Abstract
The patterns of population divergence of mid-latitude marine birds are impacted by only a few biogeographic barriers to dispersal and the effect of intrinsic factors, such as fidelity to natal colonies or wintering grounds, may become more conspicuous. Here we describe, for the first time, the phylogeographic patterns and historical demography of Bulwer’s petrel Bulweria bulwerii and provide new insights regarding the drivers of species diversification in the marine environment. We sampled Bulwer’s petrels from the main breeding colonies and used a statistical phylogeography approach based on surveying nuclear and mitochondrial loci (~ 9100 bp) to study its mechanisms of global diversification. We uncovered three highly differentiated groups including the Western Pacific, the Central Pacific and the Atlantic. The older divergence occurred within the Pacific Ocean, ca. 850,000 ya, and since then the W Pacific group has been evolving in isolation. Conversely, divergence between the Central Pacific and Atlantic populations occurred within the last 200,000 years. While the Isthmus of Panama is important in restricting gene flow between oceans in Bulwer’s petrels, the deepest phylogeographic break is within the Pacific Ocean, where oceanographic barriers are key in driving and maintaining the remarkable structure found in this highly mobile seabird. This is in contrast with the Atlantic, where no structure was detected. Further data will provide insights regarding the extent of lineage divergence of Bulwer’s petrels in the Western Pacific.
Similar content being viewed by others
Introduction
Reconstructing the diversification history of species and identifying the main drivers of population differentiation in widely distributed species is key to understand population connectivity and how biodiversity is generated and maintained, particularly in the face of a rapidly changing world. Empirical statistical phylogeographic studies have been unveiling the primary drivers of global diversification of marine species, particularly those of high latitudes1,2,3,4,5,6,7. Mechanisms of population divergence of seabirds with pan-tropical distributions however are not as well understood8,9,10,11. Historical vicariant events, such as the emergence of the Isthmus of Panama in the late Pliocene, can create phylogeographic structure in species that previously occupied a single region, effectively isolating lineages and promoting speciation12,13,14,15. Other oceanic features, such as the Eastern Pacific Barrier, are also emerging as important barriers to gene flow in a variety of taxa9,10,16, despite the ability of marine species to disperse long distances. Changes in the oceanic environment driven by historical climatic changes have also been proposed as important drivers of divergence in marine taxa1,2,5,14,17,18, although their effect in mid-latitude species is not as clear.
Cryptic, intrinsic barriers, such as strong philopatry, which is characteristic of most pelagic seabirds, phenological differences driven by variation in non-breeding distributions and adaptation to local environmental conditions, may also play an important role on population divergence, sometimes even in the presence of gene flow6,8,19,20,21.
Bulwer’s petrel Bulweria bulwerii is a useful model to study the impact of physical and non-physical barriers in the diversification history of marine species. This small (ca. 100 g), all-dark procellariiform species is distributed in the tropical and sub-tropical oceanic islands of the Pacific and Atlantic oceans. It has also been recently discovered breeding on Round I. off Mauritius in the Indian Ocean, but the population is very small22. The species occurs in deep sea year-round23,24,25. It breeds on small oceanic islands for close to five months of the year, a period during which it behaves as a central place forager due to the constraints of incubation of the single egg and chick provisioning25. Individuals perform long migrations between their breeding and non-breeding areas. In the Atlantic, wintering areas include the southern equatorial regions23,24. In the Pacific, wintering areas are not as well known, but W Pacific populations are thought to migrate SW into the Indian Ocean, while Central Pacific populations are thought to move SE22,26. Like most procellariiform seabirds, the species is philopatric27. Although the Bulwer’s petrel population in the NW Hawaiian Islands was thought to be the largest with almost 100,000 pairs26, a recent estimate of the population on the Desertas Islands, Madeira, suggests that the population of this species in Madeira might be almost as large28. Elsewhere, populations are smaller22.
Using a gene set of 13 anonymous nuclear loci and one mitochondrial locus and coalescence theory-based methods, we studied for the first time the phylogeography and historical demography of Bulwer’s petrel across its main breeding areas in the Atlantic and Pacific Oceans. We assess the level of genetic differentiation and gene flow and provide temporal estimates for population divergence, testing for a possible effect of biogeographical barriers in the evolutionary history of the species. The Atlantic and Pacific Oceans have very different geographical configurations and oceanographic characteristics which determine the complexity of the different biogeographic domains of the two basins29. In particular, if we consider their tropical regions, the Atlantic Ocean is a much smaller and enclosed basin with a single predominant bio-oceanographic domain. Conversely, there are at least three recognized biogeographical zones in the much larger Pacific basin characterized by very different biological communities29 which might promote isolation and differentiation of seabird populations.
Results
Genetic diversity and population structure
A total of 9096 bp were surveyed per individual, including 979 bp of the mitochondrial locus and between 511 and 684 bp of each of the thirteen nuclear loci sampled. (Table 1). Not all individuals sequenced well for all loci, most frequently we sampled ca. 15 individuals for the three populations in the Pacific, ca. 20 individuals for the Azores and Selvagens, 4 individuals for the Raso islet (Cabo Verde) and seven individuals from Desertas (Supplementary information Table 1).
For the total of 8117 bp sequenced for the anonymous nuclear loci, there were 118 segregating sites corresponding to an average of 1 SNP per 69 bp. The number of alleles and segregating sites varied among the nuclear loci, and levels of nucleotide diversity varied by almost an order of magnitude (Table 1). Four of the 13 nuclear loci had length polymorphisms, one of which (an 1 bp indel in the nuclear locus Bubu4) was fixed in the Japanese population. The other three were private to either the Atlantic or Pacific Oceans but were not fixed. In the mitochondrial locus, 40 segregating sites in the 102 sampled individuals defined 26 different haplotypes. Overall, NADH2 was the most polymorphic locus, showing the highest number of segregating sites, haplotypes and nucleotide diversity (π = 0.059) (Table 1). For most loci and populations, there were no significant deviations from neutral expectations as suggested by non-significant Tajima’s D and Fu’s Fs estimates (Supplementary Table 1). Nuclear and mitochondrial sequence data have been submitted to the GenBank database (accession numbers ON918736-0N920126).
A very strong pattern of population genetic structure was revealed by the mitochondrial network, with the haplotypes distributed in three clearly segregated groups and no allele sharing between them (Fig. 1a). The group formed by the haplotypes sampled in the Western Pacific was deeply divergent (thirteen substitutions apart) from the clades sampled in the Central Pacific and Atlantic Ocean, corresponding to 1.6% and 1.4% net sequence divergence, respectively. The latter two groups also did not share alleles although haplotypes were much less divergent (0.24% sequence divergence). Within the Atlantic basin, haplotypes were shared among most populations, although the distance between the two colonies farthest apart (Vila islet, Azores and Raso Islet, Cabo Verde) is 2253 km. The global estimate of Φst for this locus was high and statistically significant in agreement with the overall geographically structured distribution of the alleles (Fig. 1a).
Genetic networks. Medium-joining haplotype networks of the 14 markers surveyed, including the mitochondrial locus NADH2 (a) and the thirteen anonymous nuclear loci, as produced by NETWORK v.4.6.0.0 (https://www.fluxus-engineering.com/sharenet.htm). The size of each circle is proportional to the frequency of that haplotype in the sample. The length of each branch is proportional to the number of substitutional steps separating haplotypes.
In contrast, allele networks for the different nuclear loci showed a variable pattern of allele sharing among populations. In several networks, the most common allele was widely distributed and shared among the three main groups but there were also private alleles to the different populations, suggesting some phylogeographic structure. For two loci (Bubu 4 and 11) the alleles sampled in Japan were not shared by any other population (Fig. 1b). Global estimates of Φst for the anonymous locus varied between 0.045 (Bubu 2) and 0.722 (Bubu 4) and were significant for all loci. Population pairwise tests (based on Φst) for the mitochondrial locus suggests a significant genetic differentiation between ocean basins and also between the western and central population of Bulwer’s petrels in the Pacific Ocean (Table 2). The pairwise Φst estimates for most nuclear loci reveal a significant differentiation between the Western Pacific and remaining groups (Supplementary Table 2), and only a few suggest significant genetic differentiation between the Central Pacific and the Atlantic populations (Supplementary Table 2). There was no genetic differentiation among the different populations from the Atlantic Ocean suggested by either marker type.
The STRUCTURE analysis based on the nuclear loci suggested that K = 2 best fitted the data, as suggested by ΔK (Fig. 2, see also Supplementary Fig. 1) (Evanno et al.31) indicating two clearly different genetic clusters, one including all individuals sampled in the Japanese islands, and a second including those of the remaining populations sampled (median posterior probability = 0.99). When STRUCTURE was re-run on the Central Pacific and Atlantic populations alone, it further suggested two genetic clusters corresponding to these two groups, although the Atlantic cluster includes a few individuals with an admixed ancestry (Fig. 2), dispersed throughout the different populations.
Structure results. Bayesian assignments of co-ancestry for all sampled Bulweria bulwerii individuals based on nuclear anonymous genotypes. For all samples, the inferred best K by the Evanno et al.31 method was two clusters, in both cases.
Phylogenetic relationships among major clusters and the historical demography
The *BEAST analyses using the mitochondrial and nuclear datasets combined provided strong support for the same topology across replicate runs and suggested a basal group of Bulwer’s petrels from the Western Pacific, and a well-supported sister relationship between the Central Pacific populations and those sampled in the Atlantic Ocean (Fig. 3). The relationships of the different populations within the Atlantic clade are not well resolved.
Species tree phylogeny. Phylogeny of the different populations sampled reconstructed with a combined dataset of a mitochondrial locus (NADH2) and thirteen nuclear anonymous markers with BEAST v1.7 (http://beast.community/index.html) and visualized with FigTree v.1.3.1 (http://tree.bio.ed.ac.uk/software/figtree/). Nodal support is indicated by Bayesian posterior probabilities. Bar is in substitutions/site. Abbreviations as follow: AZO–Ilhéu da Vila, Azores; DES–Deserta Grande, Desertas Is; SEL–Selvagem Grande, Selvagens Is., CV–Raso islet, Cabo Verde; LAY–Laysan Island, Hawaii; NIH–Nihoa Island, Hawaii; JAP–Minamijima Is, Ogasawara Islands.
Implementation of the IMa2 model to three main genetic groups of Bulwer’s petrels, suggested by the BEAST analysis with the combined dataset, and the STRUCTURE analysis, obtained convergence, low autocorrelation values and posterior density curves that showed clear peaks for all demographic parameters estimated (Supplementary Fig. 2). Based on the estimated peaks for θ, we inferred a smaller effective population size for the ancestral population of all groups (Ne ~ 9000) and the largest Ne for the ancestral population of the Central Pacific and Atlantic groups, and for the current Central Pacific group, both estimated at approximately 30,000 individuals (Table 3, Supplementary Fig. 2). Regarding the timing of divergences, t0 peaked at 0.110 and t1 peaked at 0.585 (95% HPD not estimated with confidence), suggesting that the Central Pacific and Atlantic groups diverged ~ 160,000 ya, while their ancestor and the Western Pacific group diverged much earlier (~ 850,000 ya) (Table 3, Supplementary Fig. 2). Most of the migration parameter estimates peaked near zero and include zero in their HPD suggesting that these major genetic groups diverged in isolation and have since exchanged very few migrants (Table 3). The only exceptions are the estimate of the migration parameter from the Central Pacific group into the Atlantic, which peaked at 1.744 (95% HPD 0–5.934), suggesting an exchange of ~ 0.6 migrants per generation. Also, mJapan > Hawaii peaked at 0.53, suggesting a rare westerly migration of ~ 0.2 migrants per generation (Table 3, Supplementary Fig. 2).
Discussion
Our results uncovered a contrasting phylogeographic pattern between the two main ocean basins where the species occurs, including an older evolutionary split between two distinct, isolated lineages evolving in the western and central Pacific Ocean, and a third, panmitic group in the Atlantic Ocean. The remarkable genetic differentiation within the Pacific Ocean but also between ocean basins reveal that both physical and paleo-oceanographic barriers have played an important role in the evolutionary history of this highly vagile, pan-tropical species.
In the present study, the pattern of population differentiation inferred from the complete dataset suggest that the Bulwer’s petrel populations from the Atlantic and Pacific Ocean basins are genetically differentiated, as well as those breeding in the Western and Central Pacific groups. The pattern is more clear with the mitochondrial locus whereas the nuclear data suggest a pattern of intermediate polyphyly between the Central Pacific and Atlantic groups. This is likely due to the nuclear lineages incomplete sorting, in line with the general expectation that nuclear loci will rarely show strong support for monophyly in recently diverged populations11,18,32,33.
Despite the absence of obvious physical barriers between Japan and Hawaii, we found two deep evolutionary lineages of Bulwer’s Petrels breeding in these archipelagos, which have been evolving independently for many generations, historically having exchanged very few to no migrants. This pattern of isolation despite the lack of apparent geographical barriers is counterintuitive if we consider that these pelagic seabirds are extremely mobile and routinely undertake journeys of hundreds or thousands of km during incubation shifts or while migrating to their wintering grounds23,24,25. A possible explanation is that a high level of philopatry associated with adaptation to local environments during the breeding season maintains the isolation of the two main genetic clusters found in the Pacific Ocean. The Ogasawara Islands lay in the western subtropical zone of the Pacific and are mostly under the influence of the Kuroshio current, one of its most important currents, which transports warm water northwards along the eastern coast of Japan. Further north, the cold nutrient-rich Oyashio current collides with the Kuroshio current creating a highly productive ecosystem34. Archipelagos laying within important current systems, such as the Galapagos or the Cabo Verde, hold higher levels of endemisms and diverged taxa, as species tend to ecologically adapt to local environments7. The Ogasawara Islands hold large numbers of pelagic seabirds, and although some also occur in the Northwestern Hawaiian Islands35, there are records of seabird population differentiation36 and endemisms37 suggesting the presence of unique local environmental conditions promoting species diversification.
Other factors may promote population genetic structure in seabirds, including differences in non-breeding areas6,19. Although data on species at-sea distribution are scarce for the Pacific Ocean, Bulwer’s petrels from Hawaii are thought to disperse eastward to the central and eastern Pacific during the non-breeding period26. Conversely, Bulwer’s petrels from Japan and other western colonies are thought to disperse SW as there have been sightings of this species south of the Korean Peninsula38, and into the Indian Ocean22. Likely due to its much larger size, the higher oceanographic complexity of the Pacific Ocean29 offers multiple high-quality wintering areas for a pelagic species like Bulwer’s petrel. This might enhance the connectivity of the Bulwer’s populations in the Pacific Ocean, creating a much more structured phylogeographic pattern, as has been shown in other species with genetically differentiated populations occupying distinct non-breeding ranges19. Conversely, Bulwer’s petrels breeding in the Macaronesian islands of Azores, Madeira, Canaries and Cabo Verde mostly overlap in their wintering areas, in the mid-equatorial Atlantic Ocean23. In the Atlantic Ocean, gene flow among the different archipelagos might be enhanced by shared wintering grounds that will favor intermixing of the populations. Consistent with this27 report the finding of two breeders in the small population (ca. 50 breeding pairs) from Vila islet (Azores) that had been previously captured and ringed on the Desertas Islands, which currently hold the largest population of Bulwer´s petrels in the Atlantic28.
Remarkably, and contrary to the pattern found for most other seabird species breeding in Cabo Verde, which holds several endemic Procellariiformes39, the Bulwer’s petrels from this archipelago (at least those from Raso Islet) do not appear to be genetically distinct from their more northern conspecifics. Although our sample size for this population is very small, there is clearly no genetic differentiation for the loci sampled, including the faster evolving NADH2. A larger panel of nuclear SNPs should be used to study in detail current levels of population connectivity between the Atlantic archipelagos. It would be interesting in the future to include individuals from Cima islet in Cabo Verde, where the breeding phenology of the species is very different23.
There is a clear phylogeographic break between the populations of B. bulwerii on either side of the Isthmus of Panama. Petrels from the Hawaiian and Atlantic islands share no mitochondrial haplotypes, and even show significant pairwise differentiation at a few nuclear loci, in a pattern common to many other co-distributed seabirds9,10,11,39. Such replicated phylogeographic patterns across co-distributed taxa suggests similar evolutionary processes in response to common barriers, clearly indicating that the Isthmus of Panama plays a fundamental role in the phylogeography of species with historical distributions in the Atlantic and Pacific. However, in many cases, divergence times suggest that the Pacific and Atlantic populations diverged well after the emergence of the Isthmus of Panama, ~ 3 MYA. Even though continental land bridges represent obvious barriers to dispersal for taxa that do not usually fly over land, the historical demography of many species suggests that this barrier is somewhat permeable, and most species are able of rare dispersal over land11.
Interestingly, for many pelagic species including the Bulwer’s petrel, estimates of divergence times suggest that many populations separated during the last 500,000 years11,32,33,39,40. The mechanisms by which the glaciation cycles influenced the phylogeographic structure of highly vagile, tropical and sub-tropical seabirds are not well understood. However, the sequential glacial and interglacial periods of the Pleistocene had severe consequences in the global atmospheric conditions and paleo-oceanographic environments41, which likely had profound effects on seabird distribution, dispersal patterns, establishment of new populations and extinction of others42.
Conclusion
We describe patterns of genetic differentiation across the main range of Bulwer’s petrel, identify the main phylogeographic breaks and reconstruct its demographic history. Although the Isthmus of Panama has functioned as a barrier to gene flow, we found that oceanographic barriers may also represent critical drivers of diversification in highly pelagic seabird species, despite their ability to travel long distances. In a rapidly changing world, understanding the mechanisms of divergence and speciation at mid-latitudes can provide insights regarding future biological consequences of global change.
The taxonomy of petrels and shearwaters has often been obscured by the lack of obvious morphological variation even between genetically distinct groups39. Further genomic, morphological and behavioural data of Bulwer’s petrels from the Ogasawara Islands should be collected as our data argue for a re-examination of the taxonomic status of the species in the Pacific43.
Materials and methods
Sample and data collection
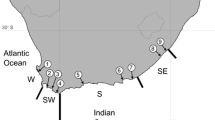
We sampled 105 Bulwer’s petrels from the main breeding localities of the species (Fig. 4). Specifically, we As we were interested in a global phylogeographic perspective, our sampling represents a compromise between the number of populations and loci sampled. Genomic DNA was extracted from blood samples kept in Queen’s lysis buffer44 solution following standard phenol–chloroform protocols. We screened for polymorphisms in thirteen anonymous nuclear loci isolated from a genomic library made originally from B. bulwerii, as described in30. We also surveyed a fragment of the NADH2 locus using primers L521545 and H106446, as the more frequently used Control Region showed a pattern of variation suggesting significant homoplasy (data not shown). Amplification of this fragment was carried out in 25 µl volume reactions containing the same final reagent concentrations as were used with the nuclear loci30, as well as the same thermocycler profile but with an annealing temperature of 58 °C. All loci were sequenced in both directions.
Main breeding areas of Bulwer’s Petrels. In colour are the populations sampled, in light grey are populations not sampled in this study. Sample origin: 1–Selvagem Grande, Selvagens Is., 2—Ilhéu da Vila, Azores; 3–Deserta Grande, Desertas Is; 4–Ilhéu Raso, Cabo Verde; 5–Laysan Is, Hawaii; 6–Nihoa Is, Hawaii; 7—Minamijima Is, Ogasawara Islands; Unsampled: 8–Marquesas Islands; 9–Round Island, Mauritius.
Sequences were assembled into contigs, aligned and edited in SEQUENCHER v.4.2 (Gene Codes Corporation), which was also used to check the validity of the base calls. Seven loci displayed heterozygous length polymorphisms, and all gaps from the data sets were trimmed. The phase of nuclear alleles with multiple heterozygous SNPs were resolved using the Bayesian algorithm implemented in PHASE v.2.1.147. Input files were prepared for PHASE using the software SeqPHASE48.
Polymorphism and population structure
Polymorphism statistics, such as the number of segregating sites, haplotypic and nucleotide diversities were determined using DNAsp v5.1049. We used ARLEQUIN v.3.5.2.250 to detect departures from neutrality for each population, by estimating Tajima’s D51 and Fu’s FS52 for each locus. The net pairwise sequence divergence between pairs of populations was calculated using MEGA 553 with the model of sequence evolution selected by the AIC in jModelTest54. Population structure among the three taxa was investigated by estimating global and pairwise Фst for all loci, using the Kimura’s-two parameter model of substitution, as implemented in ARLEQUIN. We tested the significance of results with 10,000 random permutations, considering a significance level of 0.05 after correction for false discovery rate. Median-joining haplotype networks, based on the largest blocks of non-recombining fragments, were estimated for all loci using NETWORK v.4.6.0.055 (https://www.fluxus-engineering.com/sharenet.htm).
To detect population structure without the need to define a priori the populations, we used Bayesian clustering analyses, as implemented in STRUCTURE 2.356 (https://web.stanford.edu/group/pritchardlab/structure.html). We ran two sets of analyses depending on how the data was formatted: the first set used allelic data for each of the nuclear locus, and the second used the dataset converted to SNPs. There were no differences in results between the two sets and we present results only for the former dataset. In the final analysis, In STRUCTURE, colony of origin was not used as prior information, and analyses assumed an admixture model and correlated allele frequencies among populations. We performed 10 replicate runs for each K value, which varied between 1 (single population) and 6. Each replicate was run for 1 million iterations, with a burn-in of 100,000 iterations. The value of K that best fitted the data was estimated using the method of31 implemented in STRUCTURE Harvester57. Since this method tends to recover the highest hierarchical level of population structure31, we re-ran STRUCTURE to test for further structuring, using the same run parameters as above.
Phylogenetic relationships among groups
We used the *BEAST algorithm based on the multi-species coalescent58 as implemented in the software BEAST 1.759 to estimate a species tree for the sampled taxa, using the mitochondrial and nuclear loci. The recovered topologies of the tree were the same across replicates. As most species trees methods, *BEAST requires a priori assignment of alleles to taxa. We used the well-supported and geographically structured mtDNA network for prior assignment. The input was prepared with BEAUti v1.7 (http://evomics.org/resources/software/molecular-evolution-software/beauti/). We ran *BEAST multiple times to test different priors and settings, and to assess convergence and mixing. The results shown are based on final runs which ran for 109 generations (convergence was compromised by low signal of some genes, and required very long runs), sampling every 10,000 generations, with a burn-in of 106 generations. We chose the most appropriate model of sequence evolution for each locus (see above), used a strict molecular clock, which could not be rejected for this dataset, and a Yule process for the species tree prior, and other priors were changed according to operator weight suggestions from previous runs. We monitored convergence of the sampling process using TRACER 1.560, ensuring that the ESS values were above 200 and parameter posterior distributions were unimodal. Trees were summarized in TreeAnnotator v.1.761 and visualized in FigTree 1.3.1 (http://tree.bio.ed.ac.uk/software/figtree/). We assessed nodal support using posterior probability estimates. We used a substitution rate for the NADH2 locus of 0.028 × 10–646, and the nuclear rate was allowed to be estimated relative to the mtDNA rate.
Coalescence estimates of historical demography
We inferred the demographic history of Bulwer’s Petrel with IMa262 using the full data set of twelve anonymous nuclear and NADH2 loci. IMa2 implements a coalescent-based method that uses Markov Chain Monte Carlo sampling of gene genealogies to estimate population parameters scaled by the mutation rate per locus per generation (μ), such as contemporary and ancestral effective population sizes (θ, where θ = 4Neμ, and Ne is the effective population size) migration rates (m = M/μ, where M is the effective migration rate per generation) and divergence times (t = Tμ, where T is the divergence time in years before present) among populations. Implementation of IMa2 with more than two populations requires a known sequence of splitting events, and accordingly we used our *BEAST species tree. It also assumes no recombination within loci, and we used IMgc63 to find the largest block of sequences and individuals with no evidence of recombination and used these data in our IMa2 analysis. We ran IMa2 several times, varying the prior bounds, heating schemes and number of chains, to optimize the priors, improve mixing of the chains and reach convergence of the different analyses. The final runs included 80 independent MCMC chains with a geometric heating scheme (h1 = 0.975, h2 = 0.75). All runs included 15 million generations with 500,000 generations discarded as burn-in. Trend plots, ESS values and comparisons between coupled runs were used to assess mixing and convergence, as suggested in the documentation for IMa2. To translate parameter estimates (scaled by µ) to demographic units, we used a substitution rate for NADH2 of 0.028 × 10–646, and a mean generation time of 12 years64.
Ethical statement. The volume of the blood sample collected from each petrel was 50–75 μl (approximately 0.05% vol/weight). Sampling were performed in accordance with the guidelines and regulations of each of the relevant institutions, including the Instituto das Florestas e Conservação da Biodiversidade, Portugal (permit No. 2/2012S), the Direção Regional dos Açores (permits nº4/CN/2002 and nº7/CN/2003), the US Fish and Wildlife Services and the Papahānaumokuākea Marine National Monument (permit No. PMNM-2008-029) and the Ministry of the Environment, Japan (permit No. 080428001). Sampling further followed the requirements of the Directive 2010/63/EU of the European Parliament and of the council for the protection of animals used for scientific purposes.
Data availability
DNA sequences: GenBank accession #s: ON918736–ON920126. While being curated by GenBank, the sequence data can be found temporarily at https://drive.google.com/drive/folders/1Gwhfiro2aq7aSwLTJmi2cn7Vgh2bz0UM?usp=sharing. For additional information contact corresponding author (MCS).
References
Congdon, B. C., Piatt, J. F., Martin, K. & Friesen, V. L. Mechanisms of population differentiation in Marbled Murrelets: Historical versus contemporary processes. Evolution 54, 974–986 (2000).
De Dinechin, M., Ottvall, R., Quillfeldt, P. & Jouventin, P. Speciation chronology of rockhopper penguins inferred from molecular, geological and paleoceanographic data. J. Biogeog. 36, 693–702 (2009).
Younger, J. L. et al. The challenges of detecting subtle population structure and its importance for the conservation of emperor penguins. Mol. Ecol. 26, 3883–3897 (2017).
Clucas, G. V. et al. Comparative population genomics reveals key barriers to dispersal in Southern Ocean penguins. Mol. Ecol. 27, 4680–4697 (2018).
Rexer-Huber, K. et al. Genomics detects population structure within and between ocean basins in a circumpolar seabird: the white-chinned petrel. Mol. Ecol. 28, 4552–4572 (2019).
Friesen, V. L., Burg, T. M. & McCoy, K. D. Mechanisms of population differentiation in seabirds. Mol. Ecol. 16, 1765–1785 (2007).
Friesen, V. L. Speciation in seabirds: Why are there so many species…. and why aren´t there more?. J. Ornith. 156, 27–39 (2015).
Steeves, T. E., Anderson, D. J. & Friesen, V. L. A role for nonphysical barriers to gene flow in the diversification of a highly vagile seabird, the Masked Booby Sula dactylatra. Mol. Ecol. 14, 3877–3887 (2005).
Morris-Pocock, J. A., Anderson, D. J. & Friesen, V. L. Mechanisms of global diversification in the Brown Booby (Sula leucogaster) revealed by uniting statistical phylogeographic and multilocus phylogenetic methods. Mol. Ecol. 20, 2835–2850 (2011).
Morris-Pocock, J. A., Anderson, D. J. & Friesen, V. L. Biogeographical barriers to dispersal and rare gene flow shape population genetic structure in red-footed boobies (Sula sula). J. Biogeog. 43, 2125–2135 (2016).
Silva, M. C., Matias, R., Ferreira, V., Catry, P. & Granadeiro, J. P Searching for a breeding population of Swinhoe’s Storm-petrel at Selvagem Grande, NE Atlantic, with a molecular characterization of occurring birds and relationships within the Hydrobatinae. J. Ornith. 157, 117–123 (2016).
Steeves, T. E., Anderson, D. J. & Friesen, V. L. The Isthmus of Panama: a major physical barrier to gene flow in a highly mobile pantropical seabird. J. Avian Biol. 18, 1000–1008 (2005).
Schultz, J. K. et al. Global phylogeography and seascape genetics of the lemon sharks (genus Negaprion). Mol. Ecol. 17, 5336–5348 (2008).
Jensen, M. P. et al. The evolutionary history and global phylogeography of the green turtle (Chelonia mydas). J. Biogeog. 46, 860–870 (2019).
Lombal, A. J., Odwyer, J. E., Friesen, V. L., Woehler, E. J. & Burridge, C. P. Identifying mechanism of genetic differentiation among populations in vagile species: historical factors dominate genetic differentiation in seabirds. Biol. Rev. Camb. Phil. Soc. 95, 625–651 (2020).
Briggs, J. C. The east pacific barrier and the distribution of marine shore fishes. Evolution 15, 545–554 (1961).
Techow, N. M. S. M. et al. Speciation and phylogeography of giant petrels Macronectes. Mol. Phylog. Evol. 54, 472–487 (2010).
Silva, M. C. et al. Understanding the mechanism of antitropical divergence in the seabird White-faced Storm-petrel (Procellariiformes: Pelagodroma marina) using a multi-locus approach. Mol. Ecol. 24, 3122–3137 (2015).
Rayner, M. J. et al. Contemporary and historical separation of transequatorial migration between genetically distinct seabird populations. Nat. Commun. 2, 332–337 (2011).
Danckwerts, D. K., Humeau, L., Pinet, P., McQuaid, C. D. & Le Corre, M. Extreme philopatry and genetic diversification at unprecedented scales in a seabird. Scien. Rep. 11, 6834 (2021).
Tigano, A. & Friesen, V. L. Genomics of local adaptation with gene flow. Mol. Ecol. 25, 2144–2164 (2016).
Megyesi, J. L., O´Daniel, D. L. Bulwer’s Petrel (Bulweria bulwerii), version 1.0 in Birds of of the World (ed, Billerman, S.M.), Cornell Lab of Ornithology, Ithaca, NY, USA. (2020).
Ramos, R. et al. Leapfrog migration and habitat preferences of a small oceanic seabird, Bulwer’s petrel (Bulweria bulwerii). J. Biogeog. 42, 1651–1664 (2015).
Dias, M. P., Alho, M., Granadeiro, J. P. & Catry, P. Wanderer of the deepest seas: Migratory behaviour and distribution of the highly pelagic Bulwer’s petrel. J. Ornith. 156, 955–962 (2015).
Dias, M. P. et al. Distribution and at-sea activity of a nocturnal seabird, the Bulwer’s Petrel Bulweria bulwerii, during the incubation period. Deep Sea Res. 113, 49–56 (2016).
Pyle R. L., Pyle, P. Bulwer’s Petrel. In The Birds of the Hawaiian Islands: occurrence, history, distribution and status. Version 2. B.P. Bishop Museum: Honolulu, HI, USA. (2017).
Bried, J., Andris, M., Dubois, M.-P. & Jarne, P. Decreased selectivity during mate choice in a small population of a long-lived seabird. J. Avian Biol. 52, 1–13 (2021).
Catry, P. et al. Bulwer’s petrels breeding numbers on the Desertas Islands (Madeira): Improved estimates indicate the NE Atlantic population to be much larger than previously thought. Airo 23, 10–14 (2015).
Bowen, B. W. et al. Comparative phylogeography of the ocean planet. PNAS 113, 7962–7969 (2016).
Silva, M. C., Duarte, M. A. & Coelho, M. M. Anonymous nuclear loci in the White-faced Storm-petrel Pelagodroma marina and their applicability to other procellariiform seabirds. J. Hered. 102, 362–365 (2011).
Evanno, G., Regnaut, S. & Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 14, 2611–2620 (2005).
Welch, A. J., Yoshida, A. A. & Fleischer, R. C. Mitochondrial and nuclear DNA sequences reveal recent divergence in morphologically indistinguishable petrels. Mol. Ecol. 20, 1364–1377 (2011).
Gangloff, B. et al. The evolution of north-east Atlantic gadfly petrels using statistical phylogeography. Mol. Ecol. 22, 495–507 (2013).
Martí, C., Vallerani, M., Ojamaa, P., (2017). Research for PECH committee – fisheries in Japan, European Parliament, Policy Department for Structural and Cohesion Policies, Brussels
Chiba, H., Kawakami, K., Suzuki, H. & Horikoshi, K. The distribution of seabirds in the Bonim Islands, Southern Japan. J. Yamash. Inst. Ornith. 39, 1–17 (2007).
Eda, M. et al. Genetic characteristics of the Black-footed Albatross Diomedea nigripes on the Bonin Islands and their implications for the species’demographic history and population structure. Ornith. Sci. 7, 109–116 (2008).
Kawakami, K., Eda, M., Izumi, H., Horikoshi, K. & Suzuki, H. Phylogenetic position of endangered Puffinus lherminieri bannermani. Ornith. Sci. 17, 11–18 (2018).
Gorta, S. B. Z. Southern influx of the pantropical Bulwer’s Petrel Bulweria bulwerii into the Tasman Sea. Mar. Ornith. 49, 145–150 (2021).
Taylor, R. S. et al. Cryptic species and independent origins of allochronic populations within a seabird species complex (Hydrobates spp.). Mol. Phylog. Evol. 139, 106552 (2019).
Obiol, J. F., James, H. F., Chesser, R. T., Bretagnolle, V. & González-Solís, J. Palaeoceanographic changes in the late Pliocene promoted rapid diversification in pelagic seabirds. J. Biog. 49, 171–188 (2022).
Hill, D. J., Bolton, K. P. & Haywood, A. M. Modelled ocean changes at the Plio-Pleistocene transition driven by Antarctic ice advance. Nat. Commun. 8, 14376 (2017).
Olson, S. L. A new species of shearwater of the genus Calonectris (Aves:Procellariidae) from a middle Pleistocene deposit on Bermuda. Proc. Biol. Soc. Wash. 121, 398–409 (2008).
De Queiroz, K. Species concepts and species delimitation. Syst. Biol. 56, 879–886 (2007).
Seutin, G., White, B. N. & Boag, P. T. Preservation of avian blood and tissue samples for DNA analyses. Can. J. f Zool. 69, 82–90 (1991).
Hackett, S. J. Molecular phylogenetics and biogeography of tanagers in the genus Ramphocelus (Aves). Mol. Phylog. Evol. 5, 368–382 (1996).
Drovetski, S. V. et al. Complex biogeographic history of a Holarctic passerine. Proc. Roy. Soc. B 271, 545–551 (2004).
Stephens, M., Smith, N. J. & Donnelly, P. A new statistical method for haplotype reconstruction from population data. Am. Hum. Genet. 68, 978–989 (2001).
Flot, J.-F. SEQPHASE: A web tool for interconverting phase input/output files and fasta sequence alignments. Mol. Ecol. Res. 10, 162–166 (2010).
Librado, P. & Rozas, J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinform. 25, 1451–1452 (2009).
Excoffier, L., Laval, G. & Schneider, S. Arlequin v. 3.0: An integrated software package for population genetics data analysis. Evol. Bioinform. Online 1, 47–50 (2005).
Tajima, F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123, 585–595 (1989).
Fu, Y. X. Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics 147, 915–925 (1997).
Tamura, K. et al. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739 (2011).
Posada, D. jModelTest: Phylogenetic model averaging. Mol. Biol. Evol. 25, 1253–1256 (2008).
Bandelt, H. J., Forster, P. & Röhl, A. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 16, 37–48 (1999).
Pritchard, J. K., Stephens, M. & Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 155, 945–959 (2000).
Earl, D. A. & vonHoldt, B. M. STRUCTURE HARVESTER: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Res. 4, 359–361 (2012).
Heled, J. & Drummond, A. L. Bayesian inference of species trees from multilocus data. Mol. Biol. Evol. 27, 570–580 (2010).
Drummond, A. J. & Rambaut, A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7, 214 (2007).
Rambaut, A., Drummond, A. J. Tracer v1.5. Available from http://beast.bio.ed.ac.uk/Tracer. (2007).
Rambaut, A., Drummond, A. J. TreeAnnotator v1.6.1. Available from http://beast.bio.ed.ac.uk/TreeAnnotator. (2010).
Hey, J. Isolation with migration models for more than two populations. Mol. Biol. Evol. 27, 905–920 (2010).
Woerner, A. E., Cox, M. P. & Hammer, M. F. Recombination-filtered genomic datasets by information maximization. Bioinform. 23, 1851–1853 (2007).
Bird, J. P. et al. Generation lengths of the world’s birds and their implications for extinction risk. Conserv. Biol. 34, 1252–2126 (2020).
Acknowledgements
The four samples from Raso Islet, Cabo Verde, were sent to us by the late Luís Monteiro, Universidade dos Açores. We thank Vânia Ferreira and Mauro Silva for lab assistance and data collection, and J.M. Soares and the Clube Naval from Santa Maria Island, Azores, for transportation to Vila islet. This research was funded by Fundação para a Ciência e a Tecnologia (FCT, Portugal) through projects OCEANTREE–PTDC/BIA-EVL/28565/2017, UIDB/BIA/00329/2020 granted to cE3c; UIDP/50017/2020, UIDB/50017/2020 and LA/P/0094/2020, granted to CESAM; UIDB/04292/2020 and UIDP/04292/2020, granted to MARE; project LA/P/0069/2020 granted to the Associate Laboratory ARNET and PTDC/BIABDE/67286/2006 (also funded by FEDER through the Programme COMPETE–Ref: FCOMP-01-0124-FEDER-007061) granted to JB. MCS is funded by FCT through a contract foreseen in DL 57/2016, changed by Law 57/2017. Sample collection from Japan was funded by the Tokyo Metropolitan Government and the Environment Research and Technology Development Fund (4-1402) of the Ministry of the Environment, Japan.
Author information
Authors and Affiliations
Contributions
M.C.S., P.C., J.P.G. conceived the work, P.C., J.P.G. J.B., E.F. and K.K. collected samples and contributed to manuscript writing, M.C.S. collected and analysed data, led the writing. All authors approved the last version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Silva, M.C., Catry, P., Bried, J. et al. Contrasting patterns of population structure of Bulwer’s petrel (Bulweria bulwerii) between oceans revealed by statistical phylogeography. Sci Rep 13, 1939 (2023). https://doi.org/10.1038/s41598-023-28452-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-28452-z
- Springer Nature Limited