Abstract
Dachigam National Park (DNP), in Zabarwan mountains of north-western Himalaya constitutes a region of high biodiversity with greater endemism. DNP is known for its unique micro-climate together with distinct vegetational zones providing home to variety of threatened and endemic plant, animal, and bird species. However, studies on soil microbial diversity in fragile ecosystems of north-western Himalaya in general and DNP in particular are lacking. This was thus a maiden attempt to study variations in soil bacterial diversity of DNP with respect to changing soil physico-chemical properties, vegetation, and altitude. Soil parameters depicted significant variations among different sites with highest values for temperature, OC, OM and TN being 22.2 ± 0.75 °C, 6.53 ± 0.32%, 11.25 ± 0.54%, 0.545 ± 0.04% from site-2 (low altitudinal grassland site) in summer and lowest of 5.1 ± 0.65 °C, 1.24 ± 0.26%, 2.14 ± 0.45% and 0.132 ± 0.04% at site-9 (high altitudinal mixed pine site) in winter. Bacterial CFU showed significant correlations with soil physico-chemical attributes. This study led to the isolation and identification of 92 morphologically varied bacteria with the highest (15) from site-2 and lowest (04) from site-9 which post BLAST analysis (via 16S rRNA analysis) depicted presence of only 57 distinct bacterial species under taxonomic phylum, Firmicutes and Proteobacteria. Nine species were widely spread (i.e., isolated from > 3 sites), however, most bacteria (37) were restricted to a particular site. Diversity indices ranged between 1.380 to 2.631 (Shannon–Weiner’s index); 0.747 to 0.923 (Simpson’s index) with highest values for site-2 and lowest for site-9. Index of similarity was highest (47.1%) between riverine sites (site-3 and site-4) whereas two mixed pine sites (site-9 and site-10) showed no similarity.
Similar content being viewed by others
Introduction
Bacteria are the most abundant and diverse group of organisms present in soil, catalyzing various life sustaining ecological processes on planet, Earth1,2. It is estimated that per gram of soil contains > 109 microorganisms which represents around 4000–7000 genomes3. In soil, approximately 80–90% of soil processes are mediated by bacteria, as a result of which there is a greater interest in the relation between their diversity and function in the soil ecosystems4. Soil and its associated bacterial communities may be affected by different intertwined factors which may vary within the ecosystems thereby making the communities of bacteria distinctive to a particular ecosystem5. Therefore, keeping into consideration the contribution of bacterial populations in maintaining the ecological balance, as well as their flexibility to grow and adapt under varied physico-chemical conditions, cataloguing their diversity as it exists is of vital importance.
Among various soil microorganisms, bacteria are a major class which helps in keeping the soils healthy and fertile by their roles in cycling of nutrients like carbon, nitrogen, phosphorous and sulphur1,6. The bacteria in soil are known to perform vital services for the maintenance of the soil ecosystem health by improving the aggregation and structure of soil7,8,9, cycling of soil nutrients1, decomposition of organic matter10,11, enhancing soil fertility12,13,14, nitrogen fixation15,16, protection of plants against various pathogens17 etc. In addition, the bacteria in soil with their secretions bind to soil particles forming soil micro-aggregates that leads to the improvement of soil structure and quality thereby increasing the infiltration of water in soil that in turn enhance its water holding capacity8.
The soil bacterial population which is generally considered to be about one half of the total microbial biomass varying between 103–106 discrete genomes in a gram of soil typically depends on its physical, chemical and biological conditions18,19. Hence, soil bacterial community composition, structure and function relies on a variety of abiotic and biotic factors including physico-chemical characteristics of soil, nutrient availability, over-ground vegetation and ambient environmental factors20,21,22.
Among ecosystems, the supply of nutrients differs23, leading to variations in the community structure of plants and their production24. Typically, vegetation is known to influence and improve soil attributes like aeration, infiltration rate, hydraulic conductivity, structure, and water holding capacity25. However, bacterial diversity in soils has also been observed to be affected by seasonal fluctuations in vegetation leading to the replacement of dominant soil bacterial groups26.
Among various ecosystems, the Himalayas are known to inhabit great variety of soil microorganisms including mesophilic bacteria27. Earlier bacterial investigations in this part were limited to snow and glacier samples28,29 however, only recently focus has been shifted to assess the Himalayan soil bacterial diversity30,31,32,33,34,35. Specifically, the north-western part of Himalayas that encompasses through the erstwhile state of Jammu and Kashmir to Ladakh is considered to consist different climatic zones possessing characteristic attributes such as diverse soaring heights, alpine glaciers, lush green meadows, and a series of elevational zones having varied soil textures which inhabits the richest plethora of microorganisms particularly bacteria and actinomycetes having enormous biotechnological and bioprospecting potential27,36. Although there have been several attempts by various researchers for documenting soil bacterial diversity in this region37,38,39,40,41,42, only a few have focused on the forest ecosystems43 and protected areas (Table 1) while no study has been conducted in Dachigam National Park (DNP) which harbours various distinctive and endemic plant and animal biodiversity of the western Himalayan region53,54,55,56. Therefore, this study was taken up with a sole aim to generate the first ever baseline data on the culturable soil bacterial diversity, their altitudinal, seasonal and vegetational variations in different microhabitats of lower Dachigam National Park, Kashmir.
Materials and methods
Research area
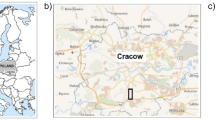
Dachigam National Park (henceforth referred to as DNP) located in the Zabarwan range of the Western Himalaya extends between 34°05′ N–34°11′ N and 74°54′ E–75°09′ E and stretches over 1677–4270 m altitude (Fig. 1). DNP roughly sprawls over 141 km2 officially comprising of two regions: lower Dachigam (26 km2) and upper Dachigam (115 km2) on the basis of altitude, forest types, and movement of its critically endangered red stag57. The current study was performed in lower DNP covering 1/3rd of its western end, and containing a deep gorge cut by Dagwan river (and its tributaries) originating in the Marsar lake (situated at about 4200 m altitude) of alpine Upper Dachigam (also called Dagwan Valley). Soils were sampled seasonally over a period of 2 years from ten different locations covering five different dominant vegetation types in lower DNP (Table 2), with each habitat type having a low and high altitudinal site so as to ascertain the influence of vegetation and altitude on the diversity of bacteria. The detailed vegetational attributes of each site is given in the table (Table 3).
Map depicting different study sites in lower DNP. This figure was generated in ArcGIS version 10.4.1 (https://www.esri.com/en-us/arcgis/products/arcgis-pro/).
Soil sampling and sampling site parameters
Soils were sampled with a sterilized soil-corer up to a depth of 15 cm in poly-ethylene (PET) bags and sterile plastic vials were used for the assessment of the soil properties and bacterial analysis respectively58. At each site, five randomly collected soil samples were taken from different locations (approximately 8–10 m) on the same day and pooled together to get a composite representative sample for the site so as to document maximum diversity of the bacteria. The soil samples kept in sterile plastic vials were then stored at 4 °C until processed within 24 h59. Geographical coordinates of every sampling site were noted employing digital GPS (Garmin 7.6).
Soil physico-chemical properties
Soil temperature was recorded on site at a depth of about 10–15 cm using a standard soil thermometer58. pH was determined by 1:2.5 (w/v) soil–water suspension using a digital pH meter60 and the percent moisture content in soil was analyzed gravimetrically61. The organic carbon and organic matter in soil was computed62 followed by the determination of soil total nitrogen by Kjeldahl method63.
Isolation, enumeration and preservation of bacteria
The isolation of the culturable bacteria in the sampled soils was carried out by standard serial dilution and spread-plate method64. 1 g of each soil samples were put in 10 mL of sterile 0.85% NSS (normal saline solution), followed by thorough mixing in a shaking incubator for 4–5 min (120 rpm) so as to obtain a dilution series for inoculation. 100 μL (0.1 mL) aliquot from each dilution was gently spreaded on agar plates (Nutrient agar, Luria Bertani agar and Reasoner’s 2A agar) in triplicates. The agar plates were kept in incubation (24–48 h) at 37 ± 2 °C and the colonies which developed over the inoculated petri-plates were counted using digital Quebec-counter for assessing the soil bacterial colony forming units (cfu/g). The well-isolated colonies from each plate with different morphologies were then randomly selected and streaked onto the fresh agar plates. Pure isolates were maintained by re-streaking via sub-culturing and nutrient agar slants (stored at 4 °C) for future use65.
Statistical analysis
In this study, all of the experiments were performed in triplicates and the results were expressed as Mean (± SD). Datasets have been subjected to Kruskal–Wallis test (p = < 0.0001), a non-parametric alternative to one-way ANOVA typically considered to be more appropriate than the traditional one-way ANOVA employing R-packages “tidyverse”, “ggpubr” and “rstatix”66. All possible pairwise comparisons were carried out by Wilcoxon’s test (p < 0.05) employing Dunn’s and Bonferroni adjustment.
Correlation between soil physico-chemical properties and bacterial CFU
Correlation test, which measures the relationship between two or more variables was employed to determine the relation of bacterial colony-forming units with the soil properties. For the correlation analysis, Kendall’s rank-based correlation was used employing “ggpubr” package in R Software66.
Identification of bacteria
The isolates were identified using morphological, Gram-staining and molecular approaches. The macro-morphological colony features of the isolated bacteria were assessed by Bergey′s manual67 followed by the Gram staining determination using Olympus 1X71 microscope. The 16S rRNA identification was carried out by extracting the DNA using QIAprep® Spin Miniprep Kit (Catalog. No. 27104, by QIAGEN laboratories) following the manufacturers protocol with slight modifications. The extracted genomic DNA was utilized as a template for 16S rRNA gene amplification.
Amplification was performed by Polymerase Chain Reaction (PCR) in a thermo-cycler (CG Palm Cycler by Genetix Biotech Asia Pvt. Ltd) with universal bacterial primers68 synthesized by IDT (Integrated DNA Technologies) yielding a PCR product of about 1.5 kb. This was carried out in a final reaction mixture volume of 50 μL and the cycling parameters comprised of 5 min initial de-naturation (94 °C) followed by 30 cycles, each of de-naturation (94 °C) for 1 min, annealing (55 °C) for 45 s, extension (72 °C) for 2 min and final extension (72 °C) for 10 min. For negative control reaction, ultrapure (MilliQ) water was taken instead of exogenous template.
The amplicons of the expected size, approximately 1500 nucleotides (1.5 kb) were observed and confirmed via gel electrophoresis on 1.5% agarose gel in 1 × Tris–Acetate-EDTA (TAE) buffer with ethidium bromide stain wherein the banding patterns were visualized using UV illumination in a GEL DOC/Bio-imaging System. A 100 base pair DNA ladder (ThermoFischer SCIENTIFIC) was taken as a standard molecular weight DNA marker. The PCR products were sent to SciGenom Labs, Kerala for purification and subsequent partial DNA sequencing. Nucleotide Basic Local Alignment Search Tool (BLASTn) was used to identify all retrieved sequences by determining the phylogenetic neighbors from the databases of National Centre for Biotechnology Information (NCBI). Evolutionary phylogenetic trees were determined by the neighbor-joining method using Maximum Composite Likelihood as a correction factor by MEGA 7 with 1000 replicate bootstrap value.
Assessment of soil bacterial diversity in DNP
The diversity of the isolated bacterial species community in terms of Shannon–Wiener’s (H′), Simpson’s index (d′), Dominance (D) and Evenness (J) was calculated using PAST 4.03 software69 whereas the similarity index of the bacterial species isolated from different study sites covering five dominant vegetational types located at varying altitudes was determined using Sorensen similarity index70.
Results
Soil physico-chemical properties
Soil temperature depicted site-wise fluctuations with the lowest of 5.1 ± 0.65 °C recorded at site-9 (HAMP) in winter season and a highest of 22.2 ± 0.75 °C at site-2 (LAG) in summer. Statistically significant differences existed in the soil temperature (Fig. 2A) as suggested by Kruskal–Wallis test (p = < 0.0001) and Wilcoxon’s test (p < 0.05) whereby mean value of site-2 i.e., LAG (13.7 ± 4.76 °C) depicted highest significance with site-3 i.e., HAR (11.3 ± 3.20 °C), site-5 i.e., HAO (11.8 ± 3.90 °C) and site-7 i.e., HAP (11.2 ± 3.42 °C). In case of temperature, eta squared based on H statistic, displayed large effect (> 0.14) and about 14.5% variance was explained by the sites. Seasonally, soil temperature (Fig. 3A) ranged between 12.3 ± 0.4 to 20.6 ± 0.8 °C (spring), 14.2 ± 0.5 to 21.7 ± 0.7 °C (summer), 11.6 ± 0.8 to 16.1 ± 0.5 °C (autumn) and 5.4 ± 0.4 to 8.6 ± 1.3 °C (winter).
Soil moisture content (MC) which is the amount of water present within the soil was found to be highly variable among different study sites with the highest percent value of 47.87 ± 6.45% at site-7 (HAP) in winter and a lowest of 6.33 ± 1.15% at site-2 (LAG) during the autumn season. However, no statistically significant variations (p = 0.11) and effect size (eta squared) was observed across the sites (Fig. 2B). The seasonal range (Fig. 3B) of percent MC in soil was found to be 27.15 ± 0.2 to 33.60 ± 0.4% (spring), 18.65 ± 0.8 to 26.75 ± 0.2% (summer), 6.95 ± 0.9 to 15.95 ± 1.2% (autumn), and 30.45 ± 0.1 to 46.65 ± 1.8% (winter).
Soil pH displayed variations from one site to another wherein the highest pH of 7.78 ± 0.12 and a lowest of 5.11 ± 0.10 was noted from site-3 (HAR) in winter and site-6 (LAO) in summer season respectively. Significant difference existed between all sites (Fig. 2C) with the site depicting highest mean pH of 7.05 ± 0.57 i.e., site-3 (HAR) showing greater differences with site-5 (HAO, 5.91 ± 0.47), site-6 (LAO, 5.68 ± 0.42), site-9 (HAMP, mean pH 6.21 ± 0.52) and site-10 (LAMP, mean pH 6.02 ± 0.51). Moreover, 37.9% of variance was explained by the sites, with respect to pH. Seasonally, the soil pH (Fig. 3C) ranged between 5.94 ± 0.22 to 7.41 ± 0.40 (spring), 5.13 ± 0.06 to 6.37 ± 0.06 (summer), 5.52 ± 0.13 to 6.75 ± 0.02 (autumn) and 6.16 ± 0.08 to 7.68 ± 0.15 (winter).
The percent soil organic carbon (SOC) varied between the sites wherein the lowest (1.24 ± 0.26%) and highest (6.53 ± 0.32%) OC levels were found at site-9 (HAMP) in winter and at site-2 (LAG) in summer season respectively. Between the sites (Fig. 2D), the mean SOC values of site-2 (LAG, 4.71 ± 1.19%) were found to show higher statistical significance with site-3 (HAR, 2.92 ± 0.74%), site-5 (HAO, 2.65 ± 0.82%), site-9 (HAMP, 2.20 ± 0.71%), and site-10 (LAMP, 2.72 ± 0.89%). Moreover, the amount variance in SOC explained by site variation was regarded large (37.1%). The seasonal range of soil OC (Fig. 3D) was 2.18 ± 0.17 to 4.85 ± 0.13% (spring), 3.25 ± 0.49 to 6.49 ± 0.06% (summer), 1.83 ± 0.06 to 3.97 ± 0.09% (autumn) and 1.56 ± 0.45 to 3.54 ± 0.26% (winter). As soil organic matter (SOM) is determined from SOC, it also exhibited the same trend between the sites and seasons with the lowest of 2.14 ± 0.45% reported from site-9 (HAMP) in winter and a highest of 11.25 ± 0.54% from site-2 (LAG) in summer season respectively. Average SOM values also were found to have higher statistically significant differences among site-2 (LAG, 8.12 ± 2.05%) and site-3 (HAR, 5.03 ± 1.28%), site-5 (HAO, 4.57 ± 1.42%), site-9 (HAMP, 3.79 ± 1.22%), and site-10 (LAMP, 4.69 ± 1.54%) with the amount variance being large (37.2%) as explained by site variation based on eta squared values (Fig. 2E). Seasonally (Fig. 3E), the percent SOM ranged between 3.77 ± 0.29 to 8.36 ± 0.23% (spring), 5.60 ± 0.84 to 11.19 ± 0.09% (summer), 3.14 ± 0.11 to 6.84 ± 0.16% (autumn) and 2.69 ± 0.77 to 6.09 ± 0.45% (winter).
The disparities in the soil total nitrogen content were observed among the sites, with the highest of 0.545 ± 0.04% recorded at site-2 (LAG) in summer and a lowest of 0.132 ± 0.04% noted from site-9 (HAMP) in winter season. Mean values of nitrogen at site-2 (LAG, 0.416 ± 0.09%) displayed greater statistically significant difference with site-5 (HAO, 0.256 ± 0.08%), site-9 (HAMP, 0.219 ± 0.07%) and site-10 (LAMP, 0.271 ± 0.09%) as per Kruskal–Wallis test (p = < 0.0001) and the Wilcoxon’s test (p < 0.05). Moreover, the amount variance in nitrogen as explained by the site variation based on eta squared values was 29.6% which is often regarded large (Fig. 2F). The seasonal variation in the percent soil nitrogen (Fig. 3F) ranged between 0.238 ± 0.02 to 0.428 ± 0.01% (spring), 0.300 ± 0.04 to 0.526 ± 0.03% (summer), 0.191 ± 0.05 to 0.382 ± 0.02% (autumn) and 0.148 ± 0.02 to 0.331 ± 0.01% (winter).
Enumeration of bacteria
Seasonal fluctuations in the bacterial colony forming units were observed with the CFU per soil gram increasing from spring to summer seasons followed by a decrease in autumn seasons and the lowest being recorded in the winter seasons. On comparatively analyzing the sites, altitudinal differences were also noted wherein sites located at low altitude had more bacterial colony count than the ones at the higher altitude and this was a trend present among all the study sites (high as well as low altitudinal sites) based on the five vegetation types. Subsequently bacterial density as CFU/g of soil was the highest (2.98 ± 0.03 × 10−7) at site-2 which is the low altitudinal Grassland site (LAG) being recorded during year 2 of study in summers whereas the lowest (1.23 ± 0.04 × 10−7) was observed in winter season of first sampling year at site-9 being the high altitudinal Mixed Pine, (HAMP) site (Table 4).
Statistically, no significant differences were noticed between the sites with respect to CFU using Kruskal–Wallis test (p = < 0.0001), and pairwise Wilcoxon’s test (Wilcoxon’s test, p > 0.05). Furthermore, the eta squared based on H statistic, suggested that in case of CFU, no large effect (< 0.036) was displayed as about 3.6% variance was explained by the sites (Fig. 4A). However, during this study, statistically significant differences with respect to CFU were observed among various seasons as determined by Kruskal–Wallis test (p = < 0.0001) and pairwise Wilcoxon’s test (p < 0.05). The eta squared based on H statistic, revealed that in case of seasonal colony-forming units (CFU), large effect (> 0.90) was displayed and about 90% variance in CFU was explained by the seasons (Fig. 4B).
Site-wise (A) and seasonal variations (B) in soil bacterial colony forming units (CFU) of lower DNP. (C) Correlation of bacterial colony forming units (CFU) with various physico-chemical properties of soil—(i) temperature, (ii) moisture content, (iii) pH, (iv) organic carbon, (v) organic matter, and (vi) total nitrogen obtained from different micro-vegetational habitats of lower DNP. (D) Taxonomic classification of bacterial strains isolated from Dachigam National Park, Kashmir.
Correlational study of soil bacterial CFU with soil physico-chemical properties
During the 2-year study period, the results of the correlations tests between the bacterial colony-forming units (CFU) and various soil physico-chemical properties, suggested that their correlations were statistically significant (Fig. 4C). Bacterial CFU depicted significant positive correlations with soil characteristics like temperature (r = 0.66), organic carbon (r = 0.55), organic matter (r = 0.55) and total nitrogen (r = 0.50) whereas significant negative correlations were displayed with moisture content (r = − 0.12) and pH (r = − 0.4) of soil, with p values for all the tested parameters being less than the significance level alpha (= 0.05).
Identification of bacteria
On the basis of morphological identification, a total of 92 morphologically different bacteria were isolated, of which the maximum isolates (15) were obtained for low altitudinal Grassland site (LAG) i.e., site-2 while as the minimum number of isolates (04) were recorded for the high altitudinal Mixed Pine, (HAMP) i.e., site-9 (Fig. S1A). All the isolated strains of bacteria depicted marked variations in macro-morphological attributes of their colonies, the detailed identification of which are given in the table (Table S1). The results of Gram staining suggested the dominance of Gram-positive (64.13%) followed by Gram-negative (23.91%) rod-shaped bacterial forms (Bacilli), existing either as a single, diplo-bacilli, or short/long chains. The presence of Gram-positive (6.52%) and Gram-negative (5.43%) round-shaped Cocci forms on the other hand were much less prevalent and arranged as cocci, diplo-cocci, or simply as bacterial clusters (Table S2).
The retrieved nucleotide sequences of the bacterial strains that were isolated from the ten (10) sampling sites in lower DNP and identified through 16S rRNA gene approach (Fig. S1B), which is a greatly conserved gene typically used for the prokaryotic species identification, were blasted (BLASTn) in the available databases of NCBI so as to find their closest neighbor. After proper percent similarity check and identification of all isolated bacterial strains, they were deposited in GenBank, NCBI and their accession numbers were availed (Table S3).
During this study, out of the total ninety-two (92) identified bacterial species, only fifty-seven (57) were found to be taxonomically different at a species level after combining all varied strains of a particular bacterial species (Fig. 4D). The overall systematic diversity of the isolated bacterial strains reflected the presence of two major taxonomic phyla of soil bacteria i.e., Firmicutes (40 species) and Proteobacteria (17 species) covering two classes (Bacilli and Gammaproteobacteria), three orders (Bacillales, Pseudomonadales and Xanthomonadales), six families (Bacillaceae, Paenibacillaceae, Staphylococcaceae, Moraxellaceae, Pseudomonadaceae and Xanthomonadaceae) and twelve bacterial genera (Bacillus, Lysinibacillus, Metabacillus, Peribacillus, Brevibacillus, Staphylococcus, Acinetobacter, Psychrobacter, Pseudomonas, Pseudoxanthomonas, Xanthomonas and Stenotrophomonas).
The evolutionary phylogenetic relationships based on 16S rRNA gene sequences of the bacterial species isolated from all of the study sites belonging to phylum, Firmicutes and Proteobacteria are given in the respective figures (Fig. 5A,B).
Assessment of soil bacterial diversity in lower DNP
The diversity of the bacteria (Table 5) isolated from ten selected study sites covering five different micro- vegetational habitats of the lower DNP was determined by calculating the diversity indices such as Shannon–Wiener’s index (H′), Simpson’s index (d′), Dominance index (D), and Evenness index (J). The Shannon–Wiener’s index (H′) ranged from 1.380 to 2.631 with the lowest observed at site-9 (HAMP) whereas the highest was recorded at site-2 (LAG). The calculated values for Simpson’s diversity (d′) revealed it to be in the range of 0.749 to 0.923 with the lowest and highest values recorded from site-9 and site-2 respectively. On the other hand, the Dominance index (D) was found to be in the range of 0.078 to 0.253 with the lowest recorded value for site-2 and the highest for site-9 respectively. Likewise, the Evenness index (J) values of bacterial diversity showed the highest recorded value of 0.994 for site-9 however, the lowest value (0.920) did not depict to follow the same trend as it was observed for site-1 (HAG).
The assessment of the soil bacterial species diversity in lower DNP ascertained that while some of the species were specific to a particular vegetation type, some others were present across various vegetation types although with differing morphological characteristics on account of being different strains of the same bacterial species (Table S4). During this study, among the fifty-seven (57) distinct bacterial strains, certain species namely B. aerius, B. licheniformis, B. mycoides, B. pumilus, B. simplex, B. thuringiensis, B. frigoritolerans, and S. pavanii showed a wider prevalence in the soils of DNP as they were found in three (03) or more sites whereas the bacteria namely S. maltophilia, was the most widespread as it was recorded from five out of the ten sites studied. However, the results clearly ascertained that about thirty-seven (37) species of bacteria identified in the present study, were found to be restricted to a particular site considering the differences in the vegetation as well as altitude at each site.
Thus, the similarity index (Table 6) of the bacteria isolated from the soils of ten study sites falling under five different vegetational types (i.e., grassland, riverine, oak, parrotiopsis and mixed pine) which were located at respective high and low altitude turned out to be—35.7% between grassland sites (HAG, site-1 and LAG, site-2); 47.1% between riverine sites (HAR, site-3 and LAR, site-4); 23.6% between oak sites (HAO, site-5 and LAO, site-6); and 22.2% between parrotiopsis sites, (HAP, site-7 and LAP, site-8). However, no similarity was found between the two mixed pine sites i.e., HAMP (site-9) and LAMP (site-10).
Discussion
National parks are key for the conservation of biodiversity by providing a safe haven for the threatened species to flourish and survive especially the endangered or endemic species71. DNP located in Zabarwan mountain range of western Himalaya is a habitat of great ecological significance as it is famous for harboring peculiar plant and animal forms comprising of more than 660 species of vascular plants, animals like critically endangered Kashmir red stag, Himalayan black bear, Himalayan brown bear, Himalayan yellow-throated marten, Himalayan gray langur, musk deer, common leopard, and about 150 bird species53. Thus, several researchers have extensively studied the national park for its phytodiversity and soil physicochemical characteristics54,72, its endangered animals55,56,73, but the national park has not been studied for its microbial populations including its bacteria in soil.
Soil is a chief component of the environment performing several vital functions as a result of which there is a continuous circulation of nutrients between various abiotic, and biotic processes. The temperature of soil influences its several operations (physical, chemical and biological) and is among the primary factors influencing such soil properties and processes which are engaged in the bacterial growth and developmental activities74,75. During the present study, soil temperature displayed significant (p = < 0.0001) differences between the study sites which can be attributed to the variations in the incoming radiation and the energy differences via the surface of the soil76, as these depend on factors like vegetation cover77, organic material content78, evaporation79, and inclination of land surface80. The soil moisture content in this study did not reflect any statistically significant differences (p = 0.11) among different sites which could be attributed to the depth differences from which the woody vegetation and grasses may obtain their soil water81, and variations in vegetation cover82,83.
In this research work, the maximum pH value (alkaline) was observed during winters whereas the lowest (slightly acidic) values were documented during the summer seasons which could be attributed to the presence of higher humus content releasing several acids in the soils84. Seasonal mean percentage of SOC and SOM showed a similar significant trend (p = < 0.0001). This could be either to the fine extensive root length of grasses per unit volume of soil, which is twenty times more in temperate grasslands in comparison to forest soils85, ratio of root to shoot which is about thirty times more in grasslands as compared to forests86, and enhanced process of organic matter decomposition87. The results of soil nitrogen suggested that the increase in soil nitrogen may be due to the higher amounts of soil organic matter88,89,90,91 and decreased N-mineralization as well as nitrification rates in forest soils with increase in altitude92 reflecting towards the temperature being the regulating factor.
Colony forming unit (CFU), which is an estimation of viable cells in a sample were determined for the seasonally collected soil samples over the period of 2 years (Spring, 2017–Winter, 2018). The results of CFU estimation revealed significant seasonal variations between the sites which can be attributed to the differences in general properties of soil, physico-chemical conditions and vegetation types as they are the major factors influencing the density, diversity, growth, and population of microorganisms including bacteria in soil93,94. Morphologically, in soil there is a prevalence of rod-shaped bacteria or bacilli followed by round/spherical-shaped bacteria or cocci and a few forms of spirillum-shaped bacteria or spirilla95 which are in accordance to the findings of this work. As has been documented here, earlier studies have also reported the pre-dominance of Gram-positive in comparison to Gram-negative bacterial forms obtained from diverse soils96,97,98.
The diversity of the culturable bacteria in different altitudinally varied micro-vegetational habitats of DNP revealed clear variations with respect to differences in soil properties as well as vegetation types which are often considered to be the vital factors affecting the density, diversity, growth and populations of bacteria in soil31,93,94,99. The vegetational diversity is considered to likely affect the soil bacterial activity, biomass as well as their composition, either in a direct manner by the production of litter and root exudates, or in an indirect way by influencing and changing the physico-chemical properties of soil100,101,102. In our study, results depicted that in Grassland vegetation, there was pre-dominance of soil bacterial species belonging to Firmicutes which are capable of surviving under harsh environments103,104. Lugo et al.105 in their study, reported species of genera Arthrobacter, Bacillus, and Pseudomonas, during their investigation on the rhizospheric bacterial diversity in a South-American grassland whereas from the Grasslands under consideration, we reported more diverse bacterial species belonging to genus, Acinetobacter, Bacillus, Lysinibacillus, Staphylococcus, Stenotrophomonas, Xanthomonas, Pseudomonas with the dominance of Bacillus species.
Riverine forests mostly comprise of moist temperate deciduous broad-leaved trees producing relatively higher quantities of litter and have enhanced decomposition rates in comparison to coniferous trees, which result in higher nutrient levels supporting more dense and diverse microbial populations106 and a similar trend has been noticed in the current study whereby the total bacterial species at Riverine vegetational sites were more than what has been recorded in Mixed Pine sites. Microbial communities have been found to be distinct not only between the forest and pasture soils107 but also among various forests108. Chim Chan et al.109 while studying the impact of vegetational covers (forests, shrubs, pastures) on the soil bacteria, concluded that while Acidobacteria dominated the broad-leaf forests, Proteobacteria and Firmicutes was prevalent in the soils of shrub and the pastures showed the predominance of alpha- and beta-Proteobacteria and Bacteroidetes. A similar observation was made in our study whereby the bacterial species under shrub vegetational type (Parrotiopsis) were mainly found to be comprised of Firmicutes (13) and Proteobacteria (05). During the present investigation, as far as the number of bacterial species in the Mixed Pine vegetational sites are concerned, the results clearly showed that among all vegetational types, the least number of bacteria were recorded from these sites. This could be attributed to the quality of the litter as well as its decomposition rates as several studies on the conifers have suggested that the litter of the coniferous tree species is rich in the amounts of acids, lignin, tannins, and other phenolic compounds, thereby making its decomposition arduous as a result of which there is a strong influence on the growth of soil microorganisms110,111,112.
As various ecological factors are considered to affect the diversity and distribution of bacteria in soil113, several studies have concluded that the regions situated at higher altitudes possess scanty vegetation constituting distinctive soil attributes which support less diverse bacterial community and structure in comparison to those regions located at lower ones supporting higher bacterial diversity31,114 which is in agreement with the findings of this study. Estimates have predicted that the values of the Shannon–Wiener’s Index (H′) typically range from 1.5 to 3.5 for ecological data115, and in this study, the H′-value ranged from > 1.3 up to as high as > 2.6 which clearly reflect the prevalence of bacterial diversity in lower DNP. On the other hand, the evenness index (J) of the bacteria in soil reflects certain pressures that might shape their community diversity and thus, its measurement is considered to be among one of the most significant attributes while assessing the impacts of various environmental factors on the diversity of bacteria in soil116. Evenness index (J) in this study didn’t reflect any significant trend within the sites which are consistent with the findings of Bryant et al.117, Fierer et al.118 and Lyngwi et al.31. Furthermore, the variations in the composition of vegetation displayed significant influence on the similarity index of the soil bacterial populations among the five studied vegetational types wherein the highest similarity (> 47%) was depicted among the Riverine vegetational sites and no similarity was recorded in between the Mixed Pine vegetational sites. This is in accordance with the study of Liu et al.99 which concluded that tree species compositions had a profound influence on the similarity coefficients of their rhizospheric soil bacterial communities.
Conclusions
The north-western part of Himalayas encompassing the Zabarwan range of Kashmir is known to consist different climatic and elevational zones having varied soil textures which serves as an access-point inhabiting the richest bio-diversity with great level of endemism. However, this region is not extensively studied as far as its microbial diversity including bacteria is concerned. In this study, the physico-chemical parameters of the soil were determined, the diversity and distribution of culturable soil bacteria from low to high (1671–1870 m.a.s.l.) elevational gradient spreading across five different vegetation types were characterized, and correlated with the soil bacterial distribution and diversity. Soil parameters such as temperature, pH, moisture content, organic carbon and matter, and nitrogen were measured seasonally at each site. The bacteria in soil were cultured on three different media (Nutrient agar, Luria Bertani agar and Reasoner’s 2A agar) and were initially characterized by morphological and Gram staining methods. A total of 92 morphologically different bacterial isolates were then subjected to 16S rRNA sequence analysis for estimating the diversity of lower DNP. The phylogenetic analysis of these 92 isolated strains varying in their macro- and micro-morphological characteristics, revealed the presence of only fifty-seven (57) different species at the molecular level with the Firmicutes being the most common bacterial group, followed by Proteobacteria. Bacterial CFUs showed a positive correlation with parameters of the soil, such as temperature (r = 0.66), organic carbon content (r = 0.55), organic matter content (r = 0.55), and total nitrogen content (r = 0.5), whereas moisture content (r = − 0.12) and pH (r = − 0.4) of soil showed a negative correlation. The results of this study clearly reflected that the altitudinal gradient, coupled with the varied vegetational types and the soil physico-chemical parameters influenced the distribution and diversity of bacteria in soil. This study therefore concludes that lower DNP, an ecologically significant biome contains a vast reservoir of soil bacteria which decreases with increasing altitude and thus provide us with the first baseline information highlighting the microbial importance from this poorly explored area in the western Himalaya, with justifying efforts for the presence and need to explore the prevalence of novel species in this vital ecosystem.
Data availability
Data regarding 16S rRNA gene sequences have been deposited in GenBank, NCBI (https://www.ncbi.nlm.nih.gov/nucleotide/) under the accession numbers given in the table (Table S3) contained in Supplementary Information.
References
Hatton, P. J., Castanha, C., Torn, M. S. & Bird, J. A. Litter type control on soil C and N stabilization dynamics in a temperate forest. Glob. Change Biol. 21(3), 1358–1367. https://doi.org/10.1111/gcb.12786 (2015).
Lladó, S., López-Mondéjar, R. & Baldrian, P. Forest soil bacteria: Diversity, involvement in ecosystem processes, and response to global change. Microbiol. Mol. Biol. Rev. 81(2), e00063–16. https://doi.org/10.1128/mmbr.00063-16 (2017).
Ranjard, L. & Richaume, A. Quantitative and qualitative microscale distribution of bacteria in soil. Res. Microbiol. 152(8), 707–716. https://doi.org/10.1016/S0923-2508(01)01251-7 (2001).
Nannipieri, P., Badalucco, L., Benbi, D. K., & Nieder, R. Handbook of processes and modelling in the soil-plant system. Biological Processes, 57–82 (2003).
Wixon, D. L. & Balser, T. C. Complexity, climate change and soil carbon: A systems approach to microbial temperature response. Syst. Res. Behav. Sci. 26(5), 601–620. https://doi.org/10.1002/sres.995 (2009).
Van Der Heijden, M. G., Bardgett, R. D. & Van Straalen, N. M. The unseen majority: Soil microbes as drivers of plant diversity and productivity in terrestrial ecosystems. Ecol. Lett. 11(3), 296–310. https://doi.org/10.1111/j.1461-0248.2007.01139.x (2008).
Tisdall, J. M. Possible role of soil microorganisms in aggregation in soils. Plant Soil 159, 115–121. https://doi.org/10.1007/BF00000100 (1994).
Ingham, E. R. Soil biology primer, Chapter 4: Soil fungus. Soil and Water Conservation 22–23 (Soil and Water Conservation Society, 2009).
Stevens, W. B., Sainju, U. M., Caesar, A. J., West, M. & Gaskin, J. F. Soil-aggregating bacterial community as affected by irrigation, tillage, and cropping system in the northern great plains. Soil Sci. 179(1), 11–20 (2014).
Islam, K. R. Lecture on Soil Physics, Personal Collection of K. Islam (Ohio State University, 2008).
López-Mondéjar, R., Zühlke, D., Becher, D., Riedel, K. & Baldrian, P. Cellulose and hemicellulose decomposition by forest soil bacteria proceeds by the action of structurally variable enzymatic systems. Sci. Rep. 6(1), 25279. https://doi.org/10.1038/srep25279 (2016).
Wardle, D. A., Nilsson, M. C. & Zackrisson, O. Fire-derived charcoal causes loss of forest humus. Science 320(5876), 629–629. https://doi.org/10.1126/science.1154960 (2008).
Shelobolina, E., Roden, E., Benzine, J. & Xiong, M. Y. Using phyllosilicate-Fe (II)-oxidizing soil bacteria to improve Fe and K plant nutrition. U.S. Patent Application 14/924,397 (Wisconsin Alumni Research Foundation, 2016).
Kumar, A., & Verma, J. P. The role of microbes to improve crop productivity and soil health. In Ecological Wisdom Inspired Restoration Engineering 249–265. https://doi.org/10.1007/978-981-13-0149-0_14 (2019).
Dick, W. Lecture on Biochemistry Process in Soil Microbiology, Personal Collection of W. Dick (The Ohio State University School of Environment and Natural Resources, 2009).
Reed, S. C., Cleveland, C. C. & Townsend, A. R. Functional ecology of free-living nitrogen fixation: A contemporary perspective. Annu. Rev. Ecol. Evol. Syst. 42, 489–512. https://doi.org/10.1146/annurev-ecolsys-102710-145034 (2011).
Sylvia, D. M., Fuhrmann, J. J., Hartel, P. G. & Zuberer, D. A. Principles and Applications of Soil Microbiology (No. QR111 S674 2005) 2nd edn. (Prentice Hall, 2005).
Torsvik, V., Daae, F. L., Sandaa, R. A. & Øvreås, L. Novel techniques for analysing microbial diversity in natural and perturbed environments. J. Biotechnol. 64(1), 53–62. https://doi.org/10.1016/s0168-1656(98)00103-5 (1998).
Roesch, L. F. W. et al. Pyrosequencing enumerates and contrasts soil microbial diversity. ISME J. 1(4), 283–290. https://doi.org/10.1038/ismej.2007.53 (2007).
Rousk, J., Brookes, P. C. & Bååth, E. The microbial PLFA composition as affected by pH in an arable soil. Soil Biol. Biochem. 42(3), 516–520. https://doi.org/10.1016/j.soilbio.2009.11.026 (2010).
Brockett, B. F., Prescott, C. E. & Grayston, S. J. Soil moisture is the major factor influencing microbial community structure and enzyme activities across seven biogeoclimatic zones in western Canada. Soil Biol. Biochem. 44(1), 9–20. https://doi.org/10.1016/j.soilbio.2011.09.003 (2012).
Urbanová, M., Šnajdr, J. & Baldrian, P. Composition of fungal and bacterial communities in forest litter and soil is largely determined by dominant trees. Soil Biol. Biochem. 84, 53–64. https://doi.org/10.1016/j.soilbio.2015.02.011 (2015).
Binkley, D. & Vitousek, P. M. Soil nutrient availability. In Plant Physiological, Field Methods and Instrumentation (eds Pearey, R. W. et al.) 75–96 (Champan and Hall, 1989).
Ruess, J. O. & Innis, G. S. A grassland nitrogen flow simulation mode. Ecology 58, 348–429. https://doi.org/10.2307/1935612 (1977).
Kumar, M., Sharma, C. M. & Rajwar, G. S. Physico-chemical properties of forest soil along altitudinal gradient in Garhwal Himalaya. J. Hill Res. 17(2), 60–64 (2004).
Smit, E. et al. Diversity and seasonal fluctuations of the dominant members of the bacterial soil community in a wheat field as determined by cultivation and molecular methods. Appl. Environ. Microbiol. 67(5), 2284–2291. https://doi.org/10.1128/AEM.67.5.2284-2291.2001 (2001).
Qazi, P. H. Bioprospecting Himalayan microbial diversity. ENVIS Newsletter on Himalayan Ecology 12(4). http://gbpihedenvis.nic.in/ENVIS%20Newsletter/vol%2012(4).pdf (2015).
Pradhan, S. et al. Bacterial biodiversity from Roopkund glacier, Himalayan Mountain ranges, India. Extremophiles 14, 377–395. https://doi.org/10.1007/s00792-010-0318-3 (2010).
Shivaji, S. et al. Bacterial diversity of soil in the vicinity of Pindari glacier, Himalayan Mountain ranges, India, using culturable bacteria and soil 16S rRNA gene clones. Extremophiles 15, 1–22. https://doi.org/10.1007/s00792-010-0333-4 (2011).
Das, J. & Dangar, T. K. Microbial population dynamics, especially stress tolerant Bacillus thuringiensis, in partially anaerobic rice field soils during post-harvest period of the Himalayan, island, brackish water and coastal habitats of India. World J. Microbiol. Biotechnol. 24, 1403–1410. https://doi.org/10.1007/s11274-007-9620-3 (2008).
Lyngwi, N. A., Koijam, K., Sharma, D. & Joshi, S. R. Cultivable bacterial diversity along the altitudinal zonation and vegetation range of tropical Eastern Himalaya. Rev. Biol. Trop. 61(1), 467–490. https://doi.org/10.15517/rbt.v61i1.11141 (2013).
Pandey, S., Singh, S., Yadav, A. N., Nain, L. & Saxena, A. K. Phylogenetic diversity and characterization of novel and efficient cellulase producing bacterial isolates from various extreme environments. Biosci. Biotechnol. Biochem. 77(7), 1474–1480. https://doi.org/10.1271/bbb.130121 (2013).
Venkatachalam, S., Gowdaman, V. & Prabagaran, S. R. Culturable and culture-independent bacterial diversity and the prevalence of cold-adapted enzymes from the Himalayan Mountain ranges of India and Nepal. Microb. Ecol. 69, 472–491. https://doi.org/10.1007/s00248-014-0476-4 (2015).
Saxena, A. K., Yadav, A. N., Kaushik, R., Tyagi, S. P., & Shukla, L. Biotechnological applications of microbes isolated from cold environments in agriculture and allied sectors. In International Conference on Low Temperature Science and Biotechnological Advances, Vol. 104 (Society of Low Temperature Biology, 2015).
Singh, R. N. et al. First high-quality draft genome sequence of a plant growth promoting and cold active enzyme producing psychrotrophic Arthrobacter agilis strain L77. Stand. Genom. Sci. 11, 1–9. https://doi.org/10.1186/s40793-016-0176-4 (2016).
Mushtaq, H. et al. Biochemical characterization and functional analysis of heat stable high potential protease of Bacillus amyloliquefaciens strain HM48 from soils of Dachigam National Park in Kashmir Himalaya. Biomolecules 11(1), 117. https://doi.org/10.3390/biom11010117 (2021).
Maharana, A. K. & Ray, P. Isolation and screening of cold active extracellular enzymes producing psychrotrophic bacteria from soil of Jammu City. Biosci. Biotechnol. Res. Asia 10(1), 267–273. https://doi.org/10.13005/bbra/1120 (2013).
Rehakova, K., Chlumska, Z. & Dolezal, J. Soil cyanobacterial and microalgal diversity in dry mountains of Ladakh, NW Himalaya, as related to site, altitude, and vegetation. Microb. Ecol. 62, 337–346. https://doi.org/10.1007/s00248-011-9878-8 (2011).
Rehakova, K. et al. Bacterial community of cushion plant Thylacospermum ceaspitosum on elevational gradient in the Himalayan cold desert. Front. Microbiol. 6, 304. https://doi.org/10.3389/fmicb.2015.00304 (2015).
Gupta, P. & Vakhlu, J. Culturable bacterial diversity and hydrolytic enzymes from Drass, a cold desert in India. Afr. J. Microbiol. Res. 9, 1866–1876. https://doi.org/10.5897/AJMR2015.7424 (2015).
Yadav, A. N. et al. Culturable diversity and functional annotation of psychrotrophic bacteria from cold desert of Leh Ladakh (India). World J. Microbiol. Biotechnol. 31, 95–108. https://doi.org/10.1007/s11274-014-1768-z (2015).
Farooq, S., Nazir, R., Ganai, B. A., Mushtaq, H. & Dar, G. J. Psychrophilic and psychrotrophic bacterial diversity of Himalayan Thajwas glacial soil, India. Biologia 77, 203–213. https://doi.org/10.1007/s11756-021-00915-6 (2022).
Ahmad, N., Johri, S., Abdin, M. Z. & Qazi, G. N. Molecular characterization of bacterial population in the forest soil of Kashmir, India. World J. Microbiol. Biotechnol. 25, 107–113. https://doi.org/10.1007/s11274-008-9868-2 (2009).
Thakur, D., Yadav, A., Gogoi, B. K. & Bora, T. C. Isolation and screening of Streptomyces in soil of protected forest areas from the states of Assam and Tripura, India, for antimicrobial metabolites. J. Mycol. Méd. 17(4), 242–249. https://doi.org/10.1016/j.mycmed.2007.08.001 (2007).
Rina, K., Hiral, P., Payal, P., Dharaiya, N. & Patel, R. K. Study on microbial diversity of Wild Ass Sanctuary, Little Rann of Kutch, Gujarat, India. ICFAI Univ. J. Life Sci. 3(1), 34–41 (2009).
Das, S., Saikia, P., Baruah, P. P. & Chakraborty, A. Isolation and identification of soil bacteria collected from Dibru-Saikhowa, the National Park and Biosphere Reserve Forest of Assam, India. Int. J. Sci. Res. (IJSR), 1937–1940 (2016).
De Mandal, S., Lalremsanga, H. T. & Kumar, N. S. Bacterial diversity of Murlen National Park located in Indo-Burman Biodiversity hotspot region: A metagenomic approach. Genom. Data 5, 25–26. https://doi.org/10.1016/j.gdata.2015.04.025 (2015).
Megha, B., Sejal, P., Puja, P. & Jasrai, Y. T. Isolation and identification of soil microflora of national parks of Gujarat, India. Int. J. Curr. Microbiol. Appl. Sci. 4(3), 421–429 (2015).
Kumar, A., Singh, R. D., Patra, A. K., Sahu, S. K. & Singh, M. Impact of oak and pine canopy cover on soil biochemical and microbial indicators of Binsar Wildlife Sanctuary in the Western Himalaya, India. J. Pure Appl. Microbiol. 11(3), 1599–1607. https://doi.org/10.22207/JPAM.11.3.47 (2017).
Dhiman, P., Mehta, J. P., Singh, P. & andSharesthBaldotra, S. S.,. Effect of prescribe fire on bacterial abundance and their enzymatic activity in burnt and unburnt soil of Chilla Forest, Raja Ji National Park, Uttarakhand, India. Plant Arch. 18(1), 1125–1128 (2018).
Behera, P. et al. Spatial and temporal heterogeneity in the structure and function of sediment bacterial communities of a tropical mangrove forest. Environ. Sci. Pollut. Res. 26, 3893–3908 (2019).
Sharma, P. & Thakur, D. Antimicrobial biosynthetic potential and diversity of culturable soil actinobacteria from forest ecosystems of Northeast India. Sci. Rep. 10(1), 1–18. https://doi.org/10.1038/s41598-020-60968-6 (2020).
Dar, G. H., Bhagat, R. C. & Khan, M. A. Biodiversity of the Kashmir Himalaya (Valley Book House, 2002).
Shameem, S. A., Kangroo, N. I. & Bhat, G. A. Comparative assessment of edaphic features and herbaceous diversity in lower Dachigam national park, Kashmir, Himalaya. J. Ecol. Nat. Environ. 3(6), 196–204 (2011).
Thakur, M., Sharma, L. K., Charoo, S. A. & Sathyakumar, S. Conflict bear translocation: Investigating population genetics and fate of bear translocation in Dachigam National Park, Jammu and Kashmir, India. PLoS One 10, e0132005. https://doi.org/10.1371/journal.pone.0132005 (2015).
Ahmad, K., Qureshi, Q., Agoramoorthy, G. & Nigam, P. Habitat use patterns and food habits of the Kashmir red deer or Hangul (Cervus elaphus hanglu) in Dachigam National Park, Kashmir, India. Ethol. Ecol. Evol. 28(1), 85–101. https://doi.org/10.1080/03949370.2015.1018955 (2016).
Jammu and Kashmir Forest Department (JKFD). Handbook of Forest Statistics (Jammu and Kashmir Forest Department, 2011).
Anderson, J. M. & Ingram, J. S. I. A Handbook of Methods 62–65 (CAB International, 1993).
Joshi, S. R., Chauhan, M. A. N. J. U., Sharma, G. D. & Mishra, R. R. Effect of deforestation on microbes, VAM fungi and their enzymatic activity in Eastern Himalaya. In Studies in Himalayan Ecobiology 141–152 (Today and Tommorows Publication, 1991).
Jackson, M. L. Soil Chemical Analysis 151–154 (Prentice-Hall, 1958). https://doi.org/10.1002/jpln.19590850311.
Gardner, W. H. Water content. Methods of soil analysis: Part 1. Phys. Mineral. Methods 5, 493–544 (1986).
Walkley, A. & Black, I. A. Estimation of soil organic carbon by the chromic acid titration method. Soil Sci. 37(1), 29–38 (1934).
Bremner, J. M. Determination of nitrogen in soil by the Kjeldahl method. J. Agric. Sci. 55(1), 1–23 (1960).
Coursey, D. G. & Eggins, H. O. W. Microorganismes responsables de l’altération de l’huile de palme pendant le stockage. Oléagineux 16, 227–233 (1961).
Kumar, R., Acharya, C. & Joshi, S. R. Isolation and analyses of uranium tolerant Serratia marcescens strains and their utilization for aerobic uranium U (VI) bioadsorption. J. Microbiol. 49, 568–574. https://doi.org/10.1007/s12275-011-0366-0 (2011).
Team, R. C. R: A language and environment for statistical computing. https://www.R-project.org (R Foundation for Statistical Computing, 2017).
Bergey, D. H. & Holt, J. G. Bergey’s Manual of Determinative Bacteriology (Lippincott Williams & Wilkins, 1994).
Gürtler, V. & Stanisich, V. A. New approaches to typing and identification of bacteria using the 16S–23S rDNA spacer region. Microbiology 142(1), 3–16 (1996).
Hammer, Ø., Harper, D. A. T. & Ryan, P. D. PAST: Paleontological statistics software package for education and data analysis. Palaeontol. Electron. 4, 1–9 (2001).
Sorensen, T. A. A method of establishing groups of equal amplitude in plant sociology based on similarity of species content and its application to analyses of the vegetation on Danish commons. Biol. Skar. 5, 1–34 (1948).
Muhumuza, M. & Balkwill, K. Factors affecting the success of conserving biodiversity in national parks: A review of case studies from Africa. Int. J. Biodivers. https://doi.org/10.1155/2013/798101 (2013).
Yaqoob, A., Yunus, M., Bhat, G. A. & Singh, D. P. Phytodiversity and seasonal variations in the soil characteristics of shrublands of Dachigam National Park, Jammu and Kashmir, India. Clim. Change Environ. Sustain. 3(2), 137–143. https://doi.org/10.5958/2320-642X.2015.00015.0 (2015).
Mir, Z. R., Noor, A., Habib, B. & Veeraswami, G. G. Seasonal population density and winter survival strategies of endangered Kashmir gray langur (Semnopithecus ajax) in Dachigam National Park, Kashmir, India. Springer Plus 4, 1–8. https://doi.org/10.1186/s40064-015-1366-z (2015).
Buchan, G. D. Soil temperature regime. In Soil and Environmental Analysis: Physical Methods (eds Smith, K. A. & Mullins, C.) 539–594 (Marcel Dekker, 2001).
Buchan, G. D. Temperature effects in soil. In Encyclopedia of Agrophysics, Encyclopedia of Earth Sciences Series (Springer, 2011).
Chiemeka, I. U. Soil temperature profile at Uturu, Nigeria. Pac. J. Sci. Technol. 11(1), 478–482 (2010).
Decker, K. L. M., Wang, D., Waite, C. & Scherbatskoy, T. Snow removal and ambient air temperature effects on forest soil temperatures in northern Vermont. Soil Sci. Soc. Am. J. 67(4), 1234–1242. https://doi.org/10.2136/sssaj2003.1234 (2003).
Abu-Hamdeh, N. H. & Reeder, R. C. Soil thermal conductivity effects of density, moisture, salt concentration, and organic matter. Soil Sci. Soc. Am. J. 64(4), 1285–1290. https://doi.org/10.2136/sssaj2000.6441285x (2000).
Lu, S., Ren, T., Gong, Y. & Horton, R. An improved model for predicting soil thermal conductivity from water content at room temperature. Soil Sci. Soc. Am. J. 71(1), 8–14. https://doi.org/10.2136/sssaj2006.0041 (2007).
Elizbarashvili, E. S., Urushadze, T. F., Elizbarashvili, M. E., Elizbarashvili, S. E. & Schaefer, M. K. Temperature regime of some soil types in Georgia. Eurasian Soil Sci. 43(4), 427–435. https://doi.org/10.1134/S1064229310040083 (2010).
Walter, H. & Burnett, J. H. Ecology of Tropical and Subtropical Vegetation Vol. 539, xviii+-539 (Oliver and Boyd, 1971).
Callaway, R. M. Positive interactions and interdependence in plant communities. Springer Science Business Media https://doi.org/10.1007/978-1-4020-6224-7 (2007).
Song, Y. et al. Effects of vegetation height and density on soil temperature variations. Chin. Sci. Bull. 58(8), 907–912. https://doi.org/10.1007/s11434-012-5596-y (2013).
Dimri, B. M., Singh, S. B., Baneriee, S. K. & Singh, B. Relation of age and dominance of tree species with soil chemical attributes in Kalimpong and Kurseong District of West Bengal. Indian For. 113(4), 307–311 (1987).
Jackson, R. B., Mooney, H. A. & Schulze, E. D. A global budget for fine root biomass, surface area, and nutrient contents. Proc. Natl. Acad. Sci. 94(14), 7362–7366. https://doi.org/10.1073/pnas.94.14.7362 (1997).
Wilson, S. D. Competition between grasses and woody plants. In Population Biology of Grasses (ed. Cheplick, G. P.) 231–254 (Cambridge University Press, 1998).
Reth, S., Reichstein, M. & Falge, E. The effect of soil water content, soil temperature, soil pH-value and the root mass on soil CO2 efflux—A modified model. Plant Soil 268, 21–33. https://doi.org/10.1007/s11104-005-0175-5 (2005).
Zinke, P. J. The pattern of influence of individual forest trees on soil properties. Ecology 43(1), 130–133 (1962).
Patric, J. H. Forest management and nutrient cycling in eastern hardwoods Vol. 324 (Forest Service, US Department of Agriculture, Northeastern Forest Experiment Station, 1975).
Mroz, G. D., Jurgensen, M. F. & Frederick, D. J. Soil nutrient changes following whole tree harvesting on three northern hardwood sites. Soil Sci. Soc. Am. J. 49(6), 1552–1557. https://doi.org/10.2136/sssaj1985.03615995004900060044x (1985).
Maggs, J. & Hewett, B. Organic C and nutrients in surface soils from some primary rainforests, derived grasslands and secondary rainforests on the Atherton Tableland in North East Queensland. Soil Res. 31(3), 343–350 (1993).
Hart, S. C. & Perry, D. A. Transferring soils from high-to low-elevation forests increases nitrogen cycling rates: Climate change implications. Glob. Change Biol. 5(1), 23–32 (1999).
Atlas, R. M. Diversity of microbial communities. Adv. Microb. Ecol., 1–47 (1984).
Dimitriu, P. A. & Grayston, S. J. Relationship between soil properties and patterns of bacterial β-diversity across reclaimed and natural boreal forest soils. Microb. Ecol. 59, 563–573. https://doi.org/10.1007/s00248-009-9590-0 (2010).
Bele, S. S. Soil Testing and Soil Microbiology 79–108 (Satyam Publishers and Distributors, 2014). https://doi.org/10.1007/s11356-018-3927-5.
Cattelan, A. J., Hartel, P. G. & Fuhrmann, J. J. Bacterial composition in the rhizosphere of nodulating and non-nodulating soybean. Soil Sci. Soc. Am. J. 62(6), 1549–1555. https://doi.org/10.2136/sssaj1998.03615995006200060011x (1998).
Silva, P. D. & Nahas, E. Bacterial diversity in soil in response to different plans, phosphate fertilizers and liming. Braz. J. Microbiol. 33, 304–310 (2002).
Begum, K. et al. Isolation and characterization of bacteria with biochemical and pharmacological importance from soil samples of Dhaka City. Dhaka Univ. J. Pharm. Sci. 16(1), 129–136. https://doi.org/10.3329/dujps.v16i1.33390 (2017).
Liu, D., Liu, Y., Fang, S. & Tian, Y. Tree species composition influenced microbial diversity and nitrogen availability in rhizosphere soil. Plant Soil Environ. 61(10), 438–443. https://doi.org/10.17221/94/2015-PSE (2015).
Chodak, M., Klimek, B., Azarbad, H. & Jaźwa, M. Functional diversity of soil microbial communities under Scots pine, Norway spruce, silver birch and mixed boreal forests. Pedobiologia 58(2–3), 81–88 (2015).
Gartzia-Bengoetxea, N., Kandeler, E., de Arano, I. M. & Arias-González, A. Soil microbial functional activity is governed by a combination of tree species composition and soil properties in temperate forests. Appl. Soil. Ecol. 100, 57–64 (2016).
Shameem, S. A., Mushtaq, H., Wani, A. A., Ahmad, N. & Hai, A. Phytodiversity of herbaceous vegetation in disturbed and undisturbed forest ecosystems of Pahalgam valley, Kashmir Himalaya, India. Br. J. Environ. Clim. Change 7(3), 148–167 (2017).
Felske, A., Wolterink, A., Van Lis, R. & Akkermans, A. D. Phylogeny of the main bacterial 16S rRNA sequences in Drentse A grassland soils (The Netherlands). Appl. Environ. Microbiol. 64(3), 871–879. https://doi.org/10.1128/aem.64.3.871-879.1998 (1998).
Chodak, M., Gołębiewski, M., Morawska-Płoskonka, J., Kuduk, K. & Niklińska, M. Soil chemical properties affect the reaction of forest soil bacteria to drought and rewetting stress. Ann. Microbiol. 65, 1627–1637. https://doi.org/10.1007/s13213-014-1002-0 (2015).
Lugo, M. A. et al. Arbuscular mycorrhizal fungi and rhizospheric bacteria diversity along an altitudinal gradient in South American Puna grassland. Microb. Ecol. 55, 705–713. https://doi.org/10.1007/s00248-007-9313-3 (2008).
Wang, Q., Wang, S., Fan, B. & Yu, X. Litter production, leaf litter decomposition and nutrient return in Cunninghamia lanceolata plantations in south China: Effect of planting conifers with broadleaved species. Plant Soil 297, 201–211. https://doi.org/10.1007/s11104-007-9333-2 (2007).
Nüsslein, K. & Tiedje, J. M. Soil bacterial community shift correlated with change from forest to pasture vegetation in a tropical soil. Appl. Environ. Microbiol. 65(8), 3622–3626. https://doi.org/10.1128/aem.65.8.3622-3626.1999 (1999).
Hackl, E., Zechmeister-Boltenstern, S., Bodrossy, L. & Sessitsch, A. Comparison of diversities and compositions of bacterial populations inhabiting natural forest soils. Appl. Environ. Microbiol. 70(9), 5057–5065. https://doi.org/10.1128/AEM.70.9.5057-5065.2004 (2004).
Chan, C. et al. Vegetation cover of forest, shrub and pasture strongly influences soil bacterial community structure as revealed by 16S rRNA gene T-RFLP analysis. FEMS Microbiol. Ecol. 64(3), 449–458. https://doi.org/10.1111/j.1574-6941.2008.00488.x (2008).
Adamczyk, B., Kitunen, V. & Smolander, A. Protein precipitation by tannins in soil organic horizon and vegetation in relation to tree species. Biol. Fertil. Soils 45(1), 55–64. https://doi.org/10.1007/s00374-008-0308-0 (2008).
Kanerva, S., Kitunen, V., Loponen, J. & Smolander, A. Phenolic compounds and terpenes in soil organic horizon layers under silver birch, Norway spruce and Scots pine. Biol. Fertil. Soils 44(4), 547–556. https://doi.org/10.1007/s00374-007-0234-6 (2008).
Ushio, M., Balser, T. C. & Kitayama, K. Effects of condensed tannins in conifer leaves on the composition and activity of the soil microbial community in a tropical montane forest. Plant Soil 365(1), 157–170. https://www.jstor.org/stable/42952341 (2013).
Lomolino, M. V. Elevation gradients of species-density: Historical and prospective views. Glob. Ecol. Biogeogr. 10(1), 3–13. https://doi.org/10.1046/j.1466-822x.2001.00229.x (2001).
Thomson, B. C. et al. Vegetation affects the relative abundances of dominant soil bacterial taxa and soil respiration rates in an upland grassland soil. Microb. Ecol. 59(2), 335–343. https://doi.org/10.1007/s00248-009-9575-z (2010).
May, R. M. Patterns of species abundance and diversity. In Ecology and Evolution of Communities (eds Cody, M. L. & Diamond, J. M.) 81–120 (Harvard University, 1975).
Kapur, M. & Jain, R. K. Microbial diversity: Exploring the unexplored. World Federation of Culture Collection Newsletter 39, 12–16 (2004).
Bryant, J. A. et al. Microbes on mountainsides: Contrasting elevational patterns of bacterial and plant diversity. Proc. Natl. Acad. Sci. 105(Suppl 1), 11505–11511 (2008).
Fierer, N. et al. Microbes do not follow the elevational diversity patterns of plants and animals. Ecology 92(4), 797–804. https://doi.org/10.1890/10-1170.1 (2011).
Acknowledgements
Authors duly acknowledge the Department of Wildlife, Government of Jammu and Kashmir for providing the required soil sampling permit order (No. WLP/Res/2017-18/493-96). Dr. Ruqeya Nazir (Assistant Professor, Centre of Research for Development, University of Kashmir) is thanked for providing the laboratory facility needed for the conduct of microbiology portion in this study. This research work received no external funding but was supported via merit fellowship to the scholar (F-DFPMS-SCH-KU/17) by the Department of Environmental Science (DST-FIST assisted), University of Kashmir.
Author information
Authors and Affiliations
Contributions
Conceptualization, H.M., A.J. and B.A.G.; methodology, H.M., A.J. and B.A.G.; software, H.M.; formal analysis, H.M.; investigation: H.M.; resources, A.J., and B.A.G.; writing—original draft, H.M.; writing—review and editing: H.M., A.J., and B.A.G.; supervision: A.J. and B.A.G. All authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Mushtaq, H., Ganai, B.A. & Jehangir, A. Exploring soil bacterial diversity in different micro-vegetational habitats of Dachigam National Park in North-western Himalaya. Sci Rep 13, 3090 (2023). https://doi.org/10.1038/s41598-023-30187-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-30187-w
- Springer Nature Limited
We’re sorry, something doesn't seem to be working properly.
Please try refreshing the page. If that doesn't work, please contact support so we can address the problem.