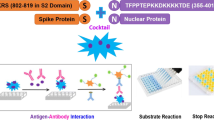
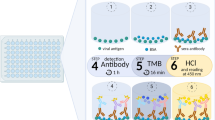
Abstract
A major challenge for developing countries during the COVID-19 pandemic is affordable and adequate monitoring of disease progression and population exposure as the primary source relevant epidemiological indicators. Serology testing enables assessing population exposure and to guide vaccination strategies but requires rigorous accuracy validation before population-wide implementation. We adapted a two-step ELISA protocol as a single-step protocol for detection of IgG against the Receptor Binding Domain (RBD) of SARS-CoV-2 spike protein and compared its diagnostic accuracy with a commercial immunoassay anti-nucleoprotein IgG. Both methods yielded adequate and comparable diagnostic accuracy after 3 weeks post-symptom onset and were implemented in a nation-wide population based serological survey during August–November 2020. Anti-RBD National seroprevalence was 23.6%, 1.3% lower, but not significantly, than for anti-N. Double positive seroprevalence was 19.7%. Anti-N single-positive seroprevalence was 3.72% and anti-RBD single-positive seroprevalence was 1.98%. Discrepancies in the positivity to either single marker may be due to different kinetics of each antibody marker as well as the heterogeneity of the sampling time in regards to local epidemic waves. Baseline single positivity prevalence will be useful to assess the serological impact of vaccination and natural infection in further serosurveillance efforts.
Similar content being viewed by others
Introduction
Developing countries have faced important challenges to respond to the COVID-19 pandemic. Access to low-cost options to assess the extent and occurrence of infection has been a key limitation for epidemic monitoring. Serology testing is used to measure previous population exposure, an approximation to population immunity and provides adequate denominators for the estimation of attack and lethality rates1.
Validating serologic tests is key to adequately inform seroprevalence. Serologic cross-reactivity between endemic HuCoV’s and SARS-CoV is well documented2,3,4; thus, the risk of false-positives as a result of previous HuCoV exposure has to be considered to validate serologic assays during the current COVID-19 pandemic5. Beyond cross-reactivity, other factors such as viral antigen and antibody kinetics, antibody class, and reagent quality could influence the performance of a serology-based test6, so rigorous validation and contextualization is required.
In general, infection by Coronaviruses initiate when the spike (S) protein interacts with its cellular receptor. The spike protein is a type I viral trimeric glycoprotein composed by an N-terminal S1 subunit and a C-terminal S2 subunit. S1 binds to the cellular receptor through the Receptor Binding Domain (RBD). The RBD is a key determinant of host cell tropism and the major target for antibody-mediated neutralization7. There is structural conservation and cross-reactivity between SARS-CoV and SARS-CoV-2 RBD’s8,9, but less conservation with the RBD of other HuCoV’s, which makes the RBD a good antigen target for developing serologic assays.
Another widely used serologic marker for SARS-CoV-2 previous infection are antibodies against the nucleoprotein (N), a predominant non-structural protein. Antibodies against N are non-neutralizing. A number of studies have described different kinetics for anti-S and anti-N IgG10,11,12, but the impact of such differences in the interpretation of serological surveys remains to be determined.
Most COVID-19 vaccines elicit an immune response against the spike protein, whereas natural infection by SARS-CoV-2 induces a general immune response against several viral proteins, indicating that the use of different serological markers at a population level can provide relevant information of the vaccination efforts. We report an adapted version of the SARS-CoV-2 anti-S IgG ELISA described by Amanat et al.13 as a single-step method for anti-RBD IgG antibody detection. We evaluated and compared its diagnostic accuracy with a commercial semi-automated method for anti-nucleoprotein IgG antibody detection in a set of 438 serum samples from pre-pandemic and confirmed COVID-19 cases. Furthermore, to define both markers baseline seroprevalence prior to the initiation of nation-wide vaccination, we used and compared both methods in parallel in 9068 serum samples from a nationwide COVID-19 serological survey implemented during August–November 2020.
Results
Serology tests validation
We collected and analyzed 438 serum samples from 391 RT-PCR confirmed COVID-19 cases, of which 43 had paired samples, one taken during acute disease and the other during convalescence, and two cases had two serial convalescent samples. The sociodemographic characteristics of control and COVID-19 donors are shown in Supplementary Figure S1.
As an initial approach to measure diagnostic accuracy of the IgG anti-RBD ELISA, we performed Receiver Operating Characteristic (ROC) curve analysis in cases and controls, according to weeks post-symptom onset (PSO) and estimated the area under the ROC curve (AUC) and 95% confidence intervals as indicators of diagnostic performance (Supplementary Table S1 and Supplementary Fig. S2). AUC did not significantly increased from > 1-week PSO (AUC = 0.94, 95% CI 0.92–0.97) to > 7-weeks PSO (AUC = 0.97, 95% CI 0.95–1.0). These results indicate that AUC for the anti-RBD IgG ELISA is critically dependent on weeks PSO, but from > 1-week PSO high specificity and sensitivity can be achieved.
We compared the AUC of anti-RBD IgG ELISA versus anti-Nucleoprotein Elecsys®. Although AUC values were slightly higher for Elecsys®, no statistically significant difference was found at any of the time PSO categories (Supplementary Table S2 and Supplementary Fig. S3). Agreement between both methods was high. In all samples, regardless of time PSO, there was a 91.1% agreement with a Cohen’s Kappa = 0.81 (P = < 0.001) (Supplementary Table S3), which corresponds to an almost perfect agreement14.
It is evident that the pattern of both methods ROC curves is different and suggested differences, particularly at higher specificities and earlier PSO (Supplementary Fig. S3). The partial AUC (pAUC) for both methods was calculated at the 1.0–0.9 specificity interval15 (Supplementary Table S4). As suggested by the differences in ROC curve pattern, pAUC was larger for Elecsys® in earlier times PSO, indicating that in terms of sensitivity, the Elecsys® system performs slightly better than the RBD ELISA at earlier times PSO.
Finally, we determined the absorbance cut-off for the anti-RBD IgG ELISA. By the time the current work was performed, we estimated a national seroprevalence of 25%, indicating that a highly specific method was required to obtain optimal positive predictive value16. Thus, we defined the absorbance cut-off as the lowest value to achieve ≥ 0.99 specificity. At 0.4 absorbance, we obtained the best sensitivity without lowering specificity below 0.99. Consistent with the ROC analysis, sensitivity, negative predictive value (NPV) and Negative Diagnostic Likelihood ratio (NDLR) were deeply affected by time PSO (Supplementary Fig. S4A-C) being reliable until > 3-weeks PSO (Table 1).
As for specificity, both methods had the same specificity (0.995), as only one out of 199 pre-pandemic samples were false-positive (different sample). As a result, Positive Predictive Value (PPV) and Positive Diagnostic Likelihood ratio (PDLR) were also optimal and largely unaffected by time PSO (Table 1 and Supplementary Fig. S4D–F).
IgG anti-N and anti-RBD comparative results in the National Health and Nutrition Survey
Anti-RBD National seroprevalence was 23.6% (CI 95% 21.5, 25.6), which is slightly lower (1.3%) than the one observed for anti-N IgG 16. National seroprevalence for both markers was 19.7% (CI 95% 18.3, 21.3) (Table 2). Anti-N single-positive seroprevalence was 3.72% (CI 95% 3.1, 4.34) and anti-RBD single-positive seroprevalence was 1.98% (CI 95% 1.58, 2.38). Among seropositives to either markers, 77.7% were double positives, 14.1% were single positive to anti-N IgG and 8.1% were single positive to anti-RBD IgG.
We analyzed the seroprevalence based on positivity to both or single markers according to socio-demographic characteristics (Table 3). The prevalence of double positives was similar across age and sex groups. By region, only Mexico City seemed to have a higher prevalence of double positives (90.4%, 95%CI 85.9, 93.6) compared to the rest of the regions that ranged from 69.7% (95% CI 60.8, 77.4%) in the Central-North to 82.4% (95% CI 77.3, 86.5%) in the Yucatan Peninsula. The prevalence of single-positive anti-N ranged from 9.8% (95% CI 6.9, 13.8%) in the age group 40–49 to 18.5% (95% CI 13.7, 24.5%) in the age group 20–29 years; 17.0% (95% CI 14.2, 30.1%) in males and 12.3% (95% CI 9.8, 15.2%) in females; across regions, Mexico City had the lowest prevalence (4.2%, 95% CI 2.3, 7.4%) and Central-North, the highest (19.6%, 95% CI 14.5, 26.0%). The prevalence of only positive to anti-RBD ranged from 6.2% (95% CI 3.8, 10.0%) in the age group 60 and older to 10.5% (95% CI 6.5, 16.6%) in 10–19 years; males and females had the same prevalence (7.8%); and across regions, Central-Pacific had the highest prevalence (12.3%, 95% CI 6.9, 20.9%) and Peninsula, the lowest (3.4%, 95% CI 2.1, 5.5%).
To identify the sociodemographic factors associated with single positivity compared to double positivity we performed a multinomial logistic regression (Table 4). Females, compared to males, had 0.66 times the relative probability of being only positive to anti-N over double-positive (p value = 0.01). Compared to Mexico City, all regions had a higher relative probability of being only positive to anti-N over double-positive and the North-Border, Central-Pacific, and Central-North had a higher relative probability of being only positive to anti-RBD over double-positive. No significant differences were observed in seropositivity to either single marker according to self-reported time post-symptom onset.
Discussion
Serosurveillance is a highly valuable approach to study epidemic dynamics and to inform public health interventions. As such, the election of the serologic method employed determines the quality of data retrieved. Aiming to validate a useful method for nation-wide serosurveillance, we have analyzed the diagnostic accuracy of an in-house, single-step adaptation of the SARS-CoV-2 RBD IgG ELISA shared by the Krammer laboratory13,17.
Once validated with a panel of pre-pandemic controls and RT-PCR-confirmed COVID-19 cases, we further compared the performance of this method with a commercial system for determining anti-N IgG in a nation-wide serosurvey. Our results indicate no significant differences in the performance of either method for serosurveillance, although slight differences in sensitivity were found, mainly related to lower sensitivity of our in-house method at early times post-symptom onset. A plausible explanation for such difference is that Elecsys® is a much more analytically sensitive electro-chemiluminescence immunoassay (ECLIA) that may detect lower quantities of antigen-bound IgG than a standard ELISA. Thus, anti-RBD ELISA test could be missing individuals recently infected, or infected several months ago, when anti-RBD response has waned and the amount of antibody is below limits of detection. In agreement with our results, ECLIA is more sensitive and has a wider linear range for detecting seroconversion against malarial antigens18.
The in-house SARS-CoV-2 RBD IgG ELISA method has several advantages for a middle-income country, such as Mexico. First, it can be implemented using standard equipment, which is usually available at serology labs. Second, reagents are low cost and can be more easily produced and /or purchased than proprietary kits, reducing the economic burden of surveillance. For the whole serosurvey, we used ~ 1 mg of recombinant RBD. Finally, adaptations can be made to increase the efficiency and reduce waste. In our case, we further simplified the original method by obviating the second confirmatory ELISA that uses the recombinant stabilized S protein, which represented a reduction in cost and time.With these changes, a validation study was performed for both tests to evaluate their diagnostic performance and to assess it in the specific context of Mexico, prior the introduction of COVID-19 vaccination. Endemic CoV’s have been present in Mexico and are frequently detected during flu season; also, Mexico is endemic for vector-borne diseases such as dengue, Zika and malaria. CoV’s and malaria have shown some degree of cross-reactivity with SARS-CoV-2 and could affect the specificity of the in-house method19,20,21; thus, a context-specific validation was needed. Given our concern with specificity, we assembled a large panel of controls from all regions of the country, using historical blood samples, allowing for a wide representation of prior viral exposures. The diagnostic accuracy of our adapted RBD IgG ELISA is comparable to what has been previously described by Indenbaum et al., in Israel who performed the adaptation of the same method shared by the Krammer lab, and showed no additional gain in sensitivity by the inclusion of IgA or IgM class detection22.
As reported by manufacturer, Elecsys® system has an overall specificity of 0.998 (95%CI 0.996–0.999)23. In an in-house validation with samples of Mexican origin, both methods displayed excellent specificity (0.995, 95% CI 0.985–1.0), ensuring high PPV in low prevalence settings (i.e. early pandemic phase, rural areas, etc.). The high specificity observed suggests that cross-reactivity in Mexico is low, increasing our confidence in the assessment, however SARS-CoV-2 serosurveillance in malaria-endemic foci along the Mexican Pacific coast, Central and South America should be interpreted carefully.
Apart from the diagnostic accuracy of each method, there is evidence that both tests provide qualitatively different information. Anti-RBD IgG titers correlate better with SARS-CoV-2 neutralization than anti-N titers10,11,24, and antibody decay seems slower for anti-S than for anti-N IgG12,25,26. Interestingly, children are often asymptomatic and in general less affected than adults27, and make more robust anti-S IgG antibody response than for the anti-N IgG SARS-CoV-228, suggesting that the sensitivity of N-based IgG immunoassays could be lower in children. Moreover, asymptomatic and mild COVID-19 adults make less robust antibody responses against SARS-CoV-210,29,30,31. A limitation of the validation approach used in this work is that all COVID-19 samples included in the panel were only from symptomatic adults, potentially underestimating false negatives derived from asymptomatic adult and children infections in large-scale serosurveillance.
Despite differences of both methods in the antibody kinetics, age differences, and analytical sensitivity differences, the results obtained at a nation-wide survey are remarkably similar for seroprevalence estimation. Nevertheless, the impact of single-marker seropositivity at an epidemiological scale is not negligible. We can think of two broad explanations of single positivity as a result of natural infection in non-vaccinated individuals:
(1) True single positivity, implying that for some biological reason, some individuals seroconvert to only one marker. As discussed, predominant seroconversion to S over N has been described in children, presumably because less viral replication favors immune response against structural antigens (i.e. Spike) over non-structural proteins such as (N)28. Although tested, we did not find any association between single S positivity and age. A matter of sample size (14 cases with single S positivity of a total of 1405 children under 9 years old) could explain lack of association.
(2) False single positivity: Individuals that seroconverted to both markers, but at the time of sampling, only one marker came out positive. In this case, different kinetics, test sensitivity or both could explain such scenario. Different kinetics of anti-N and anti-S assessed during the first pandemic waves in longitudinal cohorts for up to 8 months is well documented, indicating as a more rapid waning of anti-N antibodies than anti-S12, which could explain at least some cases of single anti-S positivity. However, our regression model failed to reveal any association with single positivity and self-reported number of months after infection.
As for test sensitivity, in our test validation we described that anti-S is slightly less sensitive than anti-N in early times PSO (not statistically significant). However, the calculation of partial AUC at high specificity values yielded significant differences in partial AUC’s (Supplementary Table S2). Thus is possible that the in-house anti-S test may be missing true positives in early phase of infection, contributing to anti-N single positivity. Not surprisingly, our regression model also failed to reveal any association between anti-N single positivity and months after infection, because seroconversion is measured in days/weeks, not months. Women have a distinctive qualitative and quantitative antibody response to influenza vaccination32. The observed differences in single-positivity to anti-RBD IgG in women are intriguing and could be the result immune response sex dimorphism. In addition, it should be noted that differences obtained in single-positivity to serological markers could also be the result from differences in the time of survey sampling and time since infection, which was different according to the phase of the epidemic wave in which each region was at the time of the survey.
Beyond the validation of the diagnostic accuracy of anti-N and anti-S IgG, this work also provides a useful baseline of seroreactivity based on natural infection. As COVID-19 vaccination coverage increases and novel SARS-CoV-2 variants are selected, seroprevalence estimation will have to take into account that most vaccines elicit anti-spike antibodies only. Thus, the combination of anti-N and anti-S serological markers may be the only approach to discriminate between natural infection and vaccination-induced seroconversion.
In conclusion, the in-house single-step anti-RBD IgG ELISA is a simple, economic, and robust assay with optimal sensitivity and specificity, with a comparable diagnostic performance in a population seroprevalence survey than the anti-N Elecsys® commercial system. As global vaccination coverage increases, combining both serological markers could be more informative in public health decisions than a single test alone.
Methods
COVID-19 cases and control serum panel for anti-RBD and N IgG test validation
Between April and August 2020, we prospectively recruited participants 18 years of age and older, with clinical presentation compatible with COVID-19 and hospitalized in preselected Mexican Institute for Social Security (IMSS) clinics of the Mexico City metropolitan area. Participants provided written informed consent. All methods were approved and performed according to the Research Ethics board of the Instituto Mexicano del Seguro Social (IMSS) guidelines (R-2020-785-065). For those who agreed to participate in the study, a serum sample was taken as soon as the RT-PCR confirmation was available. A second serum sample was taken > 3 weeks later, when possible. For validation, only RT-PCR + samples were analyzed, irrespectively of viral load. Asymptomatic individuals, or those that did not recalled their date of symptom onset were excluded. After blood draw, samples were centrifuged, and the serum was stored at − 80 °C. As non-COVID-19 samples (controls), we used a collection of 199 serum samples stored at − 80 °C, obtained during the National Health survey (ENSANUT 2018) in 2018 when no circulation of SARS-CoV-2 existed. According to the geographical origin of samples, control samples are countrywide representative, whereas COVID-19 samples derive from the Mexico City metropolitan area (MCMA) (Supplementary Fig. S1A). Age and gender distribution between cases and controls was comparable (p-value = 0.08) (Supplementary Fig. S1B–C), despite a deliberate enrichment of children samples in pre-pandemic controls to address potential antibody cross-reactivity with seasonal human coronavirus infection, which are common in the younger population33.
Anti-RBD IgG ELISA
The first step (anti-RBD screening) of the SARS-CoV-2 protocol was used as reference for a single-step in-house protocol13, and only minor modifications due to reagent and material availability were implemented. The plasmid encoding SARS-CoV-2 RBD was kindly provided by Dr. Florian Krammer at Mount Sinai Medical Center and stably transfected in CHO cells and cryopreserved. For large scale production (50 L), CHO cells were grown serially for four days at 37 °C, 5% CO2 and 130 rpm agitation in 1000-mL shake flasks (200 mL culture volume), then seeded in 5 L New Brunswick bioreactor for 3 additional days at similar conditions and with dissolved oxygen tension controlled > 30% of saturation. Finally, 3.7 L were further transferred to a 200 L Biostat CultiBag bioreactor and cultured for one more week. Fifty L of supernatant were depth-clarified and concentrated to 4 L by filtration with a Cogent M1 ultrafiltration system and a membrane cutoff of 10 kDa. Concentrated supernatant was subjected to affinity purification with Ni column in an AKTA prime chromatograph and further dialysis and sterile filtration with Millipak 20 filters. Final RBD purity was above 80% as quantified by SDS-PAGE.
RBD was immunoadsorbed to High bind 96-well ELISA flat bottomed plates (Corning CLS9018) at 2 μg/mL in PBS at 4 °C overnight and washed manually in PBS-Tween 20, followed by blocking at 4 °C in PBS-5% milk (Difco) for two hours at 4 °C. After washing, plates with 1:50 diluted serum samples were incubated at RT for two hours. After washing, plates were incubated with a 1:5,000 dilution of HRP-conjugated goat anti-human IgG (Sigma) for 1 h at RT, washed and developed with OPD in Phosphate/Citrate buffer. Absorbance was read in an ELISA plate reader (Biotek ELx808) with a 450 nm filter. Background absorbance was the average absorbance of 16 blank wells, which was subtracted to each sample well.
Anti-N IgG electro-chemiluminescence immunoassay (ECLIA)
Detection of anti-Nucleoprotein IgG was performed with the Elecsys® Anti-SARS-CoV-2 Electro-chemiluminescence immunoassay (ECLIA) (Roche Diagnostics), using the Cobas e 411 analyzer, according to manufacturer’s instructions.
Statistical analysis of diagnostic performance
Antigen-specific class-switched antibody secretion and seroconversion are highly dependent of the time since antigen exposure up to the development of functional germinal centers25. To account for the time of exposure, we categorized COVID-19 cases according to weeks PSO (Supplementary Fig. S1 and Supplementary Table S2) and evaluated diagnostic accuracy by estimating different parameters of diagnostic performance such as AUC of the ROC curve, and partial AUC using the R package pROC34. For statistical comparison of AUC and partial AUC we used the DeLong35 and Bootstrap methods36, respectively. ROC analysis allowed us to define the optical density (O.D.) cutoff for anti-RBD ELISA based on an expected specificity of ≥ 0.99.
To estimate sensitivity, specificity, predictive values and diagnostic likelihood ratios at different weeks PSO and to compare the performance of anti-RBD ELISA with the Elecsys® Anti-Nucleoprotein ECLIA, we used the statistical R package DTComPair37. Differences in sensitivity and specificity between methods were analyzed using the McNemar test38. Differences in predictive values were evaluated with the Generalized Score Statistic39. Diagnostic likelihood ratios were evaluated with the DLR regression model40. The concordance between both methods was estimated using Cohen’s Kappa statistic41 and Maxwell’s RE coefficient42 with the R package irr43.
Nationwide serological survey
The National Health and Nutrition survey (ENSANUT 2020-COVID-19) is a nationwide probabilistic household survey based on multistage and stratified sampling of participating households, implemented during August to November 2020. It included 10,216 complete household interviews and 9464 serum samples for biochemical markers and COVID-19 serology44. Participants provided written informed consent as approved by the Research ethics board of the National Institute of Public Health (CI 1679), and all methods were performed according to the Declaration of Helsinki and corresponding Institutional guidelines. All serum samples were independent of the validation sample set and were processed with the Elecsys® Anti-SARS-CoV-2 Electro-(ECLIA), whereas 9,068 were processed with the anti-RBD IgG ELISA.
Statistical analysis of anti-N and anti-RBD seroprevalence
We calculated the prevalence with 95% confidence intervals of only anti-RBD, only anti-N and both anti-RBD and anti-N, overall and by sociodemographic characteristics. Prevalence was adjusted by sensitivity and specificity of each validation test45. We grouped seropositive individuals as double-positive (anti-N and anti-RBD), positive only to anti-N, and positive only to anti-RBD. We calculated the distribution by age, sex, region, and symptoms with 95% confidence intervals. We then used a multinomial logistic regression including age, sex, and region, using the three groups as the outcome. We report the exponentiated coefficients as relative probabilities of only anti-N and only anti-RBD, using double positive as the reference group. We used the survey commands to account for the survey design. The analysis was made in Stata v14 (StataCorp).
References
World Health Organization. Population-based age-stratified seroepidemiological investigation protocol for COVID-19 virus infection. Version 1.1. 17 March, 2020. https://apps.who.int/iris/handle/10665/331656.
Che. Antigenic cross-reactivity between severe acute respiratory syndrome-associated coronavirus and human coronaviruses 229E and OC43. Journal of infectious diseases 191 (2005).
Patrick, D. M. et al. An outbreak of human coronavirus OC43 infection and serological cross-reactivity with SARS coronavirus. Can J Infect Dis Med Microbiol 17, 330–336. https://doi.org/10.1155/2006/152612 (2006).
Huang, A. T. et al. A systematic review of antibody mediated immunity to coronaviruses: kinetics, correlates of protection, and association with severity. Nat Commun 11, 4704. https://doi.org/10.1038/s41467-020-18450-4 (2020).
Woo, P. C. et al. False-positive results in a recombinant severe acute respiratory syndrome-associated coronavirus (SARS-CoV) nucleocapsid enzyme-linked immunosorbent assay due to HCoV-OC43 and HCoV-229E rectified by Western blotting with recombinant SARS-CoV spike polypeptide. J Clin Microbiol 42, 5885–5888. https://doi.org/10.1128/JCM.42.12.5885-5888.2004 (2004).
Van Walle, I., Leitmeyer, K., Broberg, E. K. & European, C.-m. l. g. Meta-analysis of the clinical performance of commercial SARS-CoV-2 nucleic acid and antibody tests up to 22 August 2020. Euro Surveill 26, doi:https://doi.org/10.2807/1560-7917.ES.2021.26.45.2001675 (2021).
Hulswit, R. J., de Haan, C. A. & Bosch, B. J. Coronavirus spike protein and tropism changes. Adv Virus Res 96, 29–57. https://doi.org/10.1016/bs.aivir.2016.08.004 (2016).
Pinto, D. et al. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature https://doi.org/10.1038/s41586-020-2349-y (2020).
Wec, A. Z. et al. Broad sarbecovirus neutralizing antibodies define a key site of vulnerability on the SARS-CoV-2 spike protein. bioRxiv, doi:https://doi.org/10.1101/2020.05.15.096511 (2020).
Roltgen, K. et al. Defining the features and duration of antibody responses to SARS-CoV-2 infection associated with disease severity and outcome. Sci Immunol. https://doi.org/10.1126/sciimmunol.abe0240 (2020).
Robbiani, D. F. et al. Convergent antibody responses to SARS-CoV-2 infection in convalescent individuals. https://doi.org/10.1101/2020.05.13.092619 (2020).
Dan, J. M. et al. Immunological memory to SARS-CoV-2 assessed for up to 8 months after infection. Science https://doi.org/10.1126/science.abf4063 (2021).
Amanat, F. et al. A serological assay to detect SARS-CoV-2 seroconversion in humans. Nat. Med. https://doi.org/10.1038/s41591-020-0913-5 (2020).
Landis, J. R. & Koch, G. G. The measurement of observer agreement for categorical data. Biometrics 33, 159–174 (1977).
McClish, D. K. Analyzing a portion of the ROC curve. Med Decis Making 9, 190–195. https://doi.org/10.1177/0272989X8900900307 (1989).
Basto-Abreu, A. et al. Nationally representative SARS-CoV-2 antibody prevalence estimates after the first epidemic wave in Mexico. Nat. Commun. 13, 589. https://doi.org/10.1038/s41467-022-28232-9 (2022).
Stadlbauer, D. et al. SARS-CoV-2 seroconversion in humans: A detailed protocol for a serological assay, antigen production, and test setup. Curr Protoc Microbiol 57, e100. https://doi.org/10.1002/cpmc.100 (2020).
Bolton, J. S. et al. Comparison of ELISA with electro-chemiluminescence technology for the qualitative and quantitative assessment of serological responses to vaccination. Malar. J. 19, 159. https://doi.org/10.1186/s12936-020-03225-5 (2020).
Emmerich, P. et al. Limited specificity of commercially available SARS-CoV-2 IgG ELISAs in serum samples of African origin. Trop. Med. Int. Health 26, 621–631. https://doi.org/10.1111/tmi.13569 (2021).
Sagara, I. et al. SARS-CoV-2 seroassay optimization and performance in a population with high background reactivity in Mali. medRxiv, doi:https://doi.org/10.1101/2021.03.08.21252784 (2021).
Lapidus, S. et al. Plasmodium infection induces cross-reactive antibodies to carbohydrate epitopes on the SARS-CoV-2 Spike protein. medRxiv, doi:https://doi.org/10.1101/2021.05.10.21256855 (2021).
Indenbaum, V. et al. Testing IgG antibodies against the RBD of SARS-CoV-2 is sufficient and necessary for COVID-19 diagnosis. PLoS ONE 15, e0241164. https://doi.org/10.1371/journal.pone.0241164 (2020).
Ltd, R. D. I. in diagnostics.roche.com (Roche Diagnostics International Ltd, 2020).
Santiago, L. et al. Determination of the concentration of IgG against the spike receptor-binding domain that predicts the viral neutralizing activity of convalescent plasma and serum against SARS-CoV-2. Biology (Basel). https://doi.org/10.3390/biology10030208 (2021).
Biram, A. & Shulman, Z. T cell help to B cells: Cognate and atypical interactions in peripheral and intestinal lymphoid tissues. Immunol. Rev. https://doi.org/10.1111/imr.12890 (2020).
Fenwick, C. et al. Changes in SARS-CoV-2 spike versus nucleoprotein antibody responses impact the estimates of infections in population-based seroprevalence studies. J Virol. https://doi.org/10.1128/JVI.01828-20 (2021).
Dong, Y. et al. Epidemiology of COVID-19 among children in China. Pediatrics https://doi.org/10.1542/peds.2020-0702 (2020).
Weisberg, S. P. et al. Distinct antibody responses to SARS-CoV-2 in children and adults across the COVID-19 clinical spectrum. Nat. Immunol. 22, 25–31. https://doi.org/10.1038/s41590-020-00826-9 (2021).
Long, Q.-X. et al. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat. Med. https://doi.org/10.1038/s41591-020-0897-1 (2020).
Long, Q.-X. et al. Clinical and immunological assessment of asymptomatic SARS-CoV-2 infections. Nat. Med. https://doi.org/10.1038/s41591-020-0965-6 (2020).
Lucas, C. et al. Delayed production of neutralizing antibodies correlates with fatal COVID-19. Nat. Med. https://doi.org/10.1038/s41591-021-01355-0 (2021).
Furman, D. et al. Systems analysis of sex differences reveals an immunosuppressive role for testosterone in the response to influenza vaccination. Proc. Natl. Acad. Sci. USA 111, 869–874. https://doi.org/10.1073/pnas.1321060111 (2014).
Su, S. et al. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol. 24, 490–502. https://doi.org/10.1016/j.tim.2016.03.003 (2016).
Robin, X. et al. pROC: An open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics 12, 77. https://doi.org/10.1186/1471-2105-12-77 (2011).
DeLong, E. R., DeLong, D. M. & Clarke-Pearson, D. L. Comparing the areas under two or more correlated receiver operating characteristic curves: A nonparametric approach. Biometrics 44, 837–845 (1988).
Hanley, J. A. & McNeil, B. J. A method of comparing the areas under receiver operating characteristic curves derived from the same cases. Radiology 148, 839–843. https://doi.org/10.1148/radiology.148.3.6878708 (1983).
DTComPair: comparison of binary diagnostic tests in a paired study design v. Version 1.0.3 (2014).
Mc, N. Q. Note on the sampling error of the difference between correlated proportions or percentages. Psychometrika 12, 153–157. https://doi.org/10.1007/BF02295996 (1947).
Leisenring, W., Alonzo, T. & Pepe, M. S. Comparisons of predictive values of binary medical diagnostic tests for paired designs. Biometrics 56, 345–351. https://doi.org/10.1111/j.0006-341x.2000.00345.x (2000).
Gu, W. & Pepe, M. S. Estimating the diagnostic likelihood ratio of a continuous marker. Biostatistics 12, 87–101. https://doi.org/10.1093/biostatistics/kxq045 (2011).
Cohen, J. A coefficient of agreement for nominal scales. Edu. Psych. Meas. 20, 37–46. https://doi.org/10.1177/001316446002000104 (1960).
Maxwell, A. E. Coefficients of agreement between observers and their interpretation. Br. J. Psychiatry. 130, 79–83. https://doi.org/10.1192/bjp.130.1.79 (1977).
irr: Various Coefficients of Interrater Reliability and Agreement v. Version: 0.84.1 (2019).
Romero-Martinez, M. et al. Metodologia de la Encuesta Nacional de Salud y Nutricion 2020 sobre Covid-19. Salud Publica Mex 63, 444–451. https://doi.org/10.21149/12580 (2021).
Rogan, W. J. & Gladen, B. Estimating prevalence from the results of a screening test. Am. J. Epidemiol. 107, 71–76. https://doi.org/10.1093/oxfordjournals.aje.a112510 (1978).
Acknowledgements
We thank Dr. Florian Krammer and co-workers of the Mount Sinai Medical Center for providing reagents for RBD recombinant production, Manuel Velazquez-Meza, Jumara Martinez-Barranca and Margarita Hernández-Salgado for technical assistance. This study was funded by the Mexican Government through the National Health and Nutrition Survey provision to the National Institute of Public Health and by the Centers for Disease Control and Prevention of the United States of America (GRANT: NU50CK000493) through the CDC-Mexico Cooperative Agreement for Surveillance, Epidemiology, and Laboratory Capacity. Funding for RBD production by Laboratorios Liomont, UNAM DGAPA PAPIIT IV-200820 and IV-200420, and CITNOVA Estado Hidalgo.
Author information
Authors and Affiliations
Contributions
C.A.A., J.M.B., M.H.A., L.A.P., O.T.R., and T.B.G. participated in the conception and design of the work; C.S., D.B., I.L.M., J.C.R., R.A.A. and R.L. participated in data acquisition. A.B.A., A.C.E., C.G.C., J.M.B. and M.C. performed data analysis and interpretation; J.M.B. drafted the work and C.A.A., M.H.A., O.T.R. and T.B.G. substantively revised it. All authors have approved the submitted version.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Martínez-Barnetche, J., Carnalla, M., Gaspar-Castillo, C. et al. Comparable diagnostic accuracy of SARS-CoV-2 Spike RBD and N-specific IgG tests to determine pre-vaccination nation-wide baseline seroprevalence in Mexico. Sci Rep 12, 18014 (2022). https://doi.org/10.1038/s41598-022-22146-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-022-22146-8
- Springer Nature Limited