Naunyn Schmiedebergs Arch. Exp. Pathol. Pharmakol. 162, 480–483 (1931).
Beaven, G. H. & Holiday, E. R. Ultraviolet absorption spectra of proteins and amino acids. In Advances in Protein Chemistry (eds Anson, M. L. et al.) (Academic Press, 1952).
Stoscheck, C. M. Quantitation of protein. In Methods Enzymol (ed. Deutscher, M. P.) (Academic Press, 1990).
Porterfield, J. Z. & Zlotnick, A. A simple and general method for determining the protein and nucleic acid content of viruses by UV absorbance. Virology 407, 281–288 (2010).
Ke, Z. et al. Structures and distributions of SARS-CoV-2 spike proteins on intact virions. Nature 588, 498–502 (2020).
Plavec, Z. et al. SARS-CoV-2 Production, Purification Methods and UV Inactivation for Proteomics and Structural Studies. Viruses 14, 1989 (2022).
James, K. T. et al. Novel high-throughput approach for purification of infectious virions. Sci. Rep. 6, 36826 (2016).
Minamikawa, T. et al. Quantitative evaluation of SARS-CoV-2 inactivation using a deep ultraviolet light-emitting diode. Sci. Rep. 11, 5070 (2021).
Hecht, E. Optics 5th edn. (Pearson Education Ltd, 2017).
Hale, G. M. & Querry, M. R. Optical constants of water in the 200-nm to 200-µm wavelength region. Appl. Opt. 12, 555–563 (1973).
Smith, R. C. & Baker, K. S. Optical properties of the clearest natural waters (200–800 nm). Appl. Opt. 20, 177–184 (1981).
Matsumoto, T., Hoshiai, T., Tatsuno, I. & Hasegawa, T. Action spectra of bacteria and purification of pollutant water at faucets using a water waveguide method. Water 14, 1394 (2022).
Kowalski, W. Ultraviolet Germicidal Irradiation Handbook (Springer, 2009). https://doi.org/10.1007/978-3-642-01999-9.
Harm, W. Biological Effects of Ultraviolet Radiation (Cambridge University Press, 1980).
Hunt, H. D. & Simpson, W. T. Spectra of simple amides in the vacuum ultraviolet. J. Am. Chem. Soc. 75, 4540–4543 (1953).
Rosenheck, K. & Doty, P. The far ultraviolet absorption spectra of polypeptide and protein solutions and their dependence on conformation. Proc. Natl. Acad. Sci. U.S.A. 47, 1775–1785 (1961).
Lo, C. W. et al. UVC disinfects SARS-CoV-2 by induction of viral genome damage without apparent effects on viral morphology and proteins. Sci. Rep. 11, 13804 (2021).
Ueki, H. et al. A 265-nanometer high-power deep-UV light-emitting diode rapidly inactivates SARS-CoV-2 aerosols. mSphere 7, e00941-21 (2022).
Walker, C. M. & Ko, G. Effect of ultraviolet germicidal irradiation on viral aerosols. Environ. Sci. Technol. 41, 5460–5465 (2007).
McDevitt, J. J., Rudnick, S. N. & Radonovich, L. J. Aerosol susceptibility of influenza virus to UV-C light. Appl. Environ. Microbiol. 78, 1666–1669 (2012).
Bohren, C. F., Huffman, D. R. & D.R,. Absorption and Scattering of Light by Small Particles (Wiley, 1983).
Bott, A. & Zdunkowski, W. Electromagnetic energy within dielectric spheres. J. Opt. Soc. Am. A 4, 1361–1365 (1987).
American Conference of Governmental Industrial Hygienists. (Accessed 2023, April 20). Ultraviolet Radiation: TLV® Physical Agents 7th Edition Documentation.
Sliney, D. H. & Stuck, B. E. A Need to Revise Human Exposure Limits for Ultraviolet UV-C Radiation. Photochem. Photobiol. 97, 485–492 (2021).
Acknowledgements
The authors gratefully thank Hiroyuki Oshiumi (Kumamoto University) and Terumasa Ikeda (Kumamoto University) for providing advice on the infection experiments. All experiments with SARS-CoV-2 were performed in a BSL3 containment facility at Kumamoto University (approval number 20). This work was supported by a Grant-in-Aid for Research at Nagoya City University (Grant Number 2121102), a Grant-in-Aid for the Research Program on Hepatitis from the Japan Agency for Medical Research and Development (AMED 20he0622003h0001), Intramural Grant of Collaborative Research on Infection at Joint Research Center for Human Retrovirus Infection at Kumamoto University, and the External Research Fund Acquisition and Activation Project of Kumamoto University.
Author information
Authors and Affiliations
Contributions
N.F. is the first author. I.T., M.T., and T.M. contributed to the design of the wavelength-tunable UV inactivation system. N.F., K.N., H.O., Y.T. and T.M. performed the SARS-CoV-2 omicron BA.2 TCID50 and qPCR experiments. I.T. and T.H. performed the E. coli inactivation experiments. K.N., Y.T. and T.H. provided expertise for virus preparation, enumeration, and infectivity assays. H.O., T.M. and M.T. constructed and analyzed the Mie scattering model that describes the enhancement in inactivation efficacy in an aerosol droplet. All authors read and approved this submitted manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Fujimoto, N., Nagaoka, K., Tatsuno, I. et al. Wavelength dependence of ultraviolet light inactivation for SARS-CoV-2 omicron variants. Sci Rep 13, 9706 (2023). https://doi.org/10.1038/s41598-023-36610-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-36610-6
- Springer Nature Limited