Abstract
This review aimed to examine the relationship between TP53 mutational status, as determined by genomic sequencing, and survival in squamous cell carcinoma of the head and neck. The databases Medline, Embase, Web of Science (core collection), Scopus and Cochrane Library were searched from inception to April 2021 for studies assessing P53 status and survival. Qualitative analysis was carried out using the REMARK criteria. A meta-analyses was performed and statistical analysis was carried out to test the stability and reliability of results. Twenty-five studies met the inclusion criteria, of which fifteen provided enough data for quantitative evaluation. TP53 mutation was associated with worse overall survival (HR 1.75 [95% CI 1.45–2.10], p < 0.001), disease-specific survival (HR 4.23 [95% CI 1.19–15.06], p = 0.03), and disease-free survival (HR 1.80 [95% CI 1.28–2.53], p < 0.001). Qualitative assessment identified room for improvement and the pooled analysis of all anatomical subsites leads to heterogeneity that may erode the validity of the observed overall effect and its subsequent extrapolation and application to individual patients. Our systematic review and meta-analysis supports the utility of TP53 mutational as a prognostic factor for survival in head and neck squamous cell carcinoma. A well designed prospective, multi-centre trial is needed to definitively answer this question.
Similar content being viewed by others
Introduction
Squamous cell carcinomas of the head and neck (HNSCC) are the most common cancers of the head and neck region constituting the 6th most common cancer worldwide (~ 1 million cases per annum1). The overarching disease of HNSCC involves tumour arising from the various subsites of the head and neck including, oral cavity, oropharynx, nasopharynx, hypopharynx, larynx and sinonasal mucosa, each displaying variability in presentation, treatment, and prognosis.
The pathogenesis of this disease is multifactorial. However, the majority are associated with tobacco and alcohol misuse, with a notable synergistic effect. A significant minority arise as a result of oncogenic viral infection, particularly Human papillomavirus (HPV; notably in the oropharynx) or Epstein Barr Virus (EBV; commonly in the nasopharynx). Despite variability between subtypes, the incidence of HNSCC is rising and, as a disease cohort, is expected to increase by 30% by 20302. In particular, oropharyngeal squamous cell carcinoma (OPSCC) has doubled in the UK, USA, and Europe in recent decades2.
HNSCC exhibits variable responses to conventional treatment. Clinical response rates correlate with survival and are inversely related to primary tumour size, presence and volume of metastatic disease in the cervical lymph nodes and pathological evidence of tumour spread through the lymph node capsule (extracapsular spread). When analysing survival across all age groups and anatomical sites, the 5-year survival for HNSCC has modestly improved from 55 to 66%3. However, a subgroup analysis highlights that survival in some anatomical subsites remains stagnant and that this observed overall survival improvement is partially attributable to the emergence of HPV-positive OPSCC4.
Despite presenting with clinico-pathological features suggestive of an aggressive phenotype, survival rates are considerably higher for patients with HPV-positive OPSCC, albeit a significant minority (15–20%) will still succumb to their disease5. Current epidemiological data suggest that poor outcome correlates tightly with cigarette smoking, hypothesised to be a cause of, and surrogate for, underlying carcinogen-induced mutational load and genetic instability. However, mainstay cisplatin-based chemoradiotherapy (CRT) results in life-changing long-term swallowing disability: up to 20% of patients undergoing CRT require long-term gastrostomy tube feeding6. Surgery followed by adjuvant therapy represents a valid alternative treatment option, but clinical decision tools are needed to achieve a consensus. Consequently, there is an urgent need to identify patients who are destined for poor outcome and those for whom treatment de-intensification, with a view to avoiding long term swallowing difficulty, is an option. In contrast, there is an urgent need for the development of new treatments to enhance survival for HPV-negative HNSCC as survival rates remain at 60%5.
Whilst data support the prognostic utility of HPV status, no data currently exist to suggest that treatment decision-making based on HPV status is safe and effective7. Moreover, the molecular mechanisms by which HPV may contribute to neoplasia development and progression in OPSCC remain poorly understood, with much of our current understanding inferred from data derived from cervical cancer research. Whether such inference is appropriate and relevant is currently unclear.
The TP53 tumour suppressor is the most commonly mutated gene in human cancer and the p53 protein it encodes plays critical roles in cell-cycle control and apoptosis in response to DNA damage and other cellular stresses8. Loss of p53 function, either through disruptive TP53 mutation or through abrogation by viral oncoproteins (in the case of HPV + disease), occurs with high frequency in HNSCC9. Given the pivotal role that p53 plays in regulating cellular response to therapeutic interventions (such as chemotherapy and radiotherapy), it is enticing to hypothesise that disruption of the gene would dictate prognostic significance. To date however, evidence regarding the role of p53 as a prognostic marker for HNSCC remains controversial10.
Structurally, p53 is a complex and multifunctional 393-residue protein. It has 3 domains: an N-terminal subunit composed of a transcription-activation domain and a proline rich domain, a central DNA-binding core domain, and a C-terminal domain involved in modulating binding behaviour of the DNA binding domain. The TP53 gene is located on chromosome 17p13.1 and is composed of 25,772 bases.
The consequences of inconsistencies in analytical approaches to identify p53 alterations and variability of cohorts has led to conflicting outcomes. Moreover, considerable variation has been identified in TP53 mutations, with consequent diverse effects on protein function and thus prognostic significance11. Loss of TP53 function has been shown to negatively affect disease outcome in other solid tumours such as bladder carcinomas, while in breast cancer, for example, TP53 mutation has been linked to improved prognosis in patients treated with chemotherapy but poor prognosis in those treated with hormone therapy12. These various responses to treatment in different cancer types appear to have a sound basis in tumour biology, and it is not unforeseeable that such differences to treatment response and TP53 mutation status may also occur in different HNSCC subtypes.
Immunohistochemistry (IHC) of p53 has been proposed as a surrogate marker for TP53 mutations in diagnostic workup of a number of cancers. However, interpretability of this technique is complicated by mutation-dependent alterations in protein stability and thus immunoreactivity. In the absence of DNA damage, p53 induces its own proteasomal degradation through transcriptional upregulation of the E3 ubiquitin ligase Mouse double minute 2 (MDM2). Thus, WT p53 is inherently unstable under usual conditions and is undetectable through IHC. In cells harbouring deleterious TP53 missense mutations, MDM2 is no longer induced, and this negative feedback loop is broken, such that p53 persists and is detectable through IHC13. This indirect strategy in identifying TP53 mutations is not suitable for detecting nonsense mutations, which result in truncated non-immunoreactive protein, or for deletions, both of which will result in the absence of p53 staining and appear indistinguishable from WT. Sequencing overcomes these limitations.
We document the outcomes of a systematic review and meta-analysis of the evidence for the prognostic relevance of p53 mutational status assessed using sequencing approaches.
Methods
This systematic review complies with PRISMA guidelines14 and closely followed the criteria of Cochrane Prognosis Methods Group15, Cochrane Handbook for Systematic Reviews of Interventions16, and Centre for reviews and Dissemination (CRD)’s guidance for undertaking reviews in healthcare17.
Protocol
In keeping with best practice, the protocol, including a priori methodology, was registered in the PROSPERO international prospective register of systematic reviews (www.crd.york.ac.uk/PROSPERO, registration number CRD42021242118), in order to minimize the risk of bias and improve the transparency, precision, and integrity of this study. The protocol adheres to PRISMA-P guidelines to ensure a rigorous approach.
Research question
This review aimed to examine the relationship between TP53 mutational status, as determined by genomic sequencing, and survival in squamous cell carcinoma of the head and neck (oral, oropharynx, nasopharynx, sinonasal, hypopharynx, larynx). Effective discrimination of clinical outcomes are hypothesised as being suitable to support the design and development of prospective studies seeking to determine the clinical utility of TP53 mutational status as a prognostic (and possibly predictive) biomarker.
Information sources and search strategy
The search strategy was developed and conducted by a medical librarian. Prior to conducting the searches, the search terms were peer reviewed by another medical librarian according to PRESS criteria18. The databases Medline (via Ovid), Embase (via Ovid), Web of Science (core collection), Scopus and Cochrane Library were searched from inception to April 2021 and limited to English language only, with variants of the following terms, which were in the title and abstract fields, as well as in the subject heading term field when these existed in the database. The Medline search is reproduced below; (see Supplementary Document S1 for the full strategies used in all databases):
-
(p53* or tp53* or pp53* or TRP53* or TP53BP1* or 53BP1* or p202*).ti,ab. OR Tumor Suppressor p53-Binding Protein 1/ or Genes, p53/ or Tumor Suppressor Protein p53/
-
AND
-
((laryn* or oropharyn* or hypopharyn* or "oral cavit*" or mouth or tongue or tonsil* or neck* or head or "sino-nasal" or sinonasal or sinus* or nasomucosa or nasopharyn* or nasal* or nose* or paranasal* or pharynx* or cheek* or lip* or gingiv* or palat*) adj3 (carcinoma* or neoplasm* or cancer* or metastas* or tumor* or tumour*)).ti,ab. OR Laryngeal Neoplasms/ or exp Pharyngeal Neoplasms/ or "head and neck neoplasms"/ or "squamous cell carcinoma of head and neck"/ or exp Nose Neoplasms/ or mouth neoplasms/ or gingival neoplasms/ or lip neoplasms/ or palatal neoplasms/ or tongue neoplasms/
Eligibility criteria
We considered all human studies that investigated the impact of p53 mutational status on patient survival in head and neck cancers. Application of DNA sequencing technique(s) was necessary for identification of p53 mutational status. Other forms of p53 status determination, such as immunohistochemistry, were excluded. All head and neck subsites were included in this study. Conference abstracts, review papers, letters to the editor and opinion pieces were excluded. Only articles published in the English language were considered.
Study selection and data extraction
Titles and abstracts were independently screened by two reviewers (SB, GN) against the agreed inclusion and exclusion criteria. Disagreements between reviewers were resolved by consensus. A data extraction tool was used for further analysis of selected full texts—this was initially performed by one reviewer (SB) and verified by a second (GN). Reasons for exclusion were recorded for any publication at full-text stage. Data extraction items included:
-
1.
Article identifiers (author, year, title).
-
2.
Study characteristics (sample size, design, population, inclusion and exclusion criteria).
-
3.
Sequencing technique.
-
4.
Anatomical subsites (oral, oropharynx, larynx, hypopharynx, sinonasal, nasopharyngeal).
-
5.
Outcome measures (survival).
-
6.
Results and conclusions.
Evaluation of quality and risk of bias
Each publication was critically appraised for both quality and risk of bias using the Reporting Recommendations for Tumor Marker Prognostic Studies (REMARK) criteria19. The REMARK criteria consists of a checklist of 20 items20, and each item can be further divided into multiple sub-categories21. To ensure a consistent interpretation and application of the REMARK criteria, the authors examined the REMARK sub-criteria in tandem and selected those of highest yield: each RCC prognostic biomarker manuscript was evaluated according to 48 separate sub-criteria for a maximum score of 20 points. A full list of the criteria and point per criteria is listed in Supplementary Table S1.
Statistical analysis
The restricted maximum-likelihood estimator for Tau2 was used to assess between-studies variance of treatment effects. Higgin’s I2 was used to assess the proportion of true variance of a weighted outcome, interpreted according to the Cochrane Collaboration, whereby 0–40% was considered as low heterogeneity, 30–60% as moderate heterogeneity, 50–90% as substantial heterogeneity and > 75% as considerable heterogeneity16. A p-value of < 0.10 was accepted as a significant Cochrane Q statistic. The generic inverse variance method was used as part of a random-effects model of hazard ratios, from studies reporting the results of multivariate Cox proportional hazards models, to provide an overall estimate of the influence of TP53 mutation status on overall survival, disease/progression-free survival, and disease-specific survival. Publication bias was assessed through funnel plots of hazard ratios against standard error, and funnel plot asymmetry was quantitatively assessed using Egger’s test. Statistical analysis was performed used the meta package on R version 4.0.0. All scripts for meta-analysis are available upon request from the corresponding author.
Results
A total of 9,229 articles were initially retrieved using the search algorithm. After title and abstract screening, 137 records were retained for full-text retrieval, and a total of 25 studies were included at full-text review (Fig. 1) of which 22 provided sufficient data to be included in the quantitative evaluation. 10 of the 25 identified studies were prospective in design, while the remainder were retrospective observational or cohort studies22,23,24,25,26,27,28,29,30,31.
Study characteristics
Table 1 summarises the main characteristics of the twenty-five selected studies, published between 1995 and 2020, that carried out p53 sequencing on 3326 tumours with associated clinical outcome data. Sample sizes ranged from 22 to 420 patients. Seventeen studies utilised Sanger sequencing technology to determine p53 status, seven used Next-Generation Sequencing (NGS), and one study used pyrosequencing. Nine studies investigated TP53 mutation in a mixed population of squamous cell carcinoma from all head and neck anatomical subsites. Eight studies restricted investigation of mutations to oral squamous cell carcinoma (OSCC), five studies to laryngeal squamous cell carcinoma (LSCC), and to single studies in oropharyngeal squamous cell carcinoma (OPSCC), nasopharyngeal carcinoma (NPC), and sinonasal squamous cell carcinoma (SNSCC) respectively. Twelve studies were conducted in Europe, six in North America, six in Asia, and one in Oceania. With the exception to Cho et al.32 and Poeta et al.31, the studies included analysis restricted to primary tumours. Cho et al.32 investigated the genomic alterations in 15 nasopharyngeal carcinoma primary tumours as well as the paired primary tumours and recurrent tumours for a further 7 nasopharyngeal carcinoma patients. Poeta et al.31 investigated patients with either newly diagnosed or recurrent tumours, with inclusion only if the treatment plan included primary surgical extirpation with curative intent.
The primary treatment modality was not explicitly reported in 7 of the 25 included studies23,28,32,33,34,35,36. Of the remaining studies, twelve investigated patients that underwent surgical resection of the tumour26,30,31,37,38,39,40,41,42,43,44,45. In one study, all patients received primary radiotherapy or chemoradiotherapy without surgery24. Five studies included patients who had undergone variable treatments22,25,27,29,46.
Qualitative analysis
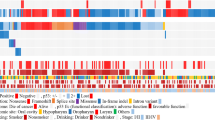
None of the publications completely fulfilled the criteria set by the REMARK guidelines. The highest scoring paper was awarded 15.16 points out of a maximum of 20 (range 6.4–15.15). Figure 2 presents each study’s fulfilment of the 20-point criterion of the REMARK guidelines with the full score included. Publication bias was assessed visually using funnel plots, identifying no obvious plot asymmetry (Supplementary Fig. S1). This finding was further supported by the results of Egger’s test of plot asymmetry (t(20) = − 1.40, p = 0.18).
Study fulfilment of the 20-point criteria as set out by the REMARK guidelines17. Green indicates the domain was fully met, amber partially met and red none. Total score provided in the rightmost column (maximum score 20).
Study outcome measures
Reporting of survival was a necessary requirement for inclusion in this systematic review, however variation in the method/number of measurements was apparent. Most studies (22/25; 88%) reported overall survival22,23,24,25,26,27,28,29,30,31,32,33,35,36,38,39,40,42,43,44,45,46, 15 studies reported disease/progression-free survival (60%)24,26,27,29,30,31,33,34,37,39,41,42,44,45,46, and 5 studies reported on disease-specific survival (20%)22,24,29,36,44.
Variation in duration of patient follow-up was also variously reported between studies. 8 of the 25 studies did not provide a specific estimate of average follow-up period23,24,25,32,33,40,43,45. Of those studies reporting a median duration, follow-up ranged from 29 months38 to 98.4 months34.
HPV status
There was inconsistency in approaches made towards inclusion and reporting of tumour HPV status. Nine studies included HPV-positive tumours in their analysis22,24,26,33,34,35,38,42,43,44,46. Two studies fully excluded HPV-positive tumours37,45, and one paper only specifically investigated HPV-positive cancers41. The remaining papers made no mention of HPV status in their inclusion criteria or methodology23,25,27,28,29,30,31,32,36,39,40.
Outcome
Table 2 summarises the principal findings of each study included in the systematic review. 8 studies reported an analysis of clinical outcomes according to location or category of TP53 mutation24,26,28,30,31,33,42,44. Lapke et al.44 defined any non-conservative mutation or mutation introducing a stop codon within the DNA binding-domain (DBD) of p53 as disruptive. These disruptive mutations were further characterised as either (i) truncated mutations associated with loss of p53 tumour suppressor activity; or (ii) DBD missense mutations resulting in a possible deleterious gain-of-function. It was found that carriage of DBD missense mutations were associated with a significant reduction in disease-specific survival and disease-free survival compared to wild type p53 (WT), whereby ‘all other mutations’ showed comparable survival characteristics to WT. Caponio et al.33 adopted a computational approach to determining the influence of specific TP53 mutations on survival outcomes, identifying DNA-binding domain mutations as a poor prognostic factor in laryngeal cancers (but not other anatomical sub-sites). Further, it was identified that mutations within the hotspot residues R175, H193 and R213 portended a poor prognosis irrespective of HNSCC subsites. In keeping with the findings of Lapke et al.44 both missense mutations and those introducing a stop codon were associated with worse overall survival when compared to WT. Both Poeta et al.31 and Fallai et al.24 characterised TP53 mutations into either disruptive or silent mutations, with disruptive mutations defined as any stop codons, frameshift mutations, and any mutations inside the L2 or L3 domains of the p53 protein resulting in a change in amino acid charge/polarity. In terms of clinical outcomes, Fallai et al.24 found no difference in survival according to type of TP53 mutation, whilst Poeta et al.31, in the largest cohort identified in this systematic review (n = 420 patients), identified a significant reduction in overall survival amongst patients with disruptive mutations. Lindenbergh-van der Plas et al.26 similarly used the classification of Poeta et al.31 classifying according to the involvement of the p53 DNA-binding domain, and additionally assessed the role of mutation type, namely truncation or missense. Affirming Poeta’s findings, this study identified significantly worse overall survival in the presence of a disruptive mutation, and found that truncating mutations (but not missense) were associated with poorer outcomes on multivariate analysis. In a modest-sized cohort, restricted to oral SCC patients, Yamazaki et al.28 showed that mutations in conserved regions of TP53 or within DNA-binding motifs exhibited poorer survival than cases with other p53 mutations. Similarly, the presence of mutations within DNA-binding regions of p53 were strongly associated with locoregional failure, nodal metastasis and distance metastasis. TP53 mutations of exon 5–8, which encode a region important in stabilisation of the protein tertiary structure and the DNA-binding domain, have also been associated with lower survival rates in laryngeal SCC. Russo et al.30 in a cohort of 81 stage III and IV laryngeal SCC, found that mutations in exon 5 were an independent prognostic factor for both disease-free survival and overall survival, and that exon 8 mutations were independently associated with overall survival but importantly not relapse.
TP53 mutation status and survival: meta-analysis
15 studies provided data from multivariate cox proportional hazards models amenable to inclusion in a meta-analysis; 11 on overall survival26,29,30,31,33,39,42,43,45,46, 8 on disease-free survival26,27,29,30,37,39,45,46 and 3 on disease-specific survival22,36,44. See Fig. 3. Random-effects models were used to quantitatively assess overall survival, disease/progression-free survival, and disease specific survival across reporting studies. Between studies, heterogeneity of outcomes was found to be significant for both disease-specific survival (tau2 = 0.96; I2 = 83.8% [95% CI 51.3; 94.6%], Q(2) = 12.36, p = 0.002) and disease-free survival (tau2 = 0.95; I2 = 51.7% [95% CI 0.0; 78.3%], Q(7) = 14.49, p = 0.04), but not for overall survival (tau2 = 0.220; I2 = 30.1% [95% CI 0.0; 65.5%], Q(10) = 14.3, p = 0.16). Random-effects models determined that overall survival (HR 1.75 [95% CI 1.45–2.10], p < 0.001), disease-specific survival (HR 4.23 [95% CI 1.19–15.06], p = 0.03), and disease-free survival (HR 1.80 [95% CI 1.28–2.53], p < 0.001) were significantly worse in patients with TP53 mutations compared to WT across reporting studies (Fig. 3).
Forest plot graphically representing the meta-analysis on the association between TP53 mutation and survival (overall, disease/progression-free, and disease specific). HR was used as effect size measure. HR > 1 suggests that P53 mutation is associated with reduced survival. Diamonds indicate the pooled HR with corresponding 95% CIs. HR hazard ratio, CI confidence interval.
Discussion
The prognostic value to alteration in the p53 gene (and its function) has been a topic of significant debate in the Head & Neck literature. Previous contrasting reports of the utility of p53 for survival discrimination may, in part, be due to variability in the analytical techniques applied to identify p53 alterations, including identified inconsistencies.
In this systematic review of the literature and associated meta-analysis, we report a comprehensive assessment of TP53 sequencing-defined mutational status as a prognostic factor for survival in patients with HNSCC. The deliberate restriction of p53 status determination to TP53 sequencing technologies alone, allows a significant improvement in understanding by addressing/countering the negative constraints that heterogeneity of mutational status determination has had on prior analyses.
A meta-analysis of the suitable results collated from 15 of the available 25 publications, demonstrated significantly worse outcome with TP53 mutation irrespective of the outcome measure. Although these results are encouraging and represent a progression in the ongoing debate, the authors acknowledge that broadly assigning a global deleterious contribution of TP53 mutation to poor outcomes irrespective of HNSCC subsite or mutation category would constitute a gross oversimplification of the clinical conundrum. Moreover, the problem of heterogeneity persists. Our qualitative analysis highlighted multiple domains of the REMARK criteria which were not sufficiently met. It was particularly notable that guidelines number 9 and 18 (relating to study design and result validation) were poorly met. The number of retrospective studies may, in part, explain this finding as many of the highest scoring papers were those with a prospective study design22,30,31.
A further tumour subsite analysis was not possible as many of the papers reporting data from mixed tumours sites failed to provide a clear breakdown based on anatomical location. Without such evidence based on each subsite, it will be difficult to translate any findings into clinical practice as the contribution of any particular head and neck subsite to the observed overall effect is unclear. Furthermore, the majority of the evidence grouped tumours of all stages, critically increasing heterogeneity, as the two most important prognostic factors in HNSCC are primary tumour size (T stage) and regional nodal status (N stage)47. It is also important to note that the included papers used a variety of treatment modalities which may influence outcome. Likewise a subgroup analysis of treatment modalities was not possible with the data available. A better appreciation for how TP53 mutation status may influence treatment response could open the potential for gene status to be used as a predictive biomarker should an interaction exist between TP53 mutation status and treatment response. Factors such as these should be taken into consideration in the planning of future studies seeking to address these questions.
A failure to address the HPV status of tumours included in various publications similarly contributes to a restriction in the applicability of prognostic determination to specific subsites48.
There is now increasing evidence to suggest that differential mutational profiles of the TP53 gene can influence prognosis in several types of tumours49,50,51. Although Neskey et al.52 was not included in this systematic review, the results are worthy of mention. This study used the TP53 evolutionary action score to determine high-risk and low-risk mutation. The results demonstrated prognostic significance with high-risk mutations conferring reduced overall and disease-free survival when compared to both low-risk mutations and WT TP53. Amongst studies that met the inclusion criteria, only 8 of the 25 studies in this review reported an analysis of clinical outcomes according to location or type of TP53 mutation24,26,28,30,31,33,42,44. Meta-analysis was thus conducted on WT vs mutated TP53, irrespective of mutation location and type.
Circulating tumour DNA (ctDNA) and cell-free DNA (cfDNA) isolated from blood may provide potential for use as blood-based biomarkers in screening, prognostication and monitoring treatment response in HNSCC. However, whilst circulating free DNA (cfDNA) represents an attractive route to explore in the field of precision and personalised oncology, there are current limitations pertaining to testing TP53 mutations in blood. The principal TP53-specific challenge is clonal haematopoiesis which may complicate the interpretation of circulating tumour DNA assays. Clonal haematopoiesis, the accumulation of somatic mutations in haematopoietic stem cells prior to clonal expansion, may result in detectable non-tumour derived mutations in the TP53 gene, with the potential to reduce the sensitivity and specificity to detect true tumour-derived cfDNA.
Despite the limitations highlighted, our meta-analysis demonstrated that TP53 mutations significantly worsen survival in HNSCC. Whilst these data are compelling, to address the highlighted restrictions in application to clinical practice, a prospective study of TP53 status by sequencing is warranted.
The ECOG-ACRIN 3132 trial (ClinicalTrials.gov Identifier: NCT02734537) offers progress towards this ambition with the incorporation of TP53 testing as patient stratification. This phase II trial aims to evaluate disease-free survival of patients with locally advanced HNSCC managed with primary surgical resection and adjuvant radiotherapy with or without cisplatin, with consideration to the influence of disruptive TP53 mutations.
The impact of TP53 mutational state on patient outcomes, stratified by critical clinical and treatment-related variables, remains of significant importance and would require a well characterised, dedicated prospective cohort to resolve understanding. Such an approach would facilitate robust characterisation of the relationship between TP53 status and outcome, in an unbiased manner, critically addressing any potential impact of disease site, stage and/or HPV status, enabling a translation towards clinical practice.
Conclusion
This review epitomises the difficulties encountered when attempting to determine the impact of TP53 mutation on outcomes in HNSCC based on retrospective data. Our qualitative assessment identified room for improvement for future studies and supports the call for high-quality prospective work to investigate our hypothesis. The pooled analysis of all anatomical subsites leads to a heterogeneity that may erode the validity of the observed overall effect and its subsequent extrapolation and application to individual patients. Furthermore, the inter-study variability with regards to HPV status creates a similar issue. Whilst this review and meta-analysis further supports the hypothesis that TP53 mutational status is of prognostic value in HNSCC, a well-designed prospective, multi-centre trial is needed to definitively answer this question prior to clinical translation.
Data availability
All scripts used for data analyses are available upon request from the corresponding author.
References
Sung, H. et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 71, 209–249. https://doi.org/10.3322/caac.21660 (2021).
Johnson, D. E. et al. Head and neck squamous cell carcinoma. Nat. Rev. Dis. Primers 6, 92. https://doi.org/10.1038/s41572-020-00224-3 (2020).
Pulte, D. & Brenner, H. Changes in survival in head and neck cancers in the late 20th and early 21st century: A period analysis. Oncology 15, 994–1001. https://doi.org/10.1634/theoncologist.2009-0289 (2010).
Chaturvedi, A. K. et al. Human papillomavirus and rising oropharyngeal cancer incidence in the United States. J. Clin. Oncol. 29, 4294–4301. https://doi.org/10.1200/jco.2011.36.4596 (2011).
Fakhry, C. et al. Improved survival of patients with human papillomavirus-positive head and neck squamous cell carcinoma in a prospective clinical trial. JNCI J. Natl. Cancer Inst. 100, 261–269. https://doi.org/10.1093/jnci/djn011 (2008).
Masterson, L. et al. De-escalation treatment protocols for human papillomavirus-associated oropharyngeal squamous cell carcinoma: A systematic review and meta-analysis of current clinical trials. Eur. J. Cancer 50, 2636–2648. https://doi.org/10.1016/j.ejca.2014.07.001 (2014).
Schache, A. G. et al. Evaluation of human papilloma virus diagnostic testing in oropharyngeal squamous cell carcinoma: Sensitivity, specificity, and prognostic discrimination. Clin. Cancer Res. 17, 6262–6271. https://doi.org/10.1158/1078-0432.ccr-11-0388 (2011).
Soussi, T. p53 alterations in human cancer: More questions than answers. Oncogene 26, 2145–2156. https://doi.org/10.1038/sj.onc.1210280 (2007).
Lawrence, M. S. et al. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature 517, 576–582. https://doi.org/10.1038/nature14129 (2015).
Tandon, S., Tudur-Smith, C., Riley, R. D., Boyd, M. T. & Jones, T. M. A systematic review of p53 as a prognostic factor of survival in squamous cell carcinoma of the four main anatomical subsites of the head and neck. Cancer Epidem. Biomark. 19, 574–587. https://doi.org/10.1158/1055-9965.epi-09-0981 (2010).
Klinakis, A. & Rampias, T. TP53 mutational landscape of metastatic head and neck cancer reveals patterns of mutation selection. EBioMedicine 58, 102905. https://doi.org/10.1016/j.ebiom.2020.102905 (2020).
Ungerleider, N. A. et al. Breast cancer survival predicted by TP53 mutation status differs markedly depending on treatment. Breast Cancer Res. 20, 115. https://doi.org/10.1186/s13058-018-1044-5 (2018).
Yemelyanova, A. et al. Immunohistochemical staining patterns of p53 can serve as a surrogate marker for TP53 mutations in ovarian carcinoma: An immunohistochemical and nucleotide sequencing analysis. Mod. Pathol. 24, 1248–1253. https://doi.org/10.1038/modpathol.2011.85 (2011).
Moher, D., Liberati, A., Tetzlaff, J., Altman, D. G. & Group, P. Preferred reporting items for systematic reviews and meta-analyses: The PRISMA statement. BMJ 339, b2535. https://doi.org/10.1136/bmj.b2535 (2009).
Riley, R. D. et al. Prognosis research: Toward evidence-based results and a Cochrane methods group. J. Clin. Epidemiol. 60, 863–865. https://doi.org/10.1016/j.jclinepi.2007.02.004 (2007).
Lefebvre, C., Manheimer, E. & Glanville, J. Cochrane Handbook for Systematic Reviews of Interventions: Cochrane Book Series 95–150 (Wiley, 2008).
Systematic Reviews. CRD’s Guidance for Undertaking Reviews in Health Care. Centre for Reviews and Dissemination (University of York, 2009). http://www.york.ac.uk/inst/crd/index_guidance.htm. Accessed 1 April 2021.
McGowan, J. et al. PRESS peer review of electronic search strategies: 2015 guideline statement. J. Clin. Epidemiol. 75, 40–46. https://doi.org/10.1016/j.jclinepi.2016.01.021 (2016).
McShane, L. M. et al. Reporting recommendations for tumor marker prognostic studies. J. Clin. Oncol. 23, 9067–9072. https://doi.org/10.1200/jco.2004.01.0454 (2005).
Sauerbrei, W., Taube, S. E., McShane, L. M., Cavenagh, M. M. & Altman, D. G. Reporting recommendations for tumor marker prognostic studies (REMARK): An abridged explanation and elaboration. JNCI J. Natl. Cancer Inst. 110, 803–811. https://doi.org/10.1093/jnci/djy088 (2018).
Iafolla, M. A. J., Picardo, S., Aung, K. & Hansen, A. R. Systematic review and REMARK scoring of renal cell carcinoma prognostic circulating biomarker manuscripts. PLoS ONE 14, e0222359. https://doi.org/10.1371/journal.pone.0222359 (2019).
Scheel, A. et al. Classification of TP53 mutations and HPV predict survival in advanced larynx cancer. Laryngoscope 126, E292–E299. https://doi.org/10.1002/lary.25915 (2016).
Bosch, F. X. et al. Head and neck tumor sites differ in prevalence and spectrum of p53 alterations but these have limited prognostic value. Int. J. Cancer 111, 530–538. https://doi.org/10.1002/ijc.11698 (2004).
Fallai, C. et al. Oropharyngeal squamous cell carcinoma treated with radiotherapy or radiochemotherapy: Prognostic role of TP53 and HPV status. Int. J. Radiat. Oncol. Biol. Phys. 75, 1053–1059. https://doi.org/10.1016/j.ijrobp.2008.12.088 (2009).
Bradford, C. R. et al. p53 mutation as a prognostic marker in advanced laryngeal carcinoma. Arch. Otolaryngol. Head Neck Surg. 123, 605–609. https://doi.org/10.1001/archotol.1997.01900060047008 (1997).
der Plas, M. L. et al. Prognostic significance of truncating TP53 Mutations in head and neck squamous cell carcinoma. Clin. Cancer Res. 17, 3733–3741. https://doi.org/10.1158/1078-0432.ccr-11-0183 (2011).
Bandoh, N. et al. Prognostic value of p53 mutations, bax, and spontaneous apoptosis in maxillary sinus squamous cell carcinoma. Cancer 94, 1968–1980. https://doi.org/10.1002/cncr.10388 (2002).
Yamazaki, Y. et al. Specific p53 mutations predict poor prognosis in oral squamous cell carcinoma. Oral. Oncol. 39, 163–169. https://doi.org/10.1016/s1368-8375(02)00064-7 (2003).
Alsner, J., Sørensen, S. B. & Overgaard, J. TP53 mutation is related to poor prognosis after radiotherapy, but not surgery, in squamous cell carcinoma of the head and neck. Radiother. Oncol. 59, 179–185. https://doi.org/10.1016/s0167-8140(01)00301-2 (2001).
Russo, A. et al. TP53 mutations and S-phase fraction but not DNA-ploidy are independent prognostic indicators in laryngeal squamous cell carcinoma. J. Cell. Physiol. 206, 181–188. https://doi.org/10.1002/jcp.20447 (2006).
Poeta, M. L. et al. TP53 mutations and survival in squamous-cell carcinoma of the head and neck. N. Engl. J. Med. 357, 2552–2561. https://doi.org/10.1056/nejmoa073770 (2007).
Cho, W. C. S. et al. Genomic characterization reveals potential biomarkers in nasopharyngeal carcinoma patients with relapse. Expert Rev. Mol. Diagn. 20, 1–11. https://doi.org/10.1080/14737159.2020.1835473 (2020).
Caponio, V. C. A. et al. Computational analysis of TP53 mutational landscape unveils key prognostic signatures and distinct pathobiological pathways in head and neck squamous cell cancer. Br. J. Cancer 123, 1302–1314. https://doi.org/10.1038/s41416-020-0984-6 (2020).
Vossen, D. M. et al. Genetic factors associated with a poor outcome in head and neck cancer patients receiving definitive chemoradiotherapy. Cancers 11, 445. https://doi.org/10.3390/cancers11040445 (2019).
Sisk, E. A. et al. Human papillomavirus and p53 mutational status as prognostic factors in head and neck carcinoma. Head Neck 24, 841–849. https://doi.org/10.1002/hed.10146 (2002).
Mineta, H. et al. p53 mutation, but not p53 overexpression, correlates with survival in head and neck squamous cell carcinoma. Br. J. Cancer 78, 1084–1090. https://doi.org/10.1038/bjc.1998.632 (1998).
Resteghini, C. et al. Prognostic role of PIK3CA and TP53 in human papillomavirus–negative oropharyngeal cancers. Tumor. J. 104, 213–220. https://doi.org/10.1177/0300891618765558 (2017).
Kozomara, R., Jović, N., Magić, Z., Branković-Magić, M. & Minić, V. p53 mutations and human papillomavirus infection in oral squamous cell carcinomas: Correlation with overall survival. J. Cranio Maxill. Surg. 33, 342–348. https://doi.org/10.1016/j.jcms.2005.05.004 (2005).
Chomchai, J. S. et al. Prognostic significance of p53 gene mutations in laryngeal cancer. Laryngoscope 109, 455–459. https://doi.org/10.1097/00005537-199903000-00021 (1999).
Miyahara, H. et al. p53 tumor suppressor gene and ras oncogene mutations in hypopharyngeal squamous cell carcinomas. Int. J. Oncol. 11, 133–137. https://doi.org/10.3892/ijo.11.1.133 (1997).
Kozomara, R. J., Brankovic-Magic, M. V., Jovic, N. R., Stosic, S. M. & Magic, Z. M. Prognostic significance of TP53 mutations in oral squamous cell carcinoma with human papilloma virus infection. Int. J. Biol. Mark. 22, 252–257. https://doi.org/10.1177/172460080702200403 (2007).
Mundi, N. et al. Genomic and human papillomavirus profiling of an oral cancer cohort identifies TP53 as a predictor of overall survival. Cancers Head Neck 4, 5. https://doi.org/10.1186/s41199-019-0045-0 (2019).
Dubot, C. et al. Comprehensive genomic profiling of head and neck squamous cell carcinoma reveals FGFR1 amplifications and tumour genomic alterations burden as prognostic biomarkers of survival. Eur. J. Cancer 91, 47–55. https://doi.org/10.1016/j.ejca.2017.12.016 (2018).
Lapke, N. et al. Missense mutations in the TP53 DNA-binding domain predict outcomes in patients with advanced oral cavity squamous cell carcinoma. Oncotarget 7, 44194–44210. https://doi.org/10.18632/oncotarget.9925 (2016).
Kobayashi, K. et al. All-exon TP53 sequencing and protein phenotype analysis accurately predict clinical outcome after surgical treatment of head and neck squamous cell carcinoma. Ann. Surg. Oncol. 26, 2294–2303. https://doi.org/10.1245/s10434-019-07287-x (2019).
Hong, A. et al. Relationships between p53 mutation, HPV status and outcome in oropharyngeal squamous cell carcinoma. Radiother. Oncol. 118, 342–349. https://doi.org/10.1016/j.radonc.2016.02.009 (2016).
Carvalho, A. L., Nishimoto, I. N., Califano, J. A. & Kowalski, L. P. Trends in incidence and prognosis for head and neck cancer in the United States: A site-specific analysis of the SEER database. Int. J. Cancer 114, 806–816. https://doi.org/10.1002/ijc.20740 (2005).
Fakhry, C. et al. Human papillomavirus and overall survival after progression of oropharyngeal squamous cell carcinoma. J. Clin. Oncol. 32, 3365–3373. https://doi.org/10.1200/jco.2014.55.1937 (2014).
Zenz, T. et al. TP53 mutation and survival in aggressive B cell lymphoma. Int. J. Cancer 141, 1381–1388. https://doi.org/10.1002/ijc.30838 (2017).
Christopoulos, P. et al. Detection of TP53 mutations in tissue or liquid rebiopsies at progression identifies ALK+ lung cancer patients with poor survival. Cancers 11, 124. https://doi.org/10.3390/cancers11010124 (2019).
Li, V. D., Li, K. H. & Li, J. T. TP53 mutations as potential prognostic markers for specific cancers: Analysis of data from the cancer genome atlas and the international agency for research on cancer TP53 database. J. Cancer Res. Clin. 145, 625–636. https://doi.org/10.1007/s00432-018-2817-z (2019).
Neskey, D. M. et al. Evolutionary action score of TP53 identifies high-risk mutations associated with decreased survival and increased distant metastases in head and neck cancer. Cancer Res. 75, 1527–1536. https://doi.org/10.1158/0008-5472.can-14-2735 (2015).
Author information
Authors and Affiliations
Contributions
Study concepts: S.B., G.N., I.R., O.J., M.L., B.O., T.J., L.M., T.F., A.S. Study design: S.B., G.N., I.R., O.J., M.L., B.O., T.J., L.M., T.F., A.S. Data acquisition: S.B., G.N., E.B. Quality control of data and algorithms: S.B., G.N., A.F., E.B., A.S., T.J., L.M., A.S. Data analysis and interpretation: S.B., G.N., A.F., T.J., L.M., A.S. Statistical analysis: S.B., A.F. Manuscript preparation: S.B., G.N. Manuscript editing: S.B., G.N., E.B., A.F., I.R., O.J., M.L., B.O., T.J., L.M., T.F., A.S. Manuscript review: S.B., G.N., E.B., A.F., I.R., O.J., M.L., B.O., T.J., L.M., T.F., A.S.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Basyuni, S., Nugent, G., Ferro, A. et al. Value of p53 sequencing in the prognostication of head and neck cancer: a systematic review and meta-analysis. Sci Rep 12, 20776 (2022). https://doi.org/10.1038/s41598-022-25291-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-022-25291-2
- Springer Nature Limited