Abstract
Grape is a globally significant fruit-bearing crop, and the grape flower bud differentiation essential to fruit production is closely related to light quality. To investigate the regulatory mechanism of grape flower bud differentiation under red‒blue light, the transcriptome and hormone content were determined at four stages of flower bud differentiation. The levels of indole-3-acetic acid (IAA) and abscisic acid (ABA) in grape flower buds at all stages of differentiation under red‒blue light were higher than those in the control. However, the levels of cytokinins (CKs) and gibberellic acid (giberellins, GAs) fluctuated continuously over the course of flower bud differentiation. Moreover, many differentially expressed genes were involved in auxin, CK, GA, and the ABA signal transduction pathways. There were significant differences in the AUX/IAA, SAUR, A-RR, and ABF gene expression levels between the red‒blue light treatment and the control buds, especially in regard to the ABF genes, the expression levels of which were completely different between the two groups. The expression of GBF4 and AI5L2 in the control was always low, while the expression under red‒blue light increased. AI5L7 and AI5L5 expression levels showed an upwards trend in the control plant buds and gradually decreased in red‒blue light treatment plant buds. Through weighted gene coexpression network analysis, we determined that the transcription factors WRK48 (WRKY family), EF110 (ERF family), ABR1, CAMTA3 (CAMTA family), and HSFA3 (HSF family) may be involved in the regulation of the GBF4 gene. This study lays a foundation for further analysis of grape flower bud differentiation regulation under red‒blue light.
Similar content being viewed by others
Introduction
Grape (Vitis vinifera L.) is a globally significant fruit-bearing crop that is cultivated on all continents. Whether it is used for making fresh food or wine, it has major economic significance and is one of the most economically profitable fruit crops1. The quality and quantity of grape fruit are the most critical standards for grape cultivation and are directly affected by flower bud differentiation2. Flower bud differentiation of grape is an unusual process, compared with 1-year-old plants3. The shoot meristem generates both nutritional and reproductive structures, and the uncommitted primordia (also called anlagen) generated by the stem meristem differentiate into tendrils or inflorescences, which complete flowering in the second year4.
Grape flower bud differentiation is a very complex biological process that is influenced by the integration of the external environment and internal factors5. In the model plant Arabidopsis thaliana, flower development involves six pathways: the photoperiod, gibberellin, vernalization, autonomous, and ageing pathways, as well as the glycometabolism pathways, regulate floral-specific genes, resulting in the physiological transformation of the vegetative meristem into floral meristem6. These pathways converge to regulate the expression of flowering-related genes, such as FLOWER LOCUS T (FT), CONSTANS (CO), SUPPRESSOR OF OVEREXPRESSION OF CO1 (SOC1), LEAFY (LFY), APETALA1 (AP1), APETALA2 (AP2), and APETALA3 (AP3), and irreversibly induce the transformation from vegetative meristem to floral meristem7. Flower development pathway genes or genes involved in photoperiod or vernalization have also been identified in the grape genome; most of these genes are flowering signal integrators, floral meristem recognition genes, and floral organ recognition genes, such as MADS-box family genes and VvFT/TFL1 family genes promoting early flowering8. The orthologue of VvFT, the Arabidopsis thaliana gene FLOWERING LOCUST, is related to seasonal flowering induction in latent buds, inflorescence and flower development9. The expression of VvLFY is related to inflorescence and flower developmen10. Homologues of VvFUL and VvAP1 appear in the early stages of lateral meristem development and are maintained in both inflorescence and tendril primordia11,12.
As an essential environmental factor, light drives photosynthesis, participates in most processes regulating the plant life cycle, and plays a key role in plant flower bud differentiation13. Light quality is an important signal affecting plant morphogenesis and flower bud formation14. Plants perceive the quality of light through photoreceptors. Photoreceptors are classified as any of the following: photosensitive pigments (Phys) (for absorption of red/far-red light), cryptochromes (Crys) (for absorption of blue/ultraviolet A (UV-A) light), phototaxis proteins, Zeitlupe (ZTL/FKF1/LKP2) family members, and the UV-B absorbing UVR8 family members15,16,17. Photosensitizers and cryptochromes are responsible for plant morphological and developmental changes18,19. Therefore, research on the effect of light quality on plant growth and development mainly focuses on red and blue light. In addition, the combination of red and blue light forms a spectral absorption peak suitable for plant photosynthesis and morphogenesis, forming fully developed flower buds and increasing the number of buds20,21. Therefore, two wavelengths (blue and red) are necessary for plants.
Although some progress has been made in studying the grape flower bud differentiation process22,23, the regulatory mechanism of grape flower bud differentiation under red‒blue light is still unclear. In the early stages of this study, we screened red‒blue light for suitablity to grape flower bud differentiation, since it is necessary to study the flower bud differentiation process under red‒blue light if we wish to understand the regulatory mechanism of flower bud differentiation under this particular type of ligh. Therefore, in this study we used the screened red‒blue light for supplementary light treatment, with conditions of no supplementary light as the control, and the hormone content at different stages of flower bud differentiation was determined by high-performance liquid chromatography (HPLC). The differentially expressed genes (DEGs) significantly involved in flower bud differentiation were analysed by comparative transcriptomics. The transcription factors involved in the regulation of critical genes in flower bud differentiation were analysed by using weighted gene coexpression network analysis (WGCNA). To explore the regulation mechanism of grape flower bud differentiation under red‒blue light. This study provides preliminarily clarification of the regulatory mechanism for grape flower bud differentiation under red‒blue light and lays the foundation for further research.
Results
Flower bud differentiation of ‘Red Globe’ grape under red–blue light
Before the transcriptome study, we conducted a morphological analysis of the floral bud development process (Fig. 1). In the first stage (April 30), the budding body was a flat green, the growth point was conical, the bud scale was not lignified, the stage one control (T1) branch diameter was less than the diameter of the stage one LED red‒blue treatment (S1) branches, and the two groups of flower buds were at the anlagen or uncommitted primordium stage (Supplementary Fig. S1). In the second stage (June 30), the buds expanded, became green, and increased in size. The stage two LED red‒blue treatment (S2) branches began to turn from green to brown. Some buds on the stage two control (T2) and S2 branches were at the stage of anlagen differentiation for forming inflorescence primordia. In the third stage (July 30), the buds became plump and brown. Seventy percent of the flower buds on the stage three control (T3) branches and 90% on the stage three LED red‒blue treatment (S3) branches were at the stage of anlagen differentiation for forming inflorescence primordia. The stage four (September 15) LED red‒blue treatment (S4) branches had hard buds that were dark brown, with semilignified scales, and both the stage four control (T4) and the S4 buds were all in the stage of anlagen differentiation for forming inflorescence primordia.
Changes in hormone content during flower bud development
Flower bud differentiation is closely related to hormones. Therefore, the levels of indole-3-acetic acid (IAA), cytokinins (CKs), gibberellic acid (giberellins, GAs), and abscisic acid (ABA) in flower buds of the red‒blue light and control plants at the four stages were determined with HPLC. The HPLC analysis showed that the levels of these hormones changed significantly in different flower bud differentiation stages. The IAA content in the control was the highest in the second stage (Fig. 2A) and then decreased with development. Interestingly, the IAA content in the red‒blue light treatment was always higher than that in the control, reaching its highest level in the third stage and the lowest in the fourth stage. As shown in Fig. 2B, the trend of CK content in the two treatments was completely different. The CK content in the flower buds increased gradually under the control while under the red‒blue light treatment it was highest in the second stage and lowest in the fourth stage. GA content fluctuated throughout the flower bud differentiation stages. The levels in the control were higher than those in the red‒blue light treatment in the first and third stages of flower bud differentiation (Fig. 2C). The ABA content in flower bud differentiation was consistently lower in the control plants than in the red‒blue light treatment plants. In the third stage, the ABA content of the control plants was the lowest, and that of the red‒blue light treatment plants was the highest, with levels that were 24.6% higher than those of the control plants (Fig. 2D).
Analysis of differentially expressed genes
We constructed a transcriptome sequencing library for the four stages of flower buds of the treatment and control groups, and 183.27 Gb clean reads were obtained. The clean reads of each sample were above 6.19 Gb, and the Q30 base percentage was above 93.28% (Supplementary Table S2). A principal component analysis (PCA) of the samples showed that the samples collected in this study had a high shared identity (Supplementary Figure S2). The DEGs were identified by using DESeq2 software, and the DEGs were found to have different expression trends in the red‒blue light treatment and the control plants (Fig. 3A–C). Compared with T1 grape buds, in T2, T3, and T4 buds 2314 common DEGs and 611, 1367, and 3072 specific expression genes were identified. There were 2262, 3328, and 3342 upregulated genes and 1896, 3290, and 4310 downregulated genes in T1 compared to T2 buds, T1 compared to T3 buds, and T1 compared to T4 buds, respectively (Fig. 3D,G). Compared with S1 buds, S2, S3, and S4 buds had 1889 common DEGs and 591, 1550, and 1613 unique expression genes. In S1 compared to S2 buds, S1 compared to S3 buds, and S1 compared to S4 buds, there were 1691, 2546, and 2116 upregulated genes and 1736, 3131, and 3161 downregulated genes, respectively (Fig. 3E,H). An analysis of T1 compared to S1 buds, T2 compared to S2 buds, T3 compared to S3 buds, and T4 compared to S4 buds showed that 1214, 1292, 1744, and 1912 genes were upregulated, and that 819, 1263, 1983, and 674 genes were downregulated at the four different maturation stages (Fig. 3F,I). In the third stage, the up- and downregulation of these genes increased significantly, indicating that these genes play a crucial regulatory role in the process of grape flower bud differentiation.
Expression analysis of DEGs in different developmental stages. (A) Clustering of DEGs. (B–C) Expression trends of DEGs in red-blue combination light treatment and control. (D) DEGs with red-blue combination light treatment development. (E) DEGs with control development. (F) DEGs between flower bud in different development stages. (G–I) Number of up- and down-DEGs.
Annotation of DEGs
We performed GO functional annotation and KEGG pathway analysis of the DEGs24. Through GO functional annotation, 6129 genes were annotated as 50 functional branches of cell components, biological processes, and molecular functions. Regarding all of the up- and downregulated DEGs, the first ten annotated GO terms were for binding (GO: 0,005,488), cellular component (GO: 0,044,464), catalytic activity (GO: 0,003,824), cellular process (GO: 0,009,987), metabolic process (GO: 0,008,152), membrane component (GO: 0,044,425), membrane (GO: 0,016,020), organelle (GO: 0,043,226), biological regulation (GO: 0,065,007) and response to stimulus (GO: 0,050,896). Among all the upregulated DEGs, 1995, 1902, 1853, 1673, 1668, 1394, 1173, 1102, 773, and 441 were annotated as these 10 GO terms. In the downregulated DEGs, the numbers were 1783, 1684, 1521, 1445, 1412, 1374, 1204, 953, 747 and 418, respectively (Supplementary Fig. S3, Supplementary Table S3).
KEGG pathway analysis showed that the plant hormone signal transduction pathway (map04075) played an essential role in the grape flower bud differentiation. The plant hormone signal transduction pathway was enriched in the downregulated DEGs for the four stages under the red‒blue light treatment and the control. In addition, other enrichment pathways included those for flavonoid biosynthesis (map00941), flavonoid and flavanol biosynthesis (map00944), circadian rhythm-plant (map04712), and glycine, serine, and threonine metabolism (map00260). The plant hormone signal transduction pathway was also significantly enriched in the upregulated genes for the four stages under both the red‒blue light treatment and the control. In addition, the most enriched pathways were those for plant‒pathogen interactions (map04626), and other enrichment pathways included those for MAPK signalling pathway-plant (map04016), glycerophospholipid metabolism (map00564), phenylpropanoid biosynthesis (map00940), and monoterpenoid biosynthesis (map00902) (Fig. 4, Supplementary Table S4). The results showed that grape flower bud differentiation is a very complex biological process. In addition to the pathway for plant hormone signal transduction, the MAPK signal transduction, phenylpropanoid biosynthesis, flavonoid biosynthesis, and circadian rhythm-plant pathways are also involved in this process.
DEGs related to plant hormone signal transduction
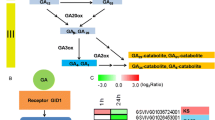
Previous studies have shown that grape flower bud differentiation is closely related to plant hormone signalling. In this study, we found that DEGs in plant hormone signal transduction are mainly concentrated in auxin, cytokinin, gibberellin, and abscisic acid pathways (Fig. 5). The 36 DEGs involved in these pathways include those involved in the auxin pathway, which are TIR1 (VIT_14s0030g0124), AUX/IAA (VIT_05s0020g01070, VIT_09s0002g05160, VIT_05s0020g04690, VIT_05s0049g01970), ARF (VIT_11s0016g00640), GH3 (VIT_07s0104g00800, VIT_03s0091g0031) and SAUR (VIT_19s0085g00010, VIT_09s0002g00670, VIT_15s0048g00530, VIT_01s0146g00180, VIT_03s0038g01150); cytokinin pathway CRE1 (VIT_12s0057g00690), B-ARR (VIT_17s0000g10100), A-ARR (VIT_08s0007g05390, VIT_17s0000g07580, VIT_01s0026g00940, VIT_13s0067g03510, VIT_18s0001g02540); those involved in the gibberellin pathway, which are GID2 (VIT_07s0129g01000), TF (VIT_07s0005g02510, VIT_14s0060g00260); and those involved in the abscisic acid pathway, which are PYR/PYL (VIT_15s0046g01050), PP2C (VIT_16s0022g02210, VIT_06s0004g05460, VIT_16s0050g02680), SnRK2 (VIT_07s0197g00080, VIT_12s0035g00310) and ABF (VIT_18s0072g00470, VIT_03s0063g00310, VIT_18s0001g10450, VIT_04s0069g01150, VIT_12s0034g00110). Further analysis showed that the expression of the TIR1, AUX/IAA, and ARF genes was relatively high in the early stage of flower bud differentiation, while the expression was downregulated in the later stage of differentiation. The expression of GH3 gradually increased during flower bud differentiation. In addition, the expression trend of the SAUR gene was completely different between the red‒blue light and the control plants. In the control plants, the expression of the A-ARR gene was high at the early stage of flower bud differentiation and then decreased, while the expression trend was opposite in the red‒blue light treatment plants. The expression trend of the TF gene was completely different in the control and red‒blue light treatment plants. In the control plants, TF gene expression levels were high at the early stage of flower bud differentiation and were low at the late stage, while in the red‒blue light treatment plants expression levels remained low throughout the flower bud differentiation stages. In the control plants, the expression of the PP2C gene increased over the course of flower bud differentiation, while in the red‒blue light treatment plants, the expression decreased. The expression trends of the ABF gene in the two groups were also different. The control plants’ expression of GBF4 (VIT_18s0072g00470) and AI5L2 (VIT_04s0069g01150) remained at a low level, while the expression in the red‒blue light treatment plants increased over the course of flower bud differentiation, reaching the highest expression levels in the third stage. Expression levels for AI5L7 (VIT_03s0063g00310) and AI5L5 (VIT_18s0001g10450) showed an upwards trend in the control plants but gradually decreased in the red‒blue light treatment plants. The results showed that these signalling genes played an important role in grape flower bud differentiation.
qRT‒PCR validates the gene expression profiles
To validate our sequencing results, DEGs involved in plant hormone signal transduction, flavonoid biosynthesis and MAPK signal transduction pathways were randomly selected for qPCR analysis. The analysed DEGs included ARR4 (VIT_01 s0026g00940), PIF3 (VIT_14s0060g00260), GBF4 (VIT_18s0072g00470), HST (VIT_09s0018g01190), C75A1 (VIT_06s0009g02840), CAMT (VIT_03s0063g00140), CML46 (VIT_14s0108g01000), YODA (VIT_02s0025g03850), and M2K5 (VIT_09s0018g01820). The qRT‒PCR results showed that the expression patterns of these genes were consistent with those in the RNA-seq data, with a correlation coefficient of R = 0.82. These results showed that the gene expression pattern revealed by RNA-seq data was reliable and could be used for further analysis (Fig. 6).
Screening through WGCNA for the transcription factors regulating grape flower bud differentiation
In this study, a total of 24 samples from four stages of grape flower bud differentiation from the red‒blue light treatment and control groups were used for WGCNA (Supplementary Fig. S4). Through an assessment of the dynamic gene changes in different developmental stages and a correlation analysis between samples, the possible transcription factors regulating flower bud differentiation were investigated. First, the expression patterns of 25,844 DEGs extracted from transcriptome sequencing were analysed by using WGCNA, and the DEGs were divided into 19 modules (Fig. 7A, Supplementary Table S5) according to the similarity of their expression patterns. These modules could be divided into two main branches (one with two of the modules and the other 17 of the modules) (Fig. 7B). The correlation between the expression patterns of each module and different stages of grape flower bud differentiation was analysed. The results showed that the modules ‘red’, ‘green’ and ‘magenta’ were highly correlated with the third stage of the fastest flower bud differentiation under the red‒blue light treatment, indicating that these modules were closely related to the flower bud differentiation of grapes (Fig. 7C). A total of 2373 genes were found in these modules, and the gene association networks of 2373 genes were constructed. According to the degree of connection, the first 50 genes were considered hub genes. Among these 50 hub genes, seven transcription factors were found, including the ERF family EF110 (VIT_18s0072g00260), ABR1 (VIT_07s0031g01980), the bHLH family BH025 (VIT_00s0824g00020), BH025 (VIT_00s0927g00010), the WRKY family WRK48 (VIT_05s0077g00730), and CAMTA3 (VIT_07s0141g00250), the NF-X1 family NFXL2 (VIT_13s0067g00920) and the bZIP family GBF4 (VIT_18s0072g00470) (Supplementary Fig. S5, Supplementary Table S6). As highly connected hub genes, these transcription factors may regulate flower bud differentiation in grape plants.
bZIP family genes are related to ABA signalling. To further analyse the transcription factors regulated by the GBF4 gene in the GA pathway, the genes related to GBF4 in the ‘green’ module with edge weight (≥ 0.4) were selected for analysis (Fig. 8, Supplementary Table S7). It was found that 42 genes were regulated by GBF4 (VIT_18s0072g00470), including the following hub transcription factors: the WRKY family WRK48 (VIT_05s0077g00730), the ERF family EF110 (VIT_18s0072g00260), ABR1 (VIT_07s0031g01980), the CAMTA family CAMTA3 (VIT_07s0141g00250), and the HSF family HSFA3 (VIT_08s0007g0390), together with other non-hub transcription factor genes that have a certain regulatory relationship with the GBF4 gene.
Disscussion
Grape yield depends on the amount of flower bud differentiation. Plant hormones are the key factors controlling the induction of flowering and interact with each other during plant developmen25,26. The types and levels of hormones in grape bud varied considerably at different developmental stages. In this study, the ‘Red Globe’ cultivar was used as the experimental material, and the supplemental treatment of red‒blue light was applied, with control conditions of no supplemental light. The four main plant hormones were identified in flower buds of both groups during four stages of flower bud differentiation. IAA was the first plant hormone discovered by researchers, and it is necessary for regular plant growth and development27. The IAA content in flower buds under red‒blue light treatment was higher than that in the control, and it showed a trend of increasing first and then decreasing over the course of flower bud differentiation, reaching its highest level at the third stage. Higher IAA content guarantees good flower bud formation, and this is consistent with the findings of previous studies28. CKs can promote the inflorescence development of lateral meristem in grape plants29. In our study, the content of CKs in flower buds increased during flower bud differentiation under the control, while the CK content decreased over the course of flower bud differentiation in the red‒blue light treatment plants. GAs are related to the initiation of flowering stage transitions and floral primordi30,31, which is a hot topic in current research32. Unlike CKs, GAs promote the differentiation of lateral meristem and inhibit the development of the inflorescence, which is beneficial to the development of the grape tendril. Grape plants with a mutation in GA-related genes are short in stature, with grape tendrils differentiated into inflorescences33. Interestingly, similar results were also observed in this study. The third stage of flower bud differentiation progressed the fastest, although this was less marked in the red‒blue light treatment plants than in the control. In addition, there was a significant difference in ABA content between normal and malformed flowers34. In this study, the ABA content in the red‒blue light treatment plants was higher than that in the control plants at the early stage of flower bud differentiation and was even 24.6% higher at the third stage, showing that ABA plays a vital role in flower bud differentiation. However, the regulation mechanism of hormone balance is complex. Like other perennial woody plants, grape flower bud differentiation requires a balance between hormones35.
In this study, 36 DEGs were annotated and identified as being involved in the auxin, cytokinin, gibberellin, and abscisic acid signal transduction pathways. In the auxin signal transduction pathway, the expression levels of TIR1, AUX/IAA, and ARF genes were relatively high at the early stage of flower bud differentiation, while they were downregulated at the late stage of differentiation, which was consistent with the findings of previous studies36. Studies have shown that ARF, combined with auxin response element AUX/IAA, plays a central role in transforming local auxin concentration into specific gene expression output, initiating flowering and regulating the development of floral organs37,38. In this study, four AUX/IAA genes and one ARF gene regulated the auxin pathway. The expression levels of the IAA17 and ARFS genes in the red‒blue light plants was substantially inconsistent with expression levels in the control plants, indicating that these genes played an essential role in responding to flower bud differentiation under red‒blue light. In the cytokinin signal transduction pathway, the ARR gene participates in the circadian rhythm mechanism under light conditions, activates the function of flowering-induced gene SOC1 to mediate the flowering of Arabidopsis39, and is highly expressed in branches with a high flowering rate in apple trees40. The expression levels of the six ARR genes identified in this study first increased and then decreased during flower bud differentiation and showed significant indigenous differences between the treatment and control groups, confirming the positive effect of ARR family genes on flower bud differentiation. GID and TF are genes that positively regulate the expression of LFY, SOC1, FT, SPL, and other genes that promote flower bud differentiation, which DELLA inhibits. The essential genes involved in flower bud differentiation in these gibberellin signal transduction pathways are also identified in this study. Unlike the results in previous studies, in this study the red–blue light treatment associated with a high flowering rate exhibited a lower expression of gibberellin than the control group, which may be due to the downregulation of gibberellin response genes without the effect of the DELLA gene41. The three signal components of PYR/PYL, PP2C, and SnRK2 constitute the core ABA signalling pathway42, and positively regulate the flowering of long-day plants43. In addition, five ABF genes were also found to be involved in grape flower bud differentiation, indicating that ABF can be phosphorylated by SnRK2, affecting flower bud differentiation44.
WGCNA has been used to analyse data from numerous species to elucidate the interaction between physiological characteristics and gene expression45,46. In this study, WGCNA was used to evaluate transcript expression and trait changes at different stages of germination and differentiation under red–blue light, and three modules, ‘red’, ‘green’, and ‘magenta’ that were highly related to flower bud differentiation were screened. In these modules, eight transcription factors were identified as hub genes, including genes from the ERF family, bHLH family, WRKY family, CAMTA family, NF-X1 family, and bZIP family, suggesting that these transcription factors may be involved in grape flower bud differentiation under red‒blue light. Previous transcriptome sequencing studies found that transcription factors such as MYB, MADS-box, WRKY, ERF, and bHLH played an essential role in flower bud differentiation47. The MYB gene can regulate floral organ development and even mediate gibberellin signal transduction in flowering responses48, while the MADS-box gene is involved in the regulation of floral meristem initiation and development49. In addition, the bZIP and ERF genes are related to floral bud differentiation hormone signal response50. The bZIP family gene GBF4 is a unique gene in the ABA signal transduction pathway. Thus far, there are few reports on this gene’s regulation of flower bud differentiation. We found that WRKY, ERF, CAMTA, and HSF family genes were involved in the transcriptional regulation of GBF4 through WGCNA. WRKY family members were the key factors in the ABA response pathway, which could accelerate flowering by regulating AP151, and HSF was involved in environmental stress response to regulate the flowering and development of plants52. The above transcription factors involved in plant hormone signal transduction regulation lacked clear evidence, but our study provided a theoretical basis for further confirming the regulatory mechanism of grape flower bud differentiation under red‒blue light.
Conclusion
Overall, the comparative transcriptome analysis of the differential genes involved in the plant hormone signal transduction pathway showed significant differences in AUX/IAA, SAUR, A-RR, and ABF between red‒blue light treatment and control buds; this was especially true for the ABF genes, which were complete different in the two groups. The GBF4 (VIT_18s0072g00470) and AI5L2 (VIT_04s0069g01150) expression in the control remained low, while the expression under the red‒blue light treatment increased with the process of flower bud differentiation. AI5L7 (VIT_03s0063g00310) and AI5L5 (VIT_18s0001g10450) showed an upwards trend in the control and gradually decreased under red‒blue light. It is speculated that the difference in ABF family gene regulation plays a vital role in the grape flower bud differentiation response to red‒blue light. The WGCNA showed that the hub transcription factors WRK48 (VIT_05s0077g00730), ERF family EF110 (VIT_18s0072g00260), ABR1 (VIT_07s0031g01980), CAMTA family CAMTA3 (VIT_07s0141g00250), and HSF family HSFA3 (VIT_08s0007g0390) may be involved in the regulation of the GBF4 gene.
Materials and methods
Plant materials
The test material was 10-year-old Vitis vinifera cv. ‘Red Globe’ grape plants. The experiment was carried out in the growing season of 2021 in the sunlight greenhouse of Helan County Gardening Industrial Park, Yinchuan City. Our previous studies have shown that a ratio of red light:blue light = 4:1 (S) can significantly increase the flowering rate of ‘Red Globe’ grape plants (unpublished). Therefore, in this study, red light:blue light = 4:1 (S) LED plant lights were selected for the treatment group plants (the wavelengths of red and blue light were 620 and 435 nm) , and conditions of no supplementary light served as a control (T). Light tubes were placed at 30 cm above the test line plants, and the leaf light intensity was maintained at 300 μmol m−2 s−1 for 14 h per day from 8:00 to 22:00. In the first stage (T1 and S1, 30 April), the second stage (T2 and S2, 15 June), the third stage (T3 and S3, 30 July), and the fourth stage (T2 and S2, 15 September) of flower bud differentiation, 26 healthy branches were selected and removed with a sharp scalpel. Twenty buds were fixed in FAA (formaldehyde-acetic acid–ethanol) for paraffin sectioning, and the other buds were immediately frozen in liquid nitrogen and stored at -80℃ until further analysis.
Physiological analysis of flower buds at different stages
Flower buds of four developmental stages were used for paraffin section making and hormone determination. Flower buds fixed in FAA were dehydrated in a gradient ethanol series and then were made transparent, wax-immersed, embedded, and patched in a dehydrator. Paraffin blocks containing flower buds were sliced with a semiautomatic slicer (Leica RM2245), stained with saffron-solid green, and sealed with neutral gum. Observation under an optical microscope (Olympus BX53) was performed using an SC180 image analysis system to obtain images53, and the flower bud differentiation stage was classified54. Auxins (IAAs), cytokinins (CKs), gibberellins (GAs), and abscisic acid (ABA) were quantified by using ultra-performance liquid chromatography spectrometry (ACQUITY H-Class, QDA), the mobile phase was methanol–water (containing 0.2% glacial acetic acid) with a volume ratio of 45:55. The flow rate was 0.25 mL min−1, the column temperature was 30 °C, and the injection volume was 10 μL55, with three biological replicates.
RNA sequencing and expression analysis
The RNA of the samples was extracted using the OminiPlant RNA Kit (DNase I) (Beijing ComWin Biotech Co., Ltd. Beijing, China). Nanodrop2000 was used to detect the concentration and purity of RNA, agarose gel electrophoresis was used to detect RNA integrity, and Agilent2100 was used to determine the RNA integrity number (RIN) value for further quantitative real-time reverse transcription-PCR (qRT‒PCR) and RNA-seq determination. The Illumina HiSeq 2500 sequencing platform was used to perform RNA-seq. Raw reads were sequenced and then were processed to obtain clean reads of high quality. The clean reads after quality control were aligned with the reference genome (http://plants.ensembl.org/Vitis_vinifera/Info/Index) using HISAT2 software (Version 2.1.0)56 to obtain the mapped reads for subsequent analysis.
GO and KEGG pathway enrichment analysis
Differential expression analysis of samples was carried out using the expression difference analysis software DESeq2 (Version 1.10.1)57. Differentially expressed genes between different samples were screened out using FPKM (Fragments Per Kilobases per Million reads) values and |log2fold changes|≥ 1, and the corrected Padjust < 0.05 as the screening standard to study the function of differential genes. We used the fastcluster (Version 1.2.4) package in R was used for cluster analysis of differentially expressed genes, hierarchical cluster analysis, and complete Gene clustering method. The DEGs in gene concentration were compared with data in the Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) databases to obtain gene functional annotation and related metabolic pathway information in different samples58.
RNA-seq data confirmation through qRT-PCR
In this study, qRT‒PCR was used to validate the RNA-Seq results. Real-time fluorescence quantitative PCR was used to verify candidate DEGs related to hormone signal transduction and other metabolic pathways. The reference gene was Actin1 (GenBank: XP_008654957.1). The primers (Supplementary Table S1) were designed by using Primer 6.0 software and were synthesized by Sangon (Shanghai, China). RNA and transcriptome sequencing samples for the same batch were conducted with qRT‒PCR using ChamQ Universal SYBR qPCR Master Mix kit (Vazyme Biotech Co., Ltd. Nanjing, China). The reaction procedure was as follows: reverse transcription at 50 °C for 5 min, predenaturation at 95 °C for 10 min, denaturation at 95 °C for 10 s, and annealing at 60 °C for 30 s, 40 cycles, and three biological repeats. The gene expression level was calculated by the 2-ΔΔCt method.
Gene network construction and visualization
To further study the hormone regulation mechanism and possible transcription factors in grape flower buds, we used the WGCNA (Version 1.63) package in R to construct the coexpression network59. The soft threshold of coexpression network clustering was selected according to the FPKM value of all genes in the sample; R2 > 0.8 was the norm, and the soft threshold was 9. All FPKM values were transformed into a topological overlap matrix (TOM), and each gene was clustered by hierarchical clustering60. The dynamic tree cutting method was used to divide genes into different coexpression modules. The minimum number of genes in each coexpression module was set at 30, and the coexpression modules with similar clustering were merged with 0.3 as the boundary. The correlation between different modules and the degree of genes in the module (module membership, MM) were calculated. Key modules related to flower bud differentiation were identified for subsequent analysis. Cytoscape (Version 3.8.0) was used to visualize the regulatory network mapping.
Ethical standards
The conducted experiments comply with the laws of China.
Data availability
Data supporting the results and conclusions are included in both the article and additional files. All the transcriptome data have been deposited in the NCBI Sequence Read Archive (SRA) under accession number: PRJNA843849 (http://www.ncbi.nlm.nih.gov/sra).
References
Zhang, P. et al. Transcriptomic and metabolomic profiling reveals the effect of LED light quality on fruit ripening and anthocyanin accumulation in cabernet sauvignon grape. Front. Nutr. 8, 790697 (2021).
Sawicki, M. et al. Distinct regulation in inflorescence carbohydrate metabolism according to grapevine cultivars during floral development. Physiol. Plant. 154, 447–467 (2015).
Gourieroux, A. M., Holzapfel, B. P., Mccully, M. E., Scollary, G. R. & Rogiers, S. Y. Vascular development of the grapevine (Vitis vinifera L.) inflorescence rachis in response to flower number, plant growth regulators and defoliation. J. Plant Res. 130, 873–883 (2017).
Guo, R. et al. Evolutionary, interaction and expression analysis of floral meristem identity genes in inflorescence induction of the second crop in two-crop-a-year grape culture system. J. Genet. 97, 439–451 (2018).
Crane, O. et al. Cytokinin-induced VvTFL1A expression may be involved in the control of grapevine fruitfulness. Planta 235, 181–192 (2012).
Blumel, M., Dally, N. & Jung, C. Flowering time regulation in crops-what did we learn from Arabidopsis?. Curr. Opin. Biotechnol. 32, 121–129 (2015).
Hu, J. et al. Transcriptome profiling of the flowering transition in saffron (Crocus sativus L.). Sci. Rep. 10, 9680 (2020).
Calonje, M., Cubas, P., Martinez-Zapater, J. M. & Carmona, M. J. Floral meristem identity genes are expressed during tendril development in grapevine. Plant Physiol. 135, 1491–1501 (2004).
Boss, P. K., Sreekantan, L. & Thomas, M. R. A grapevine TFL1 homologue can delay flowering and alter floral development when overexpressed in heterologous species. Funct. Plant Biol. 33, 31–41 (2006).
Vasconcelos, M. C., Greven, M., Winefield, C. S., Trought, M. C. T. & Raw, V. The flowering process of Vitis vinifera: A review. Am. J. Enol. Vitic. 60, 411–434 (2009).
Joly, D. et al. Expression analysis of flowering genes from seedling-stage to vineyard life of grapevine cv. Riesling. Plant Sci. 166, 1427–1436 (2004).
Carmona, M. J., Cubas, P., Calonje, M. & Martínez-Zapater, J. M. Flowering transition in grapevine (Vitis vinifera L.)This review is one of a selection of papers presented at the symposium onVitisat the XVII international botanical congress held in Vienna, Austria, 2005. Can. J. Bot. 85, 701–711 (2007).
Song, J. et al. Comparative RNA-Seq analysis on the regulation of cucumber sex differentiation under different ratios of blue and red light. Bot. Stud. 59, 21 (2018).
Modarelli, G. C. et al. The role of light quality of photoperiodic lighting on photosynthesis, flowering and metabolic profiling in Ranunculus asiaticus L.. Physiol Plant. 170, 187–201 (2020).
Hiltbrunner, A. Shedding light on the evolution of light signalling. New Phytol. 224, 1412–1414 (2019).
Wang, Q. et al. Photoactivation and inactivation of Arabidopsis cryptochrome 2. Science 354, 343–347 (2016).
Lu, X. D. et al. Red-light-dependent interaction of phyB with SPA1 promotes COP1-SPA1 dissociation and photomorphogenic development in Arabidopsis. Mol. Plant. 8, 467–478 (2015).
Hasan, M. M., Bashir, T., Ghosh, R., Lee, S. K. & Bae, H. An overview of LEDs’ effects on the production of bioactive compounds and crop quality. Molecules 22, 1420 (2017).
Park, Y. & Runkle, E. S. Blue radiation attenuates the effects of the red to far-red ratio on extension growth but not on flowering. Environ. Exp. Bot. 168, 103871 (2019).
Jeong, W. H., Chun, W. L. & Kee, Y. P. Influence of mixed LED radiation on the growth of AnnualPlants. J. Plant Biol. 49, 286–290 (2006).
Jeong, S. W., Hogewoning, S. W. & Van Ieperen, W. Responses of supplemental blue light on flowering and stem extension growth of cut chrysanthemum. Sci. Hortic. 165, 69–74 (2014).
Carmona, M. J., Calonje, M. & Martinez-Zapater, J. M. The FT/TFL1 gene family in grapevine. Plant Mol Biol 63, 637–650 (2007).
Lebon, G. et al. Sugars and flowering in the grapevine (Vitis vinifera L.). J. Exp. Bot. 59, 2565–2578 (2008).
Kanehisa, M. Toward understanding the origin and evolution of cellular organisms. Protein Sci. 28, 1947–1951 (2019).
Magar, Y.G., Ohyama, K., Noguchi, A., Amaki, W. & Furufuji, S. Effects of light quality during supplemental lighting on the flowering in an everbearing strawberry. in Xiii International Symposium on Plant Bioregulators in Fruit Production. Vol. 1206, pp. 279–284 (2018).
De Wit, M., Galvao, V. C. & Fankhauser, C. Light-mediated hormonal regulation of plant growth and development. Ann. Rev. Plant Biol. 67, 513–537 (2016).
Alabadi, D., Blazquez, M. A., Carbonell, J., Ferrandiz, C. & Perez-Amador, M. A. Instructive roles for hormones in plant development. Int. J. Dev. Biol. 53, 1597–1608 (2009).
Wang, H. et al. The influence of changes of endogenous hormones in shoot on the grapes flower bud differentiation in greenhouse. Sci. Agric. Sin. 47, 4695–4705 (2014).
Zhang, S. et al. The VAP1 gene expression in relation to GAs effect on tendrils, buds and flowers development in “Xiangfei” grapevine. Sci. Hortic. 117, 225–230 (2008).
Jing, D. et al. An integrative analysis of transcriptome, proteome and hormones reveals key differentially expressed genes and metabolic pathways involved in flower development in loquat. Int. J. Mol. Sci. 21, 5107 (2020).
Penso, G. A. et al. Development of peach flower buds under low winter chilling conditions. Agron.-Basel 10, 428–447 (2020).
Fan, S. et al. Mediation of flower induction by gibberellin and its inhibitor paclobutrazol: mRNA and miRNA integration comprises complex regulatory cross-talk in apple. Plant Cell Physiol. 59, 2288–2307 (2018).
Diaz-Riquelme, J., Martinez-Zapater, J. M. & Carmona, M. J. Transcriptional analysis of tendril and inflorescence development in grapevine (Vitis vinifera L.). PLoS ONE 9, e92339 (2014).
Liu, K. et al. Comparative transcriptomic analyses of normal and malformed flowers in sugar apple (Annona squamosa L.) to identify the differential expressed genes between normal and malformed flowers. BMC Plant Biol. 17, 170 (2017).
Li-Mallet, A., Rabot, A. & Geny, L. Factors controlling inflorescence primordia formation of grapevine: Their role in latent bud fruitfulness? A review. Botany 94, 147–163 (2016).
Villar, L. et al. Comparative transcriptomic analysis reveals novel roles of transcription factors and hormones during the flowering induction and floral bud differentiation in sweet cherry trees (Prunus avium L. cv. Bing). PLoS ONE 15, e0230110 (2020).
Liu, Z. et al. ARF2-ARF4 and ARF5 are essential for female and male gametophyte development in arabidopsis. Plant Cell Physiol. 59, 179–189 (2018).
Richter, R., Behringer, C., Zourelidou, M. & Schwechheimer, C. Convergence of auxin and gibberellin signaling on the regulation of the GATA transcription factors GNC and GNL in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 110, 13192–13197 (2013).
D’aloia, M. et al. Cytokinin promotes flowering of Arabidopsis via transcriptional activation of the FT paralogue TSF. Plant J. 65, 972–979 (2011).
Okada, K. et al. Columnar growth phenotype in apple results from gibberellin deficiency by ectopic expression of a dioxygenase gene. Tree Physiol. 40, 1205–1216 (2020).
Fleet, C. M. & Sun, T. P. A DELLAcate balance: The role of gibberellin in plant morphogenesis. Curr. Opin. Plant Biol. 8, 77–85 (2005).
Cutler, S. R., Rodriguez, P. L., Finkelstein, R. R. & Abrams, S. R. Abscisic acid: Emergence of a core signaling network. Annu. Rev. Plant Biol. 61, 651–679 (2010).
Riboni, M., Robustelli Test, A., Galbiati, M., Tonelli, C. & Conti, L. ABA-dependent control of GIGANTEA signalling enables drought escape via up-regulation of FLOWERING LOCUS T in Arabidopsis thaliana. J. Exp. Bot. 67, 6309–6322 (2016).
Qin, L. et al. Effect of exogenous spermidine on floral induction, endogenous polyamine and hormone production, and expression of related genes in ‘Fuji’ apple (Malus domestica Borkh.). Sci. Rep. 9, 12777 (2019).
Han, Z. et al. Identification of the gene network modules highly associated with the synthesis of phenolics compounds in barley by transcriptome and metabolome analysis. Food Chem. 323, 126862 (2020).
Mo, S. et al. Spatio transcriptome uncover novel insight into the Lycium ruthenicum seedling tolerant to salt stress. Ind. Crops Prod. 177, 114502 (2022).
Xia, Y. et al. Comparative transcriptome analysis of flower bud transition and functional characterization of EjAGL17 involved in regulating floral initiation in loquat. PLoS ONE 15, e0239382 (2020).
He, W. et al. Transcriptome analysis of Litsea cubeba floral buds reveals the role of hormones and transcription factors in the differentiation process. G3 (Bethesda) 8, 1103–1114 (2018).
Parenicova, L. et al. Molecular and phylogenetic analyses of the complete MADS-box transcription factor family in Arabidopsis: New openings to the MADS world. Plant Cell 15, 1538–1551 (2003).
Chen, X. et al. Comparative RNA-sequencing-based transcriptome profiling of buds from profusely flowering ‘Qinguan’ and weakly flowering ‘Nagafu no. 2’ apple varieties reveals novel insights into the regulatory mechanisms underlying floral induction. BMC Plant Biol. 18, 370 (2018).
Rushton, D. L. et al. WRKY transcription factors: Key components in abscisic acid signalling. Plant Biotechnol. J. 10, 2–11 (2012).
Liang, Y. et al. Genome-wide comparative analysis of heat shock transcription factors provides novel insights for evolutionary history and expression characterization in cotton diploid and tetraploid genomes. Front. Genet. 12, 658847 (2021).
Wang, H. B. et al. Studies on the flower bud differentiation of grape cultivars with different tolerant ability of low light in greenhouse. Acta Hortic. Sin. 43, 633–642 (2016).
Srinivasan, C. & Mullins, M. G. Physiology of flowering in the grapevine: A review. Am. J. Enol. Vitic. 32, 47–63 (1981).
Zhang, Z. et al. Transcriptome profiles reveal the crucial roles of hormone and sugar in the bud dormancy of prunus mume. Sci. Rep. 8, 5090 (2018).
Kim, D., Paggi, J. M., Park, C., Bennett, C. & Salzberg, S. L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 37, 907–915 (2019).
Maza, E. In papyro comparison of TMM (edgeR), RLE (DESeq2), and MRN normalization methods for a simple two-conditions-without-replicates RNA-Seq experimental design. Front. Genet. 7, 164 (2016).
Kanehisa, M. & Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30 (2000).
Langfelder, P. & Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 9, 559 (2008).
Zhan, J. et al. RNA sequencing of laser-capture microdissected compartments of the maize kernel identifies regulatory modules associated with endosperm cell differentiation. Plant Cell 27, 513–531 (2015).
Acknowledgements
The work was supported by National Natural Science Foundation of China (31360493), Key R & D Projects in Ningxia (2021BEF02016) and Graduate Innovation Project of Ningxia University (GIP2020-09). The collection of ‘Red Globe’ grapes in this work has been permited by Helan County Gardening Industrial Park, Yinchuan City.
Author information
Authors and Affiliations
Contributions
Y.Z., X.L. and M.Y. conceived and designed the experiments. Y.Z., X.L., M.Y., J.Z. and S.D. performed the research and analyzed the data. Y.Z. and X.L. wrote the paper. All authors inter-preted the data and contributed to drafting the manuscript. All authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Liu, X., Yuan, M., Dang, S. et al. Comparative transcriptomic analysis of transcription factors and hormones during flower bud differentiation in ‘Red Globe’ grape under red‒blue light. Sci Rep 13, 8932 (2023). https://doi.org/10.1038/s41598-023-29402-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-29402-5
- Springer Nature Limited