Abstract
The progressively increasing antimicrobial-resistant Acinetobacter baumannii infections have enforced the use of colistin as the last option for therapy, resulting in the colistin resistance evolution. This work aimed to study the pmrCAB expression in A. baumannii isolates as well as the presence of the mcr-1 gene. Colistin MICs of 100 A. baumannii isolates were measured using the broth microdilution assay. In four colistin-susceptible and four colistin-resistant isolates, the relative expression of the pmrA, pmrB, and pmrC genes was determined using reverse transcription PCR, and then selected isolates were sequenced using the Sanger technique. Finally, the mcr-1 gene was detected using conventional PCR. The colistin resistance rate among the studied isolates was 49%. The expression levels of pmrA and pmrB were statistically significantly higher in colistin-resistant isolates than in colistin-susceptible ones, while the pmrC expression had no statistically significant change. There was a weak positive correlation between colistin MICs and the expression levels of each of the pmrA and pmrB genes. By sequencing, two colistin-resistant strains with low pmrCAB expression showed insertion mutations 3277188_3277189T in pmrB and 1185149_1185150T in pmrC. Only one isolate (1%) was positive for the presence of mcr-1. We concluded that pmrCAB increased expression and/or mutations may cause colistin resistance in A. baumannii. However, increased pmrC expression may not necessarily result in colistin resistance. In Egypt, this is the first study to reveal the existence of mcr-1 in A. baumanni. This should attract attention in clinical settings due to the ultimate tendency of spreading colistin resistance.
Similar content being viewed by others
Introduction
Acinetobacter baumannii causes serious hospital-acquired infections like respiratory tract, urinary tract, bloodstream, surgical site, and wound infections. The abuse of antibiotics has led to the rise of extensively drug-resistant strains, forcing clinicians to utilize colistin as the last treatment option to fight these infections1,2.
Colistin is cyclic, positively charged peptide antibiotic with bactericidal activity against several Gram-negative bacteria due to its interaction with the negatively charged lipid A part of lipopolysaccharide (LPS). So, the bacterial outer membrane (OM) integrity is affected, resulting in cytoplasmic fluid leakage and cell death3.
Unfortunately, the surge in colistin usage in treating infections caused by multi-drug resistant A. baumannii resulted in colistin resistance spread4. The most important colistin resistance mechanisms identified in A. baumannii are (1) the LPS modification mediated by the two-component system PmrA/PmrB and (2) the LPS loss caused by lipid A impaired synthesis5,6. Recently, a plasmid-mediated colistin resistance gene has been also involved in colistin resistance7. The pmrA/pmrB encodes a two-component response regulator and sensor kinase system that helps bacteria to sense and respond to environmental factors. In addition, they influence the pmrC gene expression, which encodes lipid A phosphoethanolamine (PEtN) transferase that adds PEtN to the lipid A part of the LPS. LPS modification causes less negative charge and decreased colistin binding affinity to its target on the LPS5,8.
The mcr-1 is a plasmid-mediated gene that encodes a PEtN transferase that causes colistin resistance9. In 2015, it was found in Escherichia coli obtained from a variety of clinical, animal, and environmental specimens from China10. Furthermore, mcr-1 was discovered for the first time in P. aeruginosa and A. baumannii isolated from a variety of clinical samples in Pakistan7.
The aim of the present study was to investigate the expression of pmrCAB and mutations in A. baumannii isolates, as well as the existence of the plasmid-encoded mcr-1.
Materials and methods
We conducted a descriptive cross-sectional study from January 2020 to March 2021. The Clinical Laboratories of Cairo University hospitals in Cairo, Egypt provided 100 Acinetobacter species isolates. They were obtained from different clinical specimens, including purulent discharge, sputum, pleural fluid, urine, blood, and ascitic fluid. All isolates were transferred to the Medical Microbiology and Immunology Department, Faculty of Medicine, Cairo University for confirmation and further processing. The research was authorized by the Ethics Committee of the Institutional Review Board (Code: MD-254–2019), Faculty of Medicine, Cairo University, Egypt.
-
I.
Culture and identification of Acinetobacter isolates
Each of the collected isolates was sub-cultured on MacConkey’s medium and then incubated aerobically at 37 °C for 24 h. Identification of Acinetobacter spp. was done by conventional biochemical reactions. Acinetobacter was identified as colourless or light lavender colonies, non-motile, Gram-negative, catalase-negative, oxidase-negative, non-fermentative cocco-bacilli11.
-
II.
Molecular identification of Acinetobacter baumannii
Following the manufacturer's instructions, DNA extraction from all Acinetobacter isolates was done using the GF-1 Bacterial DNA Extraction Kit (Vivantis, Malaysia). Using PCR, amplification of the A. baumannii species-specific blaOXA-51-like gene was done12. It was performed using a ready-to-use PCR master mix (CinnaGen, Iran). Specific primers for the blaOXA-51-like gene (forward primer: TAATGCTTTGATCGGCCTTG, and reverse primer: TGGATTGCACTTCATCTTGG) were used12. The DNA amplification was performed in Rotor-Gene Q MDX (Qiagen, Germany). The cycling conditions were as follows: initial denaturation at 94 °C for 3 min, followed by 34 cycles of denaturation at 94 °C for 30 s, primer annealing at 55 °C for 30 s, and extension at 72 °C for 30 s, followed by a final extension at 72 °C for 5 min12. The amplified PCR products (353 bp) were run in agarose gel electrophoresis along with a 50 bp molecular weight ladder (Vivantis, Malaysia) and then seen under an ultra-violet (UV) transilluminator.
-
III.
Detection of colistin resistance
According to the European Committee on Antimicrobial Susceptibility Testing (EUCAST) guidelines, the colistin minimal inhibitory concentration (MIC) of each A. baumannii isolate was evaluated using the broth microdilution method with Cation-Adjusted Muller Hinton Broth (CA-MHB) (Iiofilchem, Italy)13. Using EUCAST resistance breakpoints for A. baumannii, colistin resistance was considered if MIC is > 2 mg/L13.
-
IV.
Detection of the expression of pmrA, pmrB and pmrC genes
The relative expression of pmrA, pmrB and pmrC genes was measured using qRT-PCR. RNA extraction was done using HiPurA™ Bacterial RNA Purification Kit (HiMedia, India) according to the manufacturer’s instructions. The extracted RNA was used for the synthesis of cDNA using SinaClon First Strand cDNA synthesis Kit (CinnaGen, Iran).
The expression levels of pmrA, pmrB, and pmrC genes were measured using the primer sets shown in Table 114. Quantitative RT-PCR was done using Thermo Scientific Maxima SYBR Green/ROX qPCR Master Mix (2X) (Thermofisher Scientific, US).
The prepared reaction mixtures were processed in Rotor-Gene Q MDX with software version 2.3.3 (Qiagen, Germany) for amplification and analysis. After the qRT-PCR run, data were expressed in Cycle threshold (Ct). The target genes’ expression levels were normalized using the housekeeping 16S rRNA gene expression then their analysis was finalized using the comparative 2−∆∆CT process.
-
V.
DNA sequencing of pmrA, pmrB, and pmrC genes
Eight A. baumannii isolates were selected for DNA sequencing. These isolates included two colistin-resistant isolates with high pmrCAB expression (isolates #16 and 41), two colistin-resistant isolates with low pmrCAB expression (isolates # 85 and 71), two colistin-susceptible isolates with high pmrCAB expression (isolates # 1 and 44) and two colistin-susceptible isolates with low pmrCAB expression (isolates # 4 and 43).
A portion of each of pmrA, pmrB, and pmrC genes was amplified using the primers shown in Table 115. The amplified PCR products were sent to Color Laboratory, Egypt, for Sanger sequencing. BigDye® XTerminator™ Purification Kit (Applied Biosystems, USA) was used for purifying the amplified PCR products according to the instructions of the manufacturer. Cycle DNA sequencing was done in the forward direction using BigDye™ Terminator v3.1 Cycle Sequencing Kit (Thermo Scientific, USA) according to the manufacturer’s instructions. The sequenced products were run on a 3500 Genetic Analyzer (Applied Biosystems, USA).
-
VI.
Detection of the plasmid-encoded mcr-1
All A. baumanni isolates were subjected to PCR to amplify the mcr-1 gene using the following primers: the forward primer: CGGTCAGTCCGTTTGTTC, and the reverse primer: CTTGGTCGGTCTGTAGGG5,7. The DNA amplification was performed in Rotor-Gene Q MDX (Qiagen, Germany). The amplified product (309 bp) was run in agarose gel electrophoresis along with a 50 bp molecular weight ladder (Cleaver Scientific, UK) and then seen under an ultra-violet (UV) transilluminator. The positive control used in this study was an Escherichia coli isolate carrying the mcr-1 gene previously identified by sequencing. It was supplied by the Medical Microbiology and Immunology Department, Faculty of Medicine, Cairo University.
Statistical analysis
Data were analyzed using the statistical package for the Social Sciences (SPSS) version 26 (IBM Corp., Armonk, NY, USA). Quantitative data were summarized using the mean and standard error of the mean (SEM), whereas categorical data were summarized using frequency (count) and relative frequency (percentage). For comparing quantitative variables, the non-parametric Mann–Whitney test was utilized. For comparing categorical data, the Chi-square (2) test was utilized. When the anticipated frequency was < 5, the exact test was used instead.
To calculate correlations between quantitative variables, the Spearman correlation coefficient was used. A receiver operating characteristic (ROC) curve was constructed with the area under the curve (AUC) analysis to detect the best cutoff value of RQ of pmrA, pmrB and pmrC genes for discrimination between sensitive and resistant isolates. To determine statistical significance, a probability value (P) equal to or < 0.05 was employed.
Ethics approval
This study was approved by the Research Ethics Committee of the Institutional Review Board (Code: MD-254–2019), Faculty of Medicine, Cairo University.
Results
One hundred Acinetobacter isolates were obtained from the laboratories of Cairo University hospitals. 65% of the isolates were from patients in ICUs. The isolates were from 48 male and 52 female patients with a mean age of 50.03 ± 20.72 years. Those isolates were collected from various clinical samples including purulent discharge (32), sputum (29), blood (20), urine (14), pleural fluid (3), and ascitic fluid (2). Molecular detection of the blaOXA-51-like gene confirmed that all isolates (100%) were A. baumannii. 49% of the studied A. baumannii isolates were colistin-resistant, while 51% were colistin-susceptible (Supplementary Table S1),
Expression of pmrCAB genes
Analysis of pmrCAB expression showed that pmrA expression levels were statistically significantly higher in colistin-resistant isolates (mean ± SEM = 46.84 ± 13.78) compared to the colistin-susceptible isolates (mean ± SEM = 4.56 ± 2.74) (P < 0.001) (Table 2 and Fig. 1A).
Similarly, pmrB expression levels were statistically significantly higher in colistin-resistant isolates (mean ± SEM = 64.31 ± 16.52) compared to the colistin-susceptible isolates (mean ± SEM = 5.25 ± 3.14) (P = 0.001) (Table 2 and Fig. 1B). Concerning pmrC expression levels, no significant difference was found between colistin-resistant and colistin-susceptible isolates (Table 2). Expression levels of pmrA, pmrB, and pmrC are shown in Supplementary Table S1.
The Spearman correlation coefficient test revealed a weak positive correlation between the MIC values of colistin and the expression levels of each of pmrA (r = 0.399, P < 0.001) and pmrB genes (r = 0.383, P < 0.001). However, there was insignificant correlation between pmrC expression levels and colistin MICs (r = 0.149, P = 0.139).
ROC curve analysis revealed that pmrA expression could differentiate between colistin-susceptible and colistin-resistant isolates showing sensitivity of 63.3% and a specificity of 88.2% at a cutoff value = 2.7105 (AUC 0.713, P < 0.001) (Fig. 2A).
Similarly, the expression levels of pmrB could differentiate between colistin-susceptible and colistin-resistant isolates showing sensitivity of 61.2% and a specificity of 88.2% at a cutoff value = 2.3219 (AUC = 0.699, P < 0.001) (Fig. 2B). On the other hand, the pmrC expression levels could not differentiate between colistin-susceptible and colistin-resistant isolates (AUC = 0.544, P = 0.459).
Sequencing of pmrCAB genes
The sequences of the pmrA, pmrB, and pmrC genes of the 8 isolates were compared with the reference sequence of A. baumannii ATCC 19,606 (GenBank: CP045110.1) (https://www.ncbi.nlm.nih.gov/nuccore/CP045110.1).
Insertion mutations were observed in both colistin-resistant isolates with low pmrC expression. Isolate number 71 showed 3277188_3277189T in pmrB. Isolate number 85 showed 1185149_1185150T in pmrC, in addition to two substitutions (1185196 T > G and 1185199A > C).
Detection of plasmid-mediated mcr-1 gene
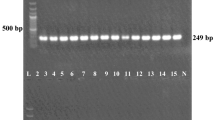
We found only one (1%) A. baumannii isolate that carried mcr-1 (Fig. 3). The colistin MIC of this isolate was 4 mg/L (Supplementary Table S1).
Discussion
A. baumannii drug resistance is spreading rapidly around the world and is impacted by various factors; one of them is antibiotic misuse and overuse16. The increasing development of carbapenem-resistant A. baumannii has made colistin one of the last therapeutic options used in treating various infections. Colistin-resistant A. baumannii strains have been reported since the reintroduction of colistin in human clinical practice17.
Our study showed that 49% A. baumannii isolates were colistin-resistant. Similar results have been found by Papathanakos et al.17 in Greece (41%) and Gerson et al.18 in Germany (48%), while a higher rate was reported by Al-Kadmy et al.19 in Iraq (76%). Lower rates were reported by Hameed et al.7 in Pakistan (9.6%) and Lukovic et al.20 in Serbia (4.3%). The high prevalence of resistance to colistin in the present work could be explained by the fact that our isolates were obtained from hospitalized patients since resistance patterns are more frequent in hospitalized patients due to the overuse of antibiotics as well as the presence of co-morbidities.
In the current study, pmrA and pmrB genes were statistically significantly overexpressed in colistin-resistant isolates compared to colistin-susceptible isolates. Similar results were reported by previous studies21,22,23. Nevertheless, Sepahvand et al.15 although found higher expression of pmrA in colistin-resistant isolates than colistin-sensitive isolates, they reported that the expression of pmrB had no significant change. In fact, overexpression of the pmrA and pmrB genes decreases lipopolysaccharide, leading to reduced permeability and colistin ineffectiveness on the A. baumannii membrane15. Additionally, Colistin resistance in A. baumannii is caused by mutations in the pmrAB genes21,24.
Alternatively, the present study showed that the pmrC expression level had no statistically significant change between colistin-susceptible and colistin-resistant isolates. Even though overexpression of pmrC is associated with reduction of colistin susceptibility and is frequently caused by mutations in the regulators pmrAB25,26, increased pmrC expression has been reported without association with increased colistin MICs18.
Moreover, we noticed a weak positive correlation between colistin MIC values and the expression levels of each of the pmrA and pmrB genes. However, there was a non-significant correlation between pmrC expression levels and colistin MICs. On the contrary, Nurtop et al.5 found a positive correlation between pmrC expression levels and colistin MIC values.
Adams et al.25 demonstrated increased pmrA expression in colistin-resistant mutants, which autoregulates the pmrCAB promoter. They also reported that partial deletion of pmrB in colistin-resistant mutant leads to reversion to a colistin-susceptible phenotype, emphasizing the role of the PmrAB system in the regulation of colistin resistance in A. baumannii25.
A paucity of studies investigated the pmrA and pmrB expression in colistin-resistant A. baumanni. However, previous research demonstrated that pmrA and pmrB mutations lead to increased pmrC expression and hence the addition of PEtN to lipid A. This observation was detected in both colistin-resistant and colistin-susceptible isolates, suggesting that this resistance mechanism may be strain-specific and that other unknown factors may be involved in resistance to colistin27. Furthermore, Nurtop et al.5 reported that the same clone isolates have different patterns of expression of pmrC, pmrA, and pmrB. This finding confirms the importance of environmental and host factors on the behavior of gene expression. Colistin use was shown in several studies to be a substantial risk factor for the emergence of chromosomal colistin resistance28.
Colistin susceptibility testing is performed by MIC determination, which is the gold standard phenotypic method, but it takes about 24 h to get the results. Molecular methods are often used in the detection of antimicrobial resistance besides phenotypic methods. Molecular methods are expected to replace phenotypic methods in many laboratories because of their higher speed and precision in identifying genetic mechanism(s) for antimicrobial resistance29. In the current study, ROC curve analysis revealed that pmrA and pmrB expressions could differentiate between colistin-susceptible and colistin-resistant isolates, with moderate sensitivity (63.3%, 61.2%) and a considerable specificity (88.2%, 88.2%) at a cutoff value of 2.7105 and 2.3219 for pmrA and pmrB respectively. On the other hand, the expression levels of pmrC could not differentiate between them. To our knowledge, this study is the first to assess the expression of pmrA, pmrB, and pmrC in the detection of colistin resistance in A. baumannii. Further studies about molecular detection of colistin resistance by detection of the expression of pmrA and pmrB are needed.
In the current study, we observed two nucleotide insertions that caused frameshift mutations in pmrB and in pmrC in the two colistin-resistant isolates with low pmrC expression. One of the two isolates also showed two substitutions in pmrC. On the other hand, previous studies reported pmrCAB mutations in association with increased pmrC expression, some of these mutations occurred during treatment with colistin5,8,18,30. The two-component pmrA/pmrB system regulates the pmrC expression and subsequently, mutations of pmrAB may increase the pmrC expression3.
The insertion and substitution mutations detected in this study in the two colistin-resistant isolates with decreased pmrC expression indicate that these pmrB/pmrC mutations may induce colistin resistance in A. baumannii without an increase in gene expression. It is unclear how amino acid mutations in the pEtN transferase influence colistin MICs, but an increase in enzyme function or activity is postulated18. Nevertheless, other colistin resistance mechanisms cannot be excluded.
The additive effect of mutations suggested that mutation accumulation could be a mechanism for increased antibiotic resistance27,31, even in the absence of increased gene expression. It can be clarified by the fact that some mutations may increase the gene function, or the activity of a given gene product (gain-of-function mutation) rather than loss of gene function32,33.
The mcr-1 gene, a plasmid-mediated colistin-resistant gene, was discovered in an E. coli isolate of animal origin in China. It was then detected nearly worldwide in bacterial isolates from animals34, humans35, and environmental sources19. Resistance transmitted by plasmids has two drawbacks. First, plasmids can bear resistance to several antibiotics. Second, plasmids can spread resistance among the bacteria at a greater rate than that which occurs via spontaneous mutation. Therefore, colistin-resistant bacteria may rapidly become widespread in the clinical setting in the lack of new antibiotics against MDR bacteria36.
In the current study, out of the 100 A. baumannii isolates only one isolate carried mcr-1. Hameed et al.7 reported a similar result in Pakistan and found that out of 62 isolates of human origin, only one isolate (1.6%) was positive for the mcr-1. A higher incidence rate was reported by Rahman and Ahmed19 in India who found that out of 100 A. baumannii isolates of human origin, 20 (20%) were positive for mcr-1 and Al-Kadmy et al.37 in Iraq found that out of 121 A. baumannii isolates of human and environmental origin, 89 (73.5%) isolates carried the mcr-1 gene. However, in Iran, Khoshnood et al.23 reported that none of the 70 identified A. baumannii isolates carried the mcr-1 resistance gene.
According to our knowledge, this study is the first to report the presence of the mcr-1 gene in a human clinical A. baumannii isolate in Egypt. This finding raises issues to investigate other mcr genes in A. baumannii in future studies. This plasmid-mediated mechanism is important because it increases the horizontal transferability of colistin resistance of A. baumannii in clinical settings. Furthermore, colistin resistance may occur because of pmrCAB overexpression and/or mutations. Further studies are recommended to confirm the biological functions of point mutations of pmr genes and their role in colistin resistance. Strategies are required to stop the spread and evolution of such genes. Our research sheds light on the complexity of colistin resistance in A. baumannii. Further research is required to correlate colistin resistance and its molecular mechanisms to strain types and to understand other colistin resistance mechanisms such as studying the lpx genes. Moreover, it is vital to establish guidelines regarding the use of colistin to inhibit the emergence of resistance.
Data availability
A. baumannii strains are available from the authors. All data are fully available without restriction. All relevant data are within the manuscript and its Supporting Information files. The following are the GenBank accession numbers for nucleotide sequences: BankIt2586606 pmrA16 ON614677; BankIt2586606 pmrA41 ON614678; BankIt2586606 pmrA85 ON614679; BankIt2586606 pmrA71 ON614680; BankIt2586606 pmrA1 ON614681; BankIt2586606 pmrA44 ON614682; BankIt2586606 pmrA4 ON614683; BankIt2586606 pmrA43 ON614684; BankIt2586606 pmrB16 ON614685; BankIt2586606 pmrB41 ON614686; BankIt2586606 pmrB85 ON614687; BankIt2586606 pmrB71 ON614688; BankIt2586606 pmrB1 ON614689; BankIt2586606 pmrB44 ON614690; BankIt2586606 pmrB4 ON614691; BankIt2586606 pmrB43 ON614692; BankIt2586606 pmrC16 ON614693; BankIt2586606 pmrC41 ON614694; BankIt2586606 pmrC85 ON614695; BankIt2586606 pmrC71 ON614696; BankIt2586606 pmrC1 ON614697; BankIt2586606 pmrC44 ON614698; BankIt2586606 pmrC4 ON614699; BankIt2586606 pmrC43 ON614700.
References
Peleg, A. Y., Seifert, H. & Paterson, D. L. Acinetobacter baumannii: emergence of a successful pathogen. Clin. Microbiol. Rev. 21(3), 538–582 (2008).
Wong, D. et al. Clinical and pathophysiological overview of Acinetobacter infections: A century of challenges. Clin. Microbiol. Rev. 30, 409–447 (2017).
Falagas, M. E. & Kasiakou, S. K. Colistin: the revival of polymyxins for the management of multidrug resistant gram-negative bacterial infections. Clin. Infect. Dis. 40(9), 1333–1341 (2005).
Qureshi, Z. A. et al. Colistin-resistant Acinetobacter baumannii: beyond carbapenem resistance. Clin. Infect. Dis. 60(9), 1295–1303 (2015).
Nurtop, E. et al. Promoters of colistin resistance in Acinetobacter baumannii infections. Microb. Drug Resist. 25, 997–1002 (2019).
Nhu, N. T. K. et al. The induction and identification of novel Colistin resistance mutations in Acinetobacter baumannii and their implications. Sci. Rep. 6, 1–8 (2016).
Hameed, F. et al. Plasmid-mediated mcr-1 gene in Acinetobacter baumannii and Pseudomonas aeruginosa: first report from Pakistan. Rev. Soc. Bras. Med. Trop. 52, e20190237 (2019).
Marano, V. et al. Identification of pmrB mutations as putative mechanism for colistin resistance in A. baumannii strains isolated after in vivo colistin exposure. Microb. Pathog. 142, 104058 (2020).
Jeannot, K., Bolard, A. & Plésiat, P. Resistance to polymyxins in Gram-negative organisms. Int. J. Antimicrob. Agents 49(5), 526–535 (2017).
Liu, Y. Y. et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect. Dis. 16(2), 161–168 (2016).
Gupta, N., Gandham, N., Jadhav, S. & Mishra, R. N. Isolation and identification of Acinetobacter species with special reference to antibiotic resistance. J. Nat. Sci. Biol. Med. 6(1), 159–162 (2015).
Pal, N., Sujatha, R. & Kumar, A. Phenotypic and genotypic identification of acinetobacter baumannii with special reference to blaOXA-51-like gene and its antimicrobial susceptibility pattern from intensive care unites in Kanpur. IJCM R. 4(5), 1154–1158 (2017).
European Committee on Antimicrobial Susceptibility Testing. Breakpoint tables for interpretation of MICs and zone diameters. Version 11.0, 2021. http://www.eucast.org.
Beceiro, A. et al. Phosphoethanolamine modification of lipid A in colistin-resistant variants of Acinetobacter baumannii mediated by the pmrAB two-component regulatory system. Antimicrob. Agents Chemother. 55, 3370–3379 (2011).
Sepahvand, S. et al. Molecular evaluation of colistin-resistant gene expression changes in Acinetobacter baumannii with real-time polymerase chain reaction. Infect. Drug Resist. 10, 455–462 (2017).
Jin, H., Qiu, F., Ji, H. J. & Lu, Q. Analysis of drug resistance in 1,861 strains of Acinetobacter baumannii. Biomed. Rep. 4, 463–466 (2016).
Papathanakos, G. et al. Colistin-resistant Acinetobacter Baumannii bacteremia: A serious threat for critically ill patients. Microorganisms 8(2), 287 (2020).
Gerson, S. et al. Diversity of amino acid substitutions in PmrCAB associated with colistin resistance in clinical Acinetobacter baumannii isolates. Int. J. Antimicrob. Agents 55(3), 105862 (2020).
Al-Kadmy, I. M. S. et al. Prevalence of genes involved in colistin resistance in Acinetobacter baumannii: First report from Iraq. Microb. Drug Resist. 26(6), 616–622 (2020).
Lukovic, B. et al. The first nationwide multicenter study of Acinetobacter baumannii recovered in Serbia: Emergence of OXA-72, OXA-23 and NDM-1-producing isolates. Antimicrob. Resist. Infect. Control 9(1), 101 (2020).
Park, Y. K., Choi, J. Y., Shin, D. & Ko, K. S. Correlation between overexpression and amino acid substitution of the PmrAB locus and colistin resistance in Acinetobacter baumannii. Int. J. Antimicrob. Agents 37(6), 525–530 (2011).
Zhang, W. et al. The role of LpxA/C/D and pmrA/B gene systems in colistin-resistant clinical strains of Acinetobacter baumannii. Front. Lab. Med. 1(2), 86–91 (2017).
Khoshnood, S., Savari, M., Montazeri, E. A. & Sheikh, A. F. Survey on genetic diversity, biofilm formation, and detection of colistin resistance genes in clinical isolates of Acinetobacter baumannii. Infect. Drug Resist. 13, 1547–1558 (2020).
Lean, S. S. et al. Prevalence and genetic characterization of polymyxin-resistant Acinetobacter baumannii isolated from a tertiary hospital in Terengganu, Malaysia. ISRN Microbiol. 953417 (2014).
Adams, M. D. et al. Resistance to colistin in Acinetobacter baumannii associated with mutations in the PmrAB two-component system. Antimicrob. Agents Chemother. 53, 3628–3634 (2009).
Olaitan, A. O., Morand, S. & Rolain, J. M. Mechanisms of polymyxin resistance: Acquired and intrinsic resistance in bacteria. Front. Microbiol. 5, 643 (2014).
Gerson, S. et al. Investigation of novel pmrB and eptA mutations in isogenic Acinetobacter baumannii isolates associated with colistin resistance and increased virulence in vivo. Antimicrob. Agents Chemother. 63(3), e01586-e1618 (2019).
Wang, Y. C. et al. Risk factors and outcome for colistin-resistant Acinetobacter nosocomialis bacteraemia in patients without previous colistin exposure. Clin. Microbiol. Infect. 21, 758–764 (2015).
Anjum, M. F., Zankari, E., Hasman, H. Molecular methods for detection of antimicrobial resistance. Microbiol. Spectr. 5(6) (2017).
Ghahraman, M. R. K. et al. Molecular characterization of lpxACD and pmrA/B two-component regulatory system in the colistin resistance Acinetobacter baumannii clinical isolates. Gene Rep. 21, 100952 (2020).
Palmieri, M. et al. Abundance of colistin-resistant, OXA-23- and ArmA-producing Acinetobacter baumannii belonging to international clone 2 in Greece. Front. Microbiol. 11, 668 (2020).
Lodish, H., Berk, A. & Zipursky, S. L. Molecular Cell Biology 4th edn (W. H. Freeman, 2000). Section 8.1, Mutations: Types and Causes.
Alberts B. et al. Molecular Biology of the Cell, 4th edn (Garland Science, 2002). Studying Gene Expression and Function.
Irrgang, A. et al. Prevalence of mcr-1 in E. coli from livestock and food in Germany, 2010–2015. PLoS ONE 11(7), e0159863 (2016).
Ye, H. et al. Diversified mcr-1-harbouring plasmid reservoirs confer resistance to colistin in human gut microbiota. MBio 7(2), e00177 (2016).
Liassine, N. et al. Very low prevalence of MCR-1/MCR-2 plasmid-mediated colistin resistance in urinary tract Enterobacteriaceae in Switzerland. Int. J. Infect. Dis. 51, 4–5 (2016).
Rahman, M. & Ahmed, S. Prevalence of colistin resistance gene mcr-1 in clinical Isolates Acinetobacter Baumannii from India. IJID 101(S1), 81 (2020).
Funding
Open access funding provided by The Science, Technology & Innovation Funding Authority (STDF) in cooperation with The Egyptian Knowledge Bank (EKB). This research was funded by the Faculty of Medicine, Cairo University. Cairo University Institute provided us with the PCR kits. https://cu.edu.eg/Home. The funders had no role in the design of the study; in the collection, analysis, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Author information
Authors and Affiliations
Contributions
N.H.O.: Supervision, Validation, Project Administration. M.S.M.: Supervision, Investigation, Validation, Writing—Review and Editing. R.Y.S.: Resources, Formal Analysis, Supervision, Writing—Review and Editing. S.M.S.: Conceptualization, Methodology, Investigation, Writing—Original Draft Preparation.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Seleim, S.M., Mostafa, M.S., Ouda, N.H. et al. The role of pmrCAB genes in colistin-resistant Acinetobacter baumannii. Sci Rep 12, 20951 (2022). https://doi.org/10.1038/s41598-022-25226-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-022-25226-x
- Springer Nature Limited