Abstract
Continuing studies imply that m6A RNA modification is involved in the development of cervical cancer (CC), but lack strong support on recurrence and diagnosis prediction. In this research, a comprehensive analysis of 33 m6A regulators was performed to fulfill them. Here, we performed diagnostic and prognosis models and identified key regulators, respectively. Then the CC patients were separated into two clusters in accordance with 33 regulators, and participants in the cluster 1 had a worse prognosis. Subsequently, the m6AScore was calculated to quantify the m6A modification pattern based on regulators and we found that patients in cluster 1 had higher m6AScore. Afterwards, immune microenvironment, cell infiltration, escape analyses and tumor burden mutation analyses were executed, and results showed that m6AScore was correlated with them, but to a limited extent. Interestingly, HLAs and immune checkpoint expression, and immunophenoscore in patients with high-m6AScores were significantly lower than those in the low-m6AScore group. These suggested the m6AScores might be used to predict the feasibility of immunotherapy in patients. Results provided a distinctive perspective on m6A modification and theoretical basis for CC diagnosis, prognosis, clinical treatment strategies, and potential mechanism exploration.
Similar content being viewed by others
Introduction
Cervical cancer (CC), which is currently the fourth most common malignancy in women worldwide, led the cause of malignant tumor deaths and a heavy social burden in developing countries1. Although early screening and effective interventions can prevent the occurrence of CC and improve the prognosis of CC, the situation is still severe, such as the high recurrence rate of CC. Apart from the recognized factor of HPV infection, its pathogenesis is not fully understood2,3. Given the deleterious influence of CC, efforts are needed to explore the potential biomarkers for the diagnosis and prognosis, as well as feasible treatment strategies.
N6-methyladenosine (m6A) RNA methylation is the most common conserved internal transcriptional and modification epigenetic modification. m6A is a dynamic process, and three kinds of essential regulators (known as “writer”, “eraser”, and “reader”) are involved in the regulation of this modification process in the human body, leading to several facets changes in RNA processing, including RNA stability, alternative splicing and translation4,5,6,7,8,9,10,11. As a hot spot in epigenetic research in recent years, the fundamental role of m6A in cancer development and prognosis may help us clarify the mechanism of CC with a novel perspective.
A growing body of literature studies has shown that the imbalance of m6A modification regulators affects a series of biomolecular events by influencing target RNAs, and ultimately affecting the occurrence, development, and prognosis of many diseases including cancers. Evidence indicates that upregulation induced by m6A methylation could contribute to the increased cancer stemness cell in colon cancer12. In addition, the abnormal expression of methyltransferase complex components in m6A modification affects both gastric cancer and liver cancer, including adjusted by FTO and WTAP instead of METTL3 and METTL14, which were considered as principal roles in m6A modification13,14. Collectively, although the mechanism of m6A modification in cancer including CC has been well studied in recent years15,16, the evidence for the importance of m6A modification importance in CC recurrence and diagnostic analysis is still lacking.
In this study, we used CC patient data from GEO and TCGA to comprehensively analyze 33 m6A regulators and their indicative roles for both CC diagnosis and prognosis. Essential targets were identified by constructing diagnostic and prognostic models of CC, and drug sensitivity analysis was carried out based on these factors. We also subsequently assessed the potential functions of m6A RNA regulators through immune prediction and enrichment analyses, and explored the clinical treatment strategies of CC.
Materials and methods
Ethical conduct of research
The authors pointed out that the Ethics Committee of Southeast University approved this study, and the informed consent was acquired from each participant recruited and all samples were used in compliance with the institution’s ethical regulations. The research design was in accordance with the Declaration of Helsinki.
Data resource
The transcriptome sequencing data (read counts and FPKM normalized) of 306 CC and three normal patients were downloaded from The Cancer Genome Atlas (TCGA) portal (https://tcga-data.nci.nih.gov/tcga/) and transformed obtained gene expression values into per kilobase million (TPM) values. The expressions of m6A modification regulators in patients from the data matrix were extracted for the subsequent analysis. The CC clinical information was downloaded from the TCGA portal. The GSE63514 and GSE6791 data were downloaded from the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/gds).
Selection of m6A RNA methylation regulators
We first aimed to m6A modification regulators from published literature17,18,19. Subsequently, we extracted the expression levels of regulators from the TCGA and cervical tissue transcription data, deleting the regulators in which expression levels were unavailable. Finally, 33 m6A modification regulators were selected as candidate molecules for this study16,20 (Table S1).
Significant differential expression gene (DEGs) analysis
To screen the regulators which play an essential role in the development of CC, differential expression analysis was performed using the R program with the “DESeq2” package. Among all the gene analyzed, FDR (false discovery rate) < 0.05 were considered as a criterion and the DEGs were obtained for subsequent analysis. Gene copy number variation (CNV) analysis was shown by cBioportal tools (http://www.cbioportal.org/). Venn plot was displayed by the venny 2.1 tool (https://bioinfogp.cnb.csic.es/tools/venny/).
Consensus clustering for subgroups identification
To investigate the function of regulators in CC, we clustered cancer tissues into two subgroups by R program with the “ConsensusClusterPlus” package. Then the principal component analysis (PCA) analysis and t-distributed stochastic neighbor embedding (t-SNE) were utilized to study the gene expression patterns in different CC subgroups. Furthermore, Kaplan–Meier analysis was drawn to assess prognosis between subgroups and compared using the log-rank test.
Pathway analysis and acquisition of gene sets
Gene Set Cancer Analysis (GSCA, bioinfo.life.hust.edu.cn/web/GSCALite/) database was used to find clues about m6A regulators in biological processes. GSEA analysis of patients in two clusters was performed using c2.cp.kegg.v7.4.symbols.gmt and c5.go.v7.4.symbols.gmt downloaded from the Gene Set Enrichment Analysis (GSEA, https://www.gsea-msigdb.org/gsea/index.jsp) database. Then, the gene sets of interest were downloaded from the GSEA database, including BIOCARTA_CELLCYCLE_PATHWAY (M17770), BIOCARTA_CASPASE_PATHWAY (M17902), REACTOME_PYROPTOSIS (M41805) and WP_FERROPTOSIS (M39768). Moreover, the ferroptosis-related gene list was also obtained from the FerrDb database (http://www.zhounan.org/ferrdb/). The genes related to programmed necroptosis21 and cuproptosis22 were identified from literature reports.
Identify important molecular markers through machine learning
To further accurately identified the critical m6A regulators affecting the diagnosis of CC, we used machine learning methods to construct a diagnostic model of CC, ranked the variables according to their importance, and visualized results by the R program.
The random forest (RF) model is a typical classifier that containing many decision trees. Patients with replacements were randomly selected from the initial dataset to assemble a sub-dataset. In this study, most of the 54 patients (28 cases and 24 normal control) from GSE63514 were used as a training set, while left participants were analyzed as a validation set. Based on the seed number 51, ROC was used to fit the optimal model using the most considerable value, and the final value selected for the RF model was mtry = 2. The fivefold cross-validation method was also applied during the analysis. RF was executed by the “randomForest” package.
The Support Vector Machines (SVM) model is an algorithm that is widely used in binary and multiple classifications. SVM core function can convert samples that were inseparable in low-dimensional into high-dimensional separable space to achieve better grouping. In this study, the tuning parameter ‘sigma’ was held constant at a value of 0.01744768, and the SVM model was performed by the “e1071” package of R.
Artificial Neural Network (ANN) is a complex network structure formed by interconnecting a series of treating units, which has been proved to be scientific and accurate in disease prediction in recent years23,24,25. In this study, we utilized the common regulators selected by RF and SVM as the input layer to construct an ANN model. Feedforward neurons generated a backpropagation during the training process, and the error rate of this backpropagation reflected the discrepancy between the model judgment and the actual patient status. During data transfer and processing, one output was produced, the result of the classification. After adjusting the weight of the input data, with the appropriate back-propagation error range as evidence, the best classification method was finally achieved. A total of 28 patients and 24 normal control from GSE63514 were used as a training set, while 19 cases and nine normal participants from GSE6791were analyzed as a validation set. The R packages “neuralnet” and “NeuralNetTools” were applied in this process.
Construction and validation of the LASSO Cox regression algorithm
To study the prognostic value of m6A RNA methylation regulators, a univariate Cox regression analysis was implemented on the regulators for subsequent model construction. Firstly, Two-thirds of the samples were randomly selected as a training set for the establishment of the model, and the remaining samples were used for model reliability verification. Secondly, the LASSO Cox regression algorithm was implemented to develop a potential risk signature. Finally, we calculated the LASSO-risk score for a signature using the following formula:
Among them, coef represents the coefficients and xi represents the relative expression value after the z-score transformation of the original expression value of each gene. In this study, this formula was used to calculate the risk score for each CC patient concerning OS and RFS, respectively. Furthermore, receiver operator characteristics (ROC) curves and area under the curve (AUC) were generated for signature validation using the R program with the “survivalROC” package, and the AUC was calculated for prediction evaluation.
The nomogram was constructed to evaluate the prediction probability of 2-, 3- and 5-year OS or RFS. The calibration curves show the 2-, 3- and 5-year OS or RFS were drawn to visualize the observed probabilities against the nomogram prediction. The R package “RMS” presented the nomogram and calibration curves. Decision curve analysis (DCA) with 2-, 3- and 5-year was performed to evaluate the suitability of the constructed model for clinical application by the “ggDCA” package.
RNA expression detection and quantitative polymerase chain reaction (qPCR)
To determine the key m6A regulators (RBM15, HNRNPA2B1, NSUN2, RBMX, CBLL1, METTL3, YTHDF3, and ZC3H13) expression pattern in CC, a total of 20 fresh CC tissue and 20 paired adjacent non-tumor tissues were acquired from patients between August 2020 and September 2021 at Zhongda Hosptial and Nanjing Maternity (Table S2). All the samples were stored well at − 80 °C with treatment by RNAlaterTM Stabilization Solution (AM7021, Thermo Fisher, US). RNA extraction protocol was described in the previous literature published26, and the primer sequences involved were listed in Table S3.
Generation of geneset scores based on PCA analysis
PCA was performed using the expression values of the 17 m6A regulators in all CC patients. Among them, both the first and second principal components were selected to participate in the calculation of m6AScore. In this study, in addition to m6AScore, a similar method was used to construct scores based on different genesets in the subsequent exploration process for comprehensive correlation analysis.
Immune correlation analysis among groups
Estimation of STromal and Immune cells in MAlignant Tumor tissues using Expression data (ESTIMATE) algorithm was executed to calculate the immune score, stromal score, estimated score, and tumor purity of each CC patient based on m6A regulators. Cell-type Identification By Estimating Relative Subsets of RNA Transcripts (CIBERSORT, http://cibersort.stanford.edu/) was utilized to calculate the abundance of immune cells. The Tumor Immune Dysfunction and Exclusion (TIDE) algorithm (http://tide.dfci.harvard.edu/) was used to infer clinical response to immunotherapy in CC with m6A regulators expression profiles. The Cancer Immunome Atlas (TCIA, https://www.tcia.at/) database was utilized for downloading the immune checkpoint inhibitor (ICI) information and immunophenoscore (IPS), an index widely used to represent the immunogenicity.
Drug sensitivity analysis
The Connectivity Map (CMap) database (https://portals.broadinstitute.org/cmap/) was utilized to list potential chemotherapeutic drugs and the half-maximal inhibitory concentration (IC50) was assessed to estimate the drug sensitivity using the “pRRophetic” package.
Bioinformatic analysis
The following PPI (Protein–protein interaction) network was analyzed using the STRING tool (http://www.string-db.org/). Correlation analysis was performed to explore the association between m6A regulators and other interested genes obtained from the TCGA-CESC database. |Cor|> 0.3 was defined as a significant criterion. PPI network and correlation network were visualized by Cytoscape v3.9.1.
Statistical analysis
All the statistical analyses were developed by R software (v4.0.5) and GraphPad Prism (v8.0.2). Spearman correlation analysis was calculated between m6A regulators and target elements. Kaplan–Meier survival analysis with log-rank test was performed to compare patients in different subgroups. The visualization of results was accomplished by R software and GraphPad Prism. P < 0.05 was considered statistically significant unless otherwise marked.
Result
A machine-learning diagnostic model derived m6A regulators in CC
All the bioinformatics analyses utilized in this study were executed as a flowchart in Fig. 1. The 33 selected m6A regulators were shown in Table S1. Before commencing the analyses, we constructed protein–protein Interaction (PPI) network and correlation network to investigate the associations between 33 m6A regulators, and the results showed that there were high functional interactions (minimum required interaction score > 0.4; Fig. S1a) among them. The CNVs analysis showed that IGF2BP2, FXR1, and NSUN2 had higher amplification frequencies, while ZC3H13 had a higher CNV deletions probability (Fig. S1b). Therefore, the expression correlations among 33 regulators demonstrated their close relationship (|R|> 0.2, P < 0.05; Fig. S1c). In addition, to explore the aberrant expression of m6A regulators in CC, we compared cancer patients and normal controls in TCGA, GSE63514 and GSE6791 datasets. As Fig. S1d showed, HNRNPA2B1, YTHDF2, RBM15, and NSUN2 were consistently up-regulated in tumor tissues in TCGA-CESC, GSE63514, and GSE6791 datasets.
The flowchart of this study. CC cervical cancer, RF Random Forest, SVM support vector machines, ANN artificial neural networks, OS overall survival, RFS recurrence-free survival, KM Kaplan–Meier; TCGA The Cancer Genome Atlas, GEO Gene Expression Omnibus, GSCA Gene Set Cancer Analysis, GSEA Gene Set Enrichment Analysis.
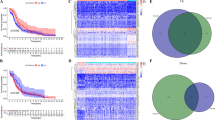
To discriminate the potential functions of m6A regulators in CC, a diagnostic model was first constructed to provide a new viewpoint of CC diagnosis and prevention. Before commencing, 70% of patients in GSE63514 were used as the training set randomly, and the rest samples were defined as the validation set. The random forest (RF) model demonstrated that RBM15, NSUN2, HNRNPA2B1, METTL3, CBLL1, ELAVL1, RBMX, ABCF1, FXR1, and YTHDF3 were the top ten elements among all regulators (Fig. 2a, Fig. S2a,b). Moreover, RBM15, HNRNPA2B1, FXR1, NSUN2, RBMX, ELAVL1, METTL3, ABCF1, CBLL1 and YTHDF3 were identified by the support vector machine (SVM) model (Fig. 2b, Fig. S2c). Based on this, ten common regulators were recognized as key factors in CC diagnostic model (Fig. 2c, Fig. S2d). ROC curves of RF and SVM models showed high accuracy (AUC = 0.946 for RF model and AUC = 0.982 for SVM model; Fig. 2d). Moreover, a nomogram containing regulators and a calibration curve was performed with good accuracy for CC risk prediction (Fig. 2e,f). Decision curve analysis (DCA) was developed showing the benefit, as well as the AUC, were significantly improved (Fig. 2g).
Diagnostic model construction and key m6A regulators identification. (a) Variable importance screening based on RF. (b) Variable importance screened via SVM. (c) Venn diagram showed the top 10 candidate regulators contained both in RF and SVM. (d) ROC curves based on machine learning methods for diagnostic probabilities. (e) The nomogram diagnostic prediction model based on ten filtered m6A regulators. (f) The calibration plots suggested the comparison between prediction and actual outcome for incidence probabilities in the nomogram model. (g) The decision curve analysis showed the net benefit in the nomogram model. (h) Establishment of CC diagnosis model with m6A factor as input layer based on ANN method. (i) ROC curves described the predictive ability of ANN model for CC incidence probabilities with the GSE63514 as train group. (j) ROC curves described the predictive ability of ANN model for CC incidence probabilities with the GSE6791 as test group.
Subsequently, to further probe the function of m6A regulators in CC diagnosis, an artificial neural network (ANN) was constructed via ten key elements mentioned above (Fig. 2h). For the training set, the ROC curve showed the ANN model had an extraordinary accuracy in CC diagnosis (AUC = 0.999; Fig. 2i), and the AUC of the test set (patients obtained from GSE6791) was 0.936 (Fig. 2j). These findings clarified that m6A regulators played an essential role in CC, which might provide a new perspective for the clinical diagnosis of CC.
Prognostic value of m6A RNA methylation regulators and a risk signature constructed with significant ones
Furthermore, we attempted to explore the prognostic effects of regulators in CC. We developed the least absolute shrinkage and selection operator (LASSO) Cox regression algorithm using 33 m6A regulators for overall survival (OS) (Fig. S3a) and recurrence-free survival (RFS) (Fig. S3b) prediction in CC, respectively. For the OS prediction model, eight regulators (FMR1, G3BP1, HNRNPA2B1, LRPPRC, METTL16, WTAP, YTHDF3, and ZC3H13) were identified, and seven factors (YTHDF1, FXR2, YTHDC2, G3BP1, IGF2BP1, RBMX, and ZC3H13) were filtered, independently. The risk score of OS model for each patient was calculated with the following formula: Riskscore = 0.014 × ZC3H13 + 0.008 × YTHDF3 + 0.007 × WTAP + 0.001 × LRPPRC + 0.008 × HNRNPA2B1 + 0.004 × G3BP1 − 0.016 × FMR1 − 0.003 × METTL16. And similarly, risk score of RFS model was calculated with the following formula: Riskscore = 0.031 × ZC3H13 + 0.011 × RBMX + 0.004 × IGF2BP1 − 0.003 × G3BP1 − 0.007 × YTHDC2 − 0.010 × FXR2 − 0.018 × YTHDF1. Afterwards, the Kaplan–Meier survival curve results confirmed that the risk signature had significant predictive power in OS prediction (P < 0.001; Fig. 3a). Similarly, consistent results were found in RFS analyses (P = 0.003; Fig. 3c). Finally, the evaluation of the LASSO regression model using the receiver operating characteristic (ROC) curves and area under the curve (AUC), and results revealed that the signature had more accurate prognostic predictability in the training set (AUCOS = 0.757 for 5-year survival rate and AUCRFS = 0.776 for 5-year recurrence-free rate; Fig. 3b,d) for CC prognosis prediction. Interestingly, our single-gene ROC analysis demonstrated that ZC3H13 had the highest AUC in both OS (AUC = 0.698; Fig. S3a) and RFS (AUC = 0.711; Fig. S3b) predictions, which indicated the potential role of ZC3H13 in patient prognosis. We then constructed a nomogram, which included the clinicopathological characteristics with ZC3H13 expression of patients, and evaluated the accuracy of the model through the calibration curve (Fig. S3c,d). An obvious trend was notable that the models had a better prognosis prediction value (Fig. 3e,f). Subsequently, DCA plots illuminated that the risk score obtained from LASSO got the highest net benefit than other clinical-pathological features for both OS (Fig. 3g) and RFS (Fig. 3h). The patients with high-risk scores exhibited reduced OS and RFS (Fig. S3e,f).
Prognostic model construction and key m6A regulators identification. (a) The Kaplan–Meier survival curves for CC patients with high- and low-risk. (b) ROC curves described the predictive ability of OS-LASSO model for 2-, 3-, and 5-year survival probabilities. (c) The Kaplan–Meier survival curves for CC patients with high- and low-risk. (d) ROC curves described the predictive ability of RFS-LASSO model for 2-, 3-, and 5-year survival probabilities. (e,f) The calibration plots suggested the comparison between prediction and actual outcome for 2-, 3-, and 5-year survival probabilities in the nomogram model for both OS (e) and RFS (f). (g,h) Decision curve analysis for the evaluation of the net benefits of riskscore, Age, HPV_Status, Grade, Genes (ZC3H13 expression) and Stage at 2-, 3-, and 5-year for both OS model (g) and RFS model (h). (i) KM analysis for patients with ZC3H13 different expression level for OS and RFS using GEPIA online tool. (j) KM analysis was performed in OS and RFS patients with different expression levels of G3BP1 using the GEPIA online tool. (k) Multivariate Kaplan–Meier survival curves for patients with different expression level of ZC3H13&G3BP1, which were selected by LASSO-Cox regression algorithm for OS and RFS prediction.
To expound on the scientificity and stability of predictive models, internal or external validation of OS and RFS models were also performed. Principal component analysis (PCA) as well as t-distributed stochastic neighbor embedding (t-SNE) processes were performed to show the patients with a different risk score based on prognostic models, and the outcome suggested that risk scores differentiated patients sufficiently (Fig. S4a,b). Moreover, several gynecological cancers contain Breast invasive carcinoma (BRCA), Ovarian serous cystadenocarcinoma (OV), Uterine Corpus Endometrial Carcinoma (UCEC), and an HPV-associated tumor, Head and Neck squamous cell carcinoma (HNSC) were selected for OS prediction model validation. In addition, the GSE44001 cohort was acquired to verify the RFS model. The consequence indicated that the prognostic model conducted could convincingly define the survival risk or recurrence-free survival risk with reasonable accuracy (Ps < 0.001, ROCs > 0.6; Fig. S4c,d).
Risk scores based on m6A regulators and the LASSO model were sufficient to forecast patient prognostic risk robustly, but several interesting factors that combined into risk scores attracted our attention. ZC3H13 and B3BP1 were common variables in both OS and RFS models, although their direction of effect on prognostic outcomes appeared to be inconsistent. Subsequently, the KM plots downloaded from GEPIA revealed that only upregulation of ZC3H13 expression was significantly associated with worse prognosis (P = 0.006 for OS and P = 0.002 for RFS; Fig. 3i), but not G3BP1 (P = 0.007 for OS and P = 0.140 for RFS; Fig. 3j). When the bivariate K-M analysis was carried out, we found that the presence of ZC3H13 significantly predicted patient outcomes (Ps < 0.01, Fig. 3k), patients with low expression of both had better prognosis, while patients with high expression of both had worse prognosis. It was evident that this trend was highly correlated with the expression level of ZC3H13. Briefly, ZC3H13 was a vital factor in CC prognosis prediction, and combined with G3BP1, can improve the predictive power.
Considering whether key factors could be used as stable CC biomarkers, we detected seven diagnostic elements and one prognostic factor in 20 pairs of population tissues. The comparison elucidated that RBM15 (P = 0.046; Fig. 4a), NSUN2 (P = 0.001), METTL3 (P = 0.001), CBLL1 (P = 0.003), RBMX (P < 0.001), and ZC3H13 (P = 0.008; Fig. 4b) were significantly up-regulated in CC tissues, while the expressions of HNRNPAB1 (P = 0.027) and YTHDF3 (P = 0.033) and showed the opposite trend.
Two CC subgroups were identified by consensus clustering and immune-associated exploration based on m6A RNA methylation regulators
To further explore the effect of m6A RNA modification in CC, we calculated cluster fitting values of k = 2 to 10 on 306 cancer samples based on the expression correlation of 33 regulators. The results, as shown in Fig. 5a and Fig. S5a, indicated that k = 2 was relatively optimal for further analysis. Based on this, the cluster 1 (n = 151) and cluster 2 (n = 155), respectively. Furthermore, to intuitively obtain the effect of two subgroups and reflect the reliability of our results, we calculated PCA and t-SNE analysis based on two subgroups. We found the clustering results could effectively distinguish the two clusters (Fig. 5b, Fig. S5b). Subsequent KM analysis results indicated that cases in cluster 1 had better prognostic status than in cluster 2 (P = 0.015 for OS and P = 0.045 for RFS; Fig. 5c). Gene Set Enrichment Analysis (GSEA) was either conducted to investigate the enrichment of the genes in two clusters. The result showed that mitochondrial drug metabolism (P450 and other enzymes) was significantly enriched in cluster 1, while cell cycle, DNA replication, nucleotide excision repair, and spliceosome-associated biological processes was found in cluster 2 (Fig. S5c).
Identification and characteristic description of consensus clustering. (a,b) Consensus clustering for k = 2 based on m6A regulators (a) and visualization by PCA plot (b). (c) Kaplan–Meier survival curves for patients in different subgroups, OS and RFS. (d) Representation of the groups character by m6AScore. (e) Analysis and comparison of tumor microenvironment in patients with different m6AScore level. (f) The comparison of proportion of 22 immune cells in 309 patients of different m6AScore group. (g) Immune function analysis and comparison in patients with different m6AScore group. (h) Immune escape analysis (Merck18, TIDE, MSI score and T cell Exclusion) and comparison in patients with different m6AScore level. (i) Tumor mutation burden between different m6AScore and TMB. (j) Box plot showed the comparison of HLA family genes expression between different m6AScore groups. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
To clarify the association between m6A regulators and clusters, an m6A-related score, named m6AScore, based on 33 key factors was calculated via PCA analysis to quantitatively describe the m6A level of each patient. Obviously, patients in cluster 1 had higher m6AScore than patients in cluster 2 (P < 0.001; Fig. 5d). After KM survival analysis, a trend was revealed that patients with higher m6AScore had better prognostic status (P = 0.003 for OS and P = 0.005 for RFS; Fig. S5d,e). Understandably, most of the 33 genes were significantly different expression in two clusters (Fig. S5f) and m6AScore groups (Fig. S5g) based on m6A regulators. These results suggested that m6AScore based on m6A regulators could also predict the prognostic risk of patients.
Few studies have delved into the association of m6A regulators and immune function comprehensively. Firstly, stromal score, immune score, and tumor purity analysis were developed via the ESTIMATE algorithm, and the differentiation of patients in different m6AScore groups was identified. The result showed that StromalScore, ImmuneScore, and total score, ESTIMATEScore, were significantly lower in the high-m6AScore group than in the low-m6AScore group (Ps < 0.05; Fig. 5e). Subsequently, we performed immune infiltration analysis using the same cases mentioned above and explored the association between different m6AScore groups (Fig. 5f). However, only the abundance of plasma cells, regulatory T cells (Treg), and eosinophils had obviously difference between low- and high-m6AScore patients. Analysis of immune function were executed subsequently and results implied that co-inhibition of antigen presenting cells (APCs), B cells, Chemokine receptors (CCRs), HLA, and T cell co-stimulation in high-m6AScore group were less than low-m6AScore group (Ps < 0.05; Fig. 5g). Hereafter, TIDE scores reflecting the patient sensitivity to immune checkpoint inhibitors (ICIs) were calculated to explore the discrepancy between high- and low-m6AScore groups. As Fig. 5h showed, Merck18 and TIDE score were reduced in high-m6AScore group, but MSI score and T cell Exclusion showed no significant difference. The tumor mutation burden (TMB) in different m6AScore groups were developed and an insignificant difference was observed (P = 0.050, Fig. 5i). The following KM curves proved that patients with high TMB level had better RFS (P < 0.001; Fig. 5h), but not OS (P = 0.152). Only when m6AScore and TMB were analyzed together, was it observed that the group with high-TMB+ low-m6AScore had the best prognostic status (P = 0.015 for OS and P < 0.001 for RFS; Fig. 5i). In general, the results of this part showed that m6AScore was significantly associated with tumor microenvironment, immune infiltration, immune function, immune escape, and TMB in CC patients. M6AScore could be used as an indicator of patients' immune status, immune escape and prognosis, but its role limited.
To precisely connect m6AScore and immune process, we shifted our focus to the HLA family mentioned above and a comparison between different m6AScore groups was performed. Unexpectedly, 18 of the 24 traits had lower levels in the high-m6AScore group, and other insignificant traits showed the same downward trend (Fig. 5j). This motivated us to explore the association between m6AScore and immune checkpoint expression. A total of 15 immune checkpoints (BTLA, CD2, CD200R, CD244, CD27, PD-L1, CD28, CD40, CD80, ICOS, KLRC1, KLRD1, LAG3, SIRPA, and TIGIT) were identified for subsequent analysis. The results clarified that CD2, CD27, LAG3, CD40, and BTLA had less abundance in high-m6AScore groups (Ps < 0.05; Fig. 6a). No distinct differences were observed in the expression abundances of TIGIT, ICOS, PD-L1, and others in between different m6AScore groups. At the end of this section, to predict the response of ICIs, we determine the association in m6AScore and immunophenoscore (IPS) in CC patients. As Fig. 6b illustrated, patients with low-m6AScore had higher PD-1 and CTLA4/PD1 scores (Ps < 0.05), elucidating patients with the high immunogenicity on ICIs. This was consistent with the result of lower expression of immune checkpoints in high-m6AScore group mentioned above. In conclusion, there were significant differences in m6AScores between immunotherapy non-responders and responders, and m6AScore could provide a new reference for individualized treatment of CC patients.
Immune checkpoint expression analysis and immunotherapy exploration based on M6AScore. (a) Expression differences in immune checkpoints (CD2, CD27, LAG3, BTLA, TIGIT, ICOS) between different m6AScore groups. (b) The comparison of the relative distribution of immunophenoscore (IPS) between different m6AScore groups.
Drug sensitivity analysis for clinical chemotherapy strategies
It was mentioned earlier that there were ten key molecules (RBM15, NSUN2, HNRNPA2B1, METTL3, CBLL1, ELAVL1, RBMX, ABCF1, FXR1, and YTHDF3) in the CC diagnostic model constructed and similarly two key elements (ZC3H13 and G3BP1) in the prognostic model (Figs. 2, 3). Based on this, we sought to explore the association of key factors with widely recognized chemotherapeutic agents. Figure 7 revealed that RBMX was strong positive correlation with Chelerythrine (R = 0.560, P < 0.001), Nelarabine (R = 0.520, P < 0.001), and Fenretinide (R = 0.437, P < 0.001); ELAVL1 was identified obvious related to Chelerythrine (R = 0.530, P < 0.001), Nelarabine (R = 0.523, P < 0.001), and Hydroxyurea (R = 0.384, P = 0.002). When the perspective turns to the prognostic factors, the results suggested that Selumetinib (R = 0.518, P < 0.001), Dabrafenib (R = 0.506, P < 0.001), and Cobimetinib (R = 0.491, P < 0.001) were filtered as the potential ZC3H13 associated drugs. As Fig. 7 showed, Chelerythrine, Nelarabine, Ifosfamide, and Selumetinib were considered as potential chemotherapeutic agents to target these factors.
Analysis of treatment strategies for diagnosis and prognosis based on m6A regulators. (a) The scatter diagram showed the association between the drugs and key regulators, which might provide new clues to uncover potential mechanisms for CC diagnosis. (b) The association between the drugs and key prognostic regulators.
In addition, we also obtained drug data through Connectivity Map (cMAP) database and performed association analysis with m6AScore to explore the clinical treatment strategies for CC patients. Comparison consequence were demonstrated that patients with low-m6AScore marked sensitivity to chemotherapeutic agents, including AKT inhibitor VIII (P < 0.001; Fig. S7a), BIRB.0796 (a p38 MAPK inhibitor; P < 0.001) and FH535 (a Wnt/β-catenin inhibitor; P < 0.001), but NVP.TAE684 (an ALK inhibitor; P < 0.001) were opposite to them. Cisplatin, Paclitaxel, and Gemcitabine, which are commonly used clinical chemotherapy drugs for malignant tumors, also had significant differences in the effects of different m6AScore patients. CC patients from low-m6AScore group were more sensitive to Cisplatin and Gemcitabine, but not to Paclitaxel. Our results demonstrated that the m6AScore calculated based on 33 m6A regulators can be used to predict the sensitivity of patients to chemotherapy drugs, which might be of great significance for clinical chemotherapy drugs.
Correlational exploration of m6A RNA modification with cell cycle and programmed death
Considering the potential molecular mechanism of m6A regulators in CC, a functional enrichment analysis was developed. By Gene Set Cancer Analysis (GSCA) database, the active of cell cycle and apoptosis were the significantly acquired pathways (Fig. 8a). Correlation analysis was subsequently fulfilled to probe potential associations of m6A regulators with gene lists of cell cycle and five programmed cell death, which included ferroptosis, pyroptosis, apoptosis, necroptosis, and cuproptosis. Figure 8b revealed a network of molecular with extensive connection (Rs > 0.300; P < 0.050). In summary, results exhibited cell cycle and cell programmed death were closely related to m6A regulators, which were the essential pathways for CC progression.
Correlation analysis of m6A regulators with cell cycle and programmed death. (a) The essential pathway enrichment analyses of 33 m6A regulators by GSCA. (b) The correlation network between m6A regulators and programmed death genes of interest, which including cell cycle, apoptosis, pyroptosis, necroptosis, ferroptosis and cuproptosis (|R|> 0.3 and P < 0.05). Solid lines represent positive correlations, dashed lines represent negative correlations. The thicker the line, the stronger the correlation between the two edges.
Discussion
In recent years, epigenetic modification has been widely studied. Existing evidence shows that epigenetic modification exists in various molecular biological processes, and it has a significant role in the occurrence and development of cancer27,28,29,30,31. As the more critical one, m6A RNA modification has also been shown to have an important direct relationship with cancers32,33,34,35,36. In this study, we demonstrated that the expression of m6A RNA modification regulators in CC was closely related to its diagnosis and prognosis. The functional enrichment results revealed the feasible key signaling pathways of m6A modification in CC, including cell cycle and cell programmed death. Next, the m6AScore was calculated via PCA algorithm and used to investigate the distinction between CC patients from different clusters obtained by consensus clustering. Fortunately, it was found that the lower the m6AScore, the better the prognosis of patients. Immune characterization and tumor microenvironment analysis subsequently showed significant differences in patients with low- or high-m6AScore, suggesting a potential association between m6A modification and immune processes. It is highly consistent with known reports37,38,39.
In the present study, we explored the diagnostic value and identified ten key regulators including RBM15, NSUN2, HNRNPA2B1, METTL3, YTHDF3, FXR1, RBMX, ELAVL1, CBLL1, and ABCF1, using RF and SVM models. Among them, RBM15 was the most crucial. Subsequent ANN results suggested that our model was accurate in both training and validation cohorts (Fig. 2). Although the current literature on m6A is numerous19,40, there are few reports identifying RBM15 as an essential biomarker for CC prognostic. The study found that RBM15 could increase the m6A level of TMBIM6 mRNA, and increase its stability after recognition by the reader protein, promoting the malignant progression of laryngeal squamous cell carcinoma41. Based on this, a hypothesis was proposed that RBM15 also has a non-negligible potential role in the occurrence and development of CC by mediating the m6A level of targets. After experimental verification (Fig. 4b), RBM15 was indeed significantly up-regulated in patients, but its in-depth molecular mechanism in CC was the focus of our future work. HNRNPA2B1 was another important factor, which was identified as an oncogene in head and neck cancer and could promote Akt/PKB signaling by upregulating the RONΔ165 isoform, thereby promoting epithelial-mesenchymal transition of head and neck cancer cells42. HNRNPA2B1 increased the stabilization of ILF3 mRNA through m6A modification, which in turn increased AKT3 expression to promote multiple myeloma progression43. Although the qPCR assay found that HNRNPA2B1 was significantly down-regulated in CC patients, which was inconsistent with literature reports and the result obtained from datasets, including TCGA-CESC, GSE63514, and GSE6791. It was consistent with them that HNRNPA2B1 was served as a risk factor in OS prognostic models, implying its complex mechanism in CC. Meanwhile, we also considered to detect the relative expression of HNRNPA2B1 again after expanding the tissue sample size.
We distinguished an eight-m6A RNA modification gene signature containing FMR1, G3BP1, HNRNPA2B1, LRPPRC, METTL16, WTAP, YTHDF3, and ZC3H13 for CC OS prediction. Similarly, seven regulators (FXR2, G3BP1, IGF2BP1, RBMX, YTHDC2, YTHDF1, and ZC3H13) were identified for RFS prediction. According to the risk obtained by the LASSO Cox model, we found that this score accurately distinguished patients with different prognostic risks. ZC3H13 and G3BP1 were served as common indicators for both OS and RFS prediction. ZC3H13 could be selected alone for CC prognosis prediction, and the ROC curve showed that its AUC is larger than other members and close to the total AUC of the model (Fig. S3a,b). We next attempted to mix both of them to predict CC prognostic risk. The KM plots suggested that the predictive risk was significant when ZC3H13 and G3BP1 were combined. For ZC3H13, studies have identified it as an important prognostic predictor in Glioblastoma44 and CC15. It is worth mentioning that our results suggested that ZC3H13 was significantly up-regulated in CC tissues (Fig. 4b), which was consistent with the reported trend15, indicating that ZC3H13 was a stable biomarker.
Consensus clustering analysis based on m6A regulators was executed to divide CC patients into two clusters with m6AScore. Patients in cluster 1 had higher m6AScores and worse prognosis, whereas patients in cluster 2 had the exact opposite. In recent years, it has been reported in the literature that m6A regulators can participate in regulating the occurrence, development, and treatment of various tumors by affecting immune-related processes, including but not limited to immune response45,46, immune checkpoint expression47,48, and immune escape49,50. Immune cell infiltration analysis and tumor microenvironment analysis clarified that the immune status and microenvironment were significantly different in m6AScore groups. The level of immune cell infiltration, immune TIDE, and TME score in the patients from high-m6AScore group were clear lower than those in patients with low-m6AScore. In subsequent analysis of immune infiltration, immune escape and TMB, the results showed that m6AScore correlated significantly with these features, but very limited. We speculated that the m6AScore constructed based on the abundance of m6A regulators did not show a strong correlation with the immune infiltration and immune escape status of patients, but the m6A-regulated target genes were directly related to the immune process51,52,53. Therefore, the m6AScore showed a weak correlation with immune status. In addition, the information loss caused by dimensionality reduction during the construction of m6AScore may also weaken the association to a certain extent. Although no significant differences were observed in TMB and MSI in different m6AScore groups, we found that the HLA and immune checkpoint expressions of patients with low-m6AScore were significantly lower than those in patients with high-m6AScore. Immunotherapy analysis also found that patients with low-m6AScore had better treatment benefits. For such patients, immunotherapy is a scientifically effective protective measure. For patients with high-m6AScore, immunotherapy might not be a high-benefit approach, possibly due to the high proportion of patients with advanced cancer. The pattern of low expression of immune checkpoints in patients with worse prognosis was previously reported in studies54, which was consistent with our results. As for patients with high-m6AScore, NVP.TAE684 and Paclitaxel were more suitable for them.
There have been many literatures on the m6A molecules in CC, some of which are similar but not the same. Pan’s study15 analyzed 13 m6A regulators in the TCGA-CESC dataset and identified ZC3H13, YTHDF1, and YTHDC1 as OS-related factors. Consistently, we also identified ZC3H13 as a key factor on patient OS prediction, and both of our studies found ZC3H13 to be the most essential influencing factor (observed from the coefficients). Neither YTHDF1 nor YTHDC1 were in our model, and we presume the reason for the discrepancy may be the difference in the number of included independent variables, which would result in non-essential variables not being stably retained in the model. Furthermore, the expression trend of METTL3 was inconsistent with our experimental results. In addition to individual differences in the population, it was our conjecture that the small sample size causes biased results. In addition, complex mechanisms between RNAs and proteins may also lead to different outcomes. Zhang’s research16 explored the expression patterns of m6A molecules in CC and comprehensively analyzed the connection with immune-related processes. Although expression patterns were explored for both, we ultimately constructed m6AScore based on m6A regulator expression values rather than differential genes. This was the main difference between our two studies and the main reason for the difference in the results that follow. Most importantly, this study also constructed a cervical cancer diagnostic model and RFS prognostic model, and proposed chemotherapy regimens for the identified key targets, while complementing the shortcomings of immunotherapy in the study.
The limitations of this study should be considered when interpreting the results. The dataset we used when building the diagnostic model was GSE63514, and the validation dataset was GSE6791. However, most of this study is based on the TCGA database, which is a completely different group. The reason was that in the diagnostic model, we needed the population of different groups in the data to be as balanced as possible, and TCGA was difficult to meet this requirement (306 CC patients and 3 normal samples). In addition, the necessary survival data in the prognostic model was also difficult to obtain in the GEO dataset; there is not yet a public database that can simultaneously meet the requirements of both. We are currently constructing a balanced CC follow-up cohort and hope to fill this gap in future studies. Another important point is that the experiments on the expression of key m6A regulators in the study only did qPCR, and there was a lack of evidence from a large number of immunochemistry results. This deficiency will also be improved and published in the future work.
Conclusion
In conclusion, our findings supported a systematic analysis that m6A regulators executed vital functions in the diagnosis, prognosis, immune microenvironment, and treatment of CC. And these mechanisms not yet completely elucidated today might be achieved by immune biological process, cell cycle, and cell programmed death. This study also offered a theoretical basis for CC clinical treatment.
Data availability
The datasets generated and analyzed during the current study are available in the TCGA (https://portal.gdc.cancer.gov/) and GEO (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE63514; https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE6791) database. The other data, algorithms and code that support the findings of this study are available on request from the corresponding author. The algorithms and code were not publicly available due to privacy or ethical restrictions.
References
Ferlay, J. C. et al. Cancer statistics for the year 2020: An overview. Int. J. Cancer. https://doi.org/10.1002/ijc.33588 (2020).
Wallace, N. A. Catching HPV in the homologous recombination cookie jar. Trends Microbiol. 28, 191–201. https://doi.org/10.1016/j.tim.2019.10.008 (2020).
Zhu, F. C. et al. Efficacy, immunogenicity and safety of the AS04-HPV-16/18 vaccine in Chinese women aged 18–25 years: End-of-study results from a phase II/III, randomised, controlled trial. Cancer Med. 8, 6195–6211. https://doi.org/10.1002/cam4.2399 (2019).
Flamand, M. N. & Meyer, K. D. m6A and YTHDF proteins contribute to the localization of select neuronal mRNAs. Nucleic Acids Res. https://doi.org/10.1093/nar/gkac251 (2022).
Tan, Z. et al. RNA N6-methyladenosine demethylase FTO promotes pancreatic cancer progression by inducing the autocrine activity of PDGFC in an m(6)A-YTHDF2-dependent manner. Oncogene. https://doi.org/10.1038/s41388-022-02306-w (2022).
Yang, H. et al. Hypoxia inducible lncRNA-CBSLR modulates ferroptosis through m6A-YTHDF2-dependent modulation of CBS in gastric cancer. J. Adv. Res. 37, 91–106. https://doi.org/10.1016/j.jare.2021.10.001 (2022).
Zhu, Y. et al. METTL3-mediated m6A modification of STEAP2 mRNA inhibits papillary thyroid cancer progress by blocking the Hedgehog signaling pathway and epithelial-to-mesenchymal transition. Cell Death Dis. 13, 358. https://doi.org/10.1038/s41419-022-04817-6 (2022).
Liu, Z. X., Li, L. M., Sun, H. L. & Liu, S. M. Link between m6A modification and cancers. Front. Bioeng. Biotechnol. 6, 89. https://doi.org/10.3389/fbioe.2018.00089 (2018).
Sun, T., Wu, R. & Ming, L. The role of m6A RNA methylation in cancer. Biomed. Pharmacother. 112, 108613. https://doi.org/10.1016/j.biopha.2019.108613 (2019).
Oerum, S., Meynier, V., Catala, M. & Tisne, C. A comprehensive review of m6A/m6Am RNA methyltransferase structures. Nucleic Acids Res. 49, 7239–7255. https://doi.org/10.1093/nar/gkab378 (2021).
An, Y. & Duan, H. The role of m6A RNA methylation in cancer metabolism. Mol. Cancer 21, 14. https://doi.org/10.1186/s12943-022-01500-4 (2022).
Zhang, Y. et al. m(6)A modification-mediated CBX8 induction regulates stemness and chemosensitivity of colon cancer via upregulation of LGR5. Mol. Cancer 18, 185. https://doi.org/10.1186/s12943-019-1116-x (2019).
Yang, S. et al. m(6)A mRNA demethylase FTO regulates melanoma tumorigenicity and response to anti-PD-1 blockade. Nat. Commun. 10, 2782. https://doi.org/10.1038/s41467-019-10669-0 (2019).
Chen, Y. et al. WTAP facilitates progression of hepatocellular carcinoma via m6A-HuR-dependent epigenetic silencing of ETS1. Mol. Cancer 18, 127. https://doi.org/10.1186/s12943-019-1053-8 (2019).
Pan, J., Xu, L. & Pan, H. Development and validation of an m6A RNA methylation regulator-based signature for prognostic prediction in cervical squamous cell carcinoma. Front. Oncol. 10, 1444. https://doi.org/10.3389/fonc.2020.01444 (2020).
Zhang, W. et al. m6A regulator-mediated tumour infiltration and methylation modification in cervical cancer microenvironment. Front. Immunol. 13, 888650. https://doi.org/10.3389/fimmu.2022.888650 (2022).
Cong, P. et al. Identification of the role and clinical prognostic value of target genes of m6A RNA methylation regulators in glioma. Front. Cell Dev. Biol. 9, 709022. https://doi.org/10.3389/fcell.2021.709022 (2021).
Jin, Y. et al. Analysis of m6A-related signatures in the tumor immune microenvironment and identification of clinical prognostic regulators in adrenocortical carcinoma. Front. Immunol. 12, 637933. https://doi.org/10.3389/fimmu.2021.637933 (2021).
Yue, L., Zhang, R., Chen, S. & Duan, G. Risk score prediction model of prognosis in GC patients by age and gender combined with m6A modification genes FTO and RBM15. Front. Cell Dev. Biol. 10, 710708. https://doi.org/10.3389/fcell.2022.710708 (2022).
Li, H. et al. ALKBH1 promotes lung cancer by regulating m6A RNA demethylation. Biochem. Pharmacol. 189, 114284. https://doi.org/10.1016/j.bcp.2020.114284 (2021).
Zhao, Z. et al. Necroptosis-related lncRNAs: Predicting prognosis and the distinction between the cold and hot tumors in gastric cancer. J. Oncol. 2021, 6718443. https://doi.org/10.1155/2021/6718443 (2021).
Tsvetkov, P. et al. Copper induces cell death by targeting lipoylated TCA cycle proteins. Science 375, 1254–1261. https://doi.org/10.1126/science.abf0529 (2022).
Jung, M. et al. Artificial intelligence system shows performance at the level of uropathologists for the detection and grading of prostate cancer in core needle biopsy: An independent external validation study. Mod. Pathol. https://doi.org/10.1038/s41379-022-01077-9 (2022).
Rashid, J. et al. An augmented artificial intelligence approach for chronic diseases prediction. Front. Public Health 10, 860396. https://doi.org/10.3389/fpubh.2022.860396 (2022).
Granata, V. et al. CT-based radiomics analysis to predict histopathological outcomes following liver resection in colorectal liver metastases. Cancers (Basel) 14, 14071648. https://doi.org/10.3390/cancers14071648 (2022).
Wang, S. et al. Fanconi anemia pathway genes advance cervical cancer via immune regulation and cell adhesion. Front. Cell Dev. Biol. 9, 734794. https://doi.org/10.3389/fcell.2021.734794 (2021).
Anobile, D. P. et al. Splicing deregulation, microRNA and Notch aberrations: Fighting the three-headed dog to overcome drug resistance in malignant mesothelioma. Expert Rev. Clin. Pharmacol. https://doi.org/10.1080/17512433.2022.2074835 (2022).
Deng, P. et al. Loss of KDM4B impairs osteogenic differentiation of OMSCs and promotes oral bone aging. Int. J. Oral Sci. 14, 24. https://doi.org/10.1038/s41368-022-00175-3 (2022).
Galbraith, K. & Snuderl, M. DNA methylation as a diagnostic tool. Acta Neuropathol. Commun. 10, 71. https://doi.org/10.1186/s40478-022-01371-2 (2022).
Markou, A. et al. DNA methylation analysis of tumor suppressor genes in liquid biopsy components of early stage NSCLC: A promising tool for early detection. Clin. Epigenet. 14, 61. https://doi.org/10.1186/s13148-022-01283-x (2022).
Yang, S., Huang, Y. & Zhao, Q. Epigenetic alterations and inflammation as emerging use for the advancement of treatment in non-small cell lung cancer. Front. Immunol. 13, 878740. https://doi.org/10.3389/fimmu.2022.878740 (2022).
Chen, Y. et al. N(6)-methyladenosine-modified TRAF1 promotes sunitinib resistance by regulating apoptosis and angiogenesis in a METTL14-dependent manner in renal cell carcinoma. Mol. Cancer 21, 111. https://doi.org/10.1186/s12943-022-01549-1 (2022).
Liu, Y. et al. LncRNA-PACERR induces pro-tumour macrophages via interacting with miR-671-3p and m6A-reader IGF2BP2 in pancreatic ductal adenocarcinoma. J. Hematol. Oncol. 15, 52. https://doi.org/10.1186/s13045-022-01272-w (2022).
Li, E. et al. METTL3 promotes homologous recombination repair and modulates chemotherapeutic response in breast cancer by regulating the EGF/RAD51 axis. Elife 11, 75231. https://doi.org/10.7554/eLife.75231 (2022).
Jin, S. et al. The m6A demethylase ALKBH5 promotes tumor progression by inhibiting RIG-I expression and interferon alpha production through the IKKepsilon/TBK1/IRF3 pathway in head and neck squamous cell carcinoma. Mol. Cancer 21, 97. https://doi.org/10.1186/s12943-022-01572-2 (2022).
Lin, C. et al. The N(6)-methyladenosine modification of circALG1 promotes the metastasis of colorectal cancer mediated by the miR-342-5p/PGF signalling pathway. Mol. Cancer 21, 80. https://doi.org/10.1186/s12943-022-01560-6 (2022).
Grenov, A., Hezroni, H., Lasman, L., Hanna, J. H. & Shulman, Z. YTHDF2 suppresses the plasmablast genetic program and promotes germinal center formation. Cell Rep. 39, 110778. https://doi.org/10.1016/j.celrep.2022.110778 (2022).
Chen, Q., Li, H., Liu, Y. & Zhao, M. Epigenetic regulation of immune and inflammatory responses in rheumatoid arthritis. Front. Immunol. 13, 881191. https://doi.org/10.3389/fimmu.2022.881191 (2022).
Chen, Y. et al. Identification of N6-methyladenosine-related lncRNAs as a prognostic signature in glioma. Front. Oncol. 12, 789283. https://doi.org/10.3389/fonc.2022.789283 (2022).
Zhao, Z., Ju, Q., Ji, J., Li, Y. & Zhao, Y. N6-methyladenosine methylation regulator RBM15 is a potential prognostic biomarker and promotes cell proliferation in pancreatic adenocarcinoma. Front. Mol. Biosci. 9, 842833. https://doi.org/10.3389/fmolb.2022.842833 (2022).
Wang, X. et al. RBM15 facilitates laryngeal squamous cell carcinoma progression by regulating TMBIM6 stability through IGF2BP3 dependent. J. Exp. Clin. Cancer Res. 40, 80. https://doi.org/10.1186/s13046-021-01871-4 (2021).
Gupta, A. et al. The HNRNPA2B1-MST1R-Akt axis contributes to epithelial-to-mesenchymal transition in head and neck cancer. Lab. Investig. 100, 1589–1601. https://doi.org/10.1038/s41374-020-0466-8 (2020).
Jiang, F. et al. HNRNPA2B1 promotes multiple myeloma progression by increasing AKT3 expression via m6A-dependent stabilization of ILF3 mRNA. J. Hematol. Oncol. 14, 54. https://doi.org/10.1186/s13045-021-01066-6 (2021).
Wang, L. C. et al. M6A RNA methylation regulator HNRNPC contributes to tumorigenesis and predicts prognosis in glioblastoma multiforme. Front. Oncol. 10, 536875. https://doi.org/10.3389/fonc.2020.536875 (2020).
Feng, J. et al. Global profiling reveals common and distinct N6-methyladenosine (m6A) regulation of innate immune responses during bacterial and viral infections. Cell Death Dis. 13, 234. https://doi.org/10.1038/s41419-022-04681-4 (2022).
Li, X. et al. FTO regulates arecoline-exposed oral cancer immune response through PD-L1. Cancer Sci. https://doi.org/10.1111/cas.15332 (2022).
Ni, Z. et al. JNK signaling promotes bladder cancer immune escape by regulating METTL3-mediated m6A modification of PD-L1 mRNA. Cancer Res. 82, 1789–1802. https://doi.org/10.1158/0008-5472.CAN-21-1323 (2022).
Xiong, W. et al. N6-methyladenosine regulator-mediated immue patterns and tumor microenvironment infiltration characterization in glioblastoma. Front. Immunol. 13, 819080. https://doi.org/10.3389/fimmu.2022.819080 (2022).
Fan, Y. et al. m(6)A regulator-mediated methylation modification patterns and characterisation of tumour microenvironment infiltration in non-small cell lung cancer. J. Inflamm. Res. 15, 1969–1989. https://doi.org/10.2147/JIR.S356841 (2022).
Peng, L. et al. Lipopolysaccharide facilitates immune escape of hepatocellular carcinoma cells via m6A modification of lncRNA MIR155HG to upregulate PD-L1 expression. Cell Biol. Toxicol. https://doi.org/10.1007/s10565-022-09718-0 (2022).
Hu, C. et al. Multifaceted roles of the N(6)-methyladenosine RNA methyltransferase METTL3 in cancer and immune microenvironment. Biomolecules 12, 1042. https://doi.org/10.3390/biom12081042 (2022).
Li, W., Hao, Y., Zhang, X., Xu, S. & Pang, D. Targeting RNA N(6)-methyladenosine modification: A precise weapon in overcoming tumor immune escape. Mol. Cancer 21, 176. https://doi.org/10.1186/s12943-022-01652-3 (2022).
Zhou, M. et al. The emerging roles and mechanism of m6a in breast cancer progression. Front. Genet. 13, 983564. https://doi.org/10.3389/fgene.2022.983564 (2022).
Zhang, B. et al. m(6)A regulator-mediated methylation modification patterns and tumor microenvironment infiltration characterization in gastric cancer. Mol. Cancer 19, 53. https://doi.org/10.1186/s12943-020-01170-0 (2020).
Funding
This work was supported by Grants from Natural Science Foundation of China (81872684), the Fundamental Research Funds for the Central Universities, Southeast University “Zhongying Young Scholars” Project, Natural Science Foundation of Jiangsu Province (BK20190357), and Postgraduate Research & Practice Innovation Program of Jiangsu Province (KYCX21_0162).
Author information
Authors and Affiliations
Contributions
X.Z. and S.W. designed this study, performed mainly data analysis and wrote the article. B.D., S.W., and W.Y. downloaded data from TCGA and GEO database, as well as accomplished data treatment before data analysis. Q.X., D.M., S.X., B.Y., and S.S. contributed to the population tissue collection and critical article revision for research content. J.H. and H.L. provided help in experiment and administrative support. The final manuscript was read and approved by all authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Wang, S., Ding, B., Wang, S. et al. Gene signature of m6A RNA regulators in diagnosis, prognosis, treatment, and immune microenvironment for cervical cancer. Sci Rep 12, 17667 (2022). https://doi.org/10.1038/s41598-022-22211-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-022-22211-2
- Springer Nature Limited