Abstract
Improving maternal nutrition during pregnancy/lactation is a promising strategy to maximise the intestinal health of piglets undergoing abrupt weaning under commercial production conditions. This experiment investigated the effects of maternal supplementation of a casein hydrolysate and yeast β-glucan (CH-YBG) from day 83 of gestation until weaning (day 28) on sow faecal microbial populations and measures of piglet gastrointestinal health parameters at weaning. Sows (n = 10 sows/group) were assigned to: (1) control diet, and (2) control diet + CH-YBG. Maternal supplementation increased the abundance of the phylum Firmicutes, including members Lactobacillus in the sows faeces, with a concomitant increase in the caecal abundance of Lactobacillus in the weaned piglets compared to the controls. Piglets weaned from the supplemented sows had increased villus height in the duodenum (P < 0.05) and increased villus height to crypt depth ratio in the jejunum, as well as a decreased expression of the proinflammatory cytokine genes (IL6/TNF/TGFB), the tight junction gene CLDN3 and the mucin gene MUC2 in the duodenum/jejunum compared to the controls (P < 0.05). In conclusion, maternal CH-YBG supplementation during pregnancy/lactation improved microbial, structural, and inflammatory measures of gastrointestinal health of piglets at weaning. This is a promising strategy to alleviate the challenges that occur with early abrupt weaning in commercial pig production.
Similar content being viewed by others
Introduction
Commercial pigs are subjected to significant levels of stress at weaning. This causes an initial reduction in feed intake, contributing to adverse gut morphological and functional changes such as villous atrophy and crypt hyperplasia. This in turn, increases epithelial permeability and inflammation1,2. Alongside an immature immune system, these factors favour the proliferation of pathogenic bacteria, including, enterotoxigenic Escherichia coli, thus increasing overall susceptibility to infection and post-weaning diarrhoea3. Supplementation of dietary zinc oxide (ZnO) at pharmacological doses improves growth and controls the proliferation of pathogenic bacteria post-weaning. However, the EU have begun the phasing out of pharmacological doses of ZnO (Commission Implementing Decision of 26.6.2017, C (2017) 4, 529 Final) alongside further restrictions on the use of infeed medication (Regulation (EU) 2019/6) in weaner pig diets by 2022 due to their association with antimicrobial resistance.
Recent research regarding the enhancement of performance and gut health, in pigs, has focussed on post-weaning dietary manipulation. However, a more effective alternative may be supplementing the diet of the gestating and/or lactating sow to enhance the pre-weaning health status of the piglet as this may have a positive influence on post-weaning performance4,5. The gastrointestinal tract (GIT) of the piglet is colonized immediately after birth by bacteria originating from both the sow and the environment, and these microbes play a fundamental role in the development of a healthy GIT and immune system6.
The digestion or enzymatic hydrolysis of milk proteins has been shown to yield physiologically important bioactive peptides that have a wide range of biological activities7,8. Casein hydrolysates reduced inflammation in an ex-vivo model9, however in-vivo they are susceptible to breakdown in the stomach. To prevent this and preserve their bioactivity, hydrolysates can be naturally encapsulated with a yeast β-glucan. The combination of 5 kDa retentate (5kDaR), derived from moderate degree of hydrolysis (11–16%) of bovine milk casein with a yeast β-glucan in the weaning pig diet was associated with an improvement in overall performance and gut health parameters10. As well as acting as a microencapsulating agent, β-glucans have been extensively studied for their antioxidant, immunological, and anti-inflammatory effects11,12,13. Thus, in the present study, it was hypothesised that maternal supplementation with a casein hydrolysate + yeast β-glucan (CH-YBG) from day 83 of gestation would alter the GIT microbiota of the sow and modulate microbial populations, the inflammatory response and aspects of intestinal health of her progeny at weaning. Thus, the objective of this experiment is to investigate the effects of maternal dietary supplementation of a CH-YBG from day 83 of gestation until weaning (day 28) on sow faecal microbial populations, and measures of piglet gastrointestinal health parameters at weaning.
Materials and methods
Experimental design and diets
A total of 20 crossbred pregnant gilts (Large White X Landrace genetic lines) were randomly assigned to one of the two dietary groups: (T1) basal gestation and lactation diet (control) and (T2) basal gestation and lactation diet supplemented with a combination of a CH-YBG from day 83 of gestation until weaning (day 28). The gestation and lactation diet were top-dressed each morning (09.00 h) with the experimental supplement (2 g/day; 1 g casein hydrolysate + 1 g yeast β-glucan) to ensure consumption. The casein hydrolysate (CH) used in the current study was produced from the hydrolysis of sodium caseinate (NaCas, ≈ 90% w/w protein, Kerry Food Ingredients, Listowel, Ireland) derived from bovine and has been reported previously by Mukhopadhya et al.9. The yeast β-glucan was derived from Saccharomyces cerevisiae (Biothera Pharmaceuticals, Inc., Eagan, MI, USA) and the optimum concentrations of a casein hydrolysate and a yeast β-glucan (CH-YBG) were established from previous studies14,15. The gestation diet contained 140 g/kg of crude protein (CP), 13.5 MJ/kg of digestible energy (DE) and 4.4 g/kg of standardised ileal digestible (SID) lysine. The lactation diet contained 190 g/kg of CP, 14.5 MJ/kg of DE and 8.5 g/kg of SID lysine. All amino acid requirements were met relative to lysine16. The ingredient composition of the diets is presented in Table 1.
From day 83 of gestation until day 110, the gilts were housed in groups of ten and the experimental supplement (CH-YBG) was top-dressed on the gestation diet prior to feeding each morning (09.00 h) to ensure consumption. The gilts were housed in individual stalls at feeding. The gilts received 2.5 kg/day of gestation diet from day 83 until day 110 of gestation. From day 110, gilts received specific amounts of feed in the following quantities: 2 kg/day of diet from day 110 of gestation until the day of farrowing (day 0) and then the feed supply was increased by 1 kg/day until day 3 post-farrowing and by 0.5 kg/day until day 6 post-farrowing. Afterwards, the sows were allowed semi-ad libitum consumption of the standard lactation diet, which was adjusted for each sow depending on daily intake until weaning at day 28. The sows were fed in 2 equal meals provided at 09.00 and 15.00 h.
The sows and piglets were individually housed in farrowing pens (2.2 × 2.4 m) with crates, slatted floors, and heat pads for piglets. The farrowing room temperature was maintained at 20 °C. The sows were individually fed and had ad libitum access to drinking water throughout the experimental period.
On the expected farrowing date, fresh sow faecal samples (approximately 10 g) were collected from the gilts into sterile containers (Sarstedt, Wexford, Ireland) and immediately frozen ( 20 °C) for subsequent 16 s rRNA sequencing.
Management of piglets
All farrowings were supervised. Between 6 and 12 h after the birth of the last piglet, litter size was adjusted by cross-fostering piglets within sow diets to ensure that sows nursed a similar number of piglets (n = 12), and this was maintained throughout the suckling period until weaning. The piglets body weight (BW) was recorded at birth and weaning, and the average daily gain (ADG) calculated. The piglets received an intramuscular injection of Fe-dextran (Ferdex 100; Medion Farma Jaya) on day 7 after birth. No creep feed was offered to the piglets throughout the lactation period, and piglets did not have access to the sows’ feed.
Collection of piglet tissue and digesta samples at weaning
On the day of weaning (day 28), one male piglet per sow of average birth weight (which was identified at parturition) was euthanized after a lethal injection with pentobarbitone sodium, Euthatal solution, 200 mg/ml, (Merial Animal Health, Essex, UK) at a rate of 0.71 ml/kg BW to the cranial vena cava to humanely euthanize the animal. Euthanasia was performed by a competent person in a separate room away from sight and sound of the other pigs. The entire intestinal tract was immediately removed. Sections from the duodenum (located 10 cm distal from the stomach), the jejunum (60 cm from the stomach) and the ileum (15 cm from the caecum) were excised and fixed in 10% neutral-buffered formalin. Digesta from the caecum and colon was collected and stored in sterile containers (Sarstedt, Wexford, Ireland) and immediately frozen at − 20 °C for subsequent 16 s rRNA sequencing. In addition, tissue samples were taken from the duodenum, jejunum and ileum for the purpose of measuring the expression of cytokines, digestive enzymes, nutrient transporters, mucins and tight junctions using QPCR. Tissue sections (1 cm) from the duodenum, jejunum, and ileum were cut out, dissected along the mesentery, emptied, and rinsed using sterile phosphate buffered saline (Oxoid, Hampshire, UK). The tissue sections were stripped of the overlying smooth muscle before storage in RNAlater® (5 ml) solution (Applied Biosystems, Foster City, CA, USA) overnight at 4 °C. The RNAlater® was removed before storing the samples at − 80 °C.
Gut morphological analysis
Preserved duodenal, jejunal and ileal tissue samples were prepared using standard paraffin embedding techniques as previously described18. All samples were sectioned at a thickness of 5 μm and stained with haematoxylin and eosin. The villus height (VH) and crypt depth (CD) were measured in the stained sections (4× objective) using a light microscope fitted with an image analyser (Image-Pro Plus; Media Cybernetics, Oxon, UK). Measurements of fifteen correctly orientated and intact villi and crypts were taken for each segment. The VH was measured from the crypt-villus junction to the tip of the villus, and CD was measured from the crypt-villus junction to the base.
Gene expression in the small intestine
RNA extraction and cDNA synthesis
The total RNA was extracted from the duodenal, jejunal and ileal tissue using the TRI Reagent (Sigma-Aldrich, St. Louis, MO, USA) according to the manufacturer’s instructions. This crude RNA extract was further purified using the GenElute™ Mammalian Total RNA Miniprep kit (Sigma-Aldrich) which incorporated a DNase step using an on-column DNase 1 Digestion set (Sigma-Aldrich). The total RNA was quantified using a Nanodrop-ND1000 spectrophotometer (Thermo Scientific) and the purity was assessed from the ratio of the absorbance at 260 nm and 280 nm. The RNA integrity was assessed using an Agilent 2100 Bioanalyzer using an RNA 6000 Nano LabChip kit (Agilent Technologies, Santa Clara, CA, USA). All samples had a 260:280 ratio > 2.0 and an RNA integrity number (RIN) > 8.0. The total RNA (2 μg) was reverse transcribed using a High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems) and oligo (dT) primers in a final reaction volume of 40 μl, according to the manufacturer’s instructions. The cDNA was then adjusted to a volume of 360 μl with nuclease-free water.
Quantitative real-time Polymerase Chain Reaction (QPCR)
The quantitative PCR (QPCR) reaction mix (20 μl) contained GoTaq qPCR Master Mix (10 μl) (Promega, Madison, WI), forward and reverse primers (1.2 μl) (5 μM), nuclease-free water (3.8 μl) and cDNA (5 μl). All QPCR reactions were performed in duplicate on the 7500 ABI Prism Sequence detection System (Applied Biosystems, Foster City, CA). The cycling conditions included a denaturation step of 95 °C for 10 min followed by 40 cycles of 95 °C for 15 s and 60 °C for 1 min. All primers were designed using the Primer Express Software (Applied Biosystems, Foster City, CA) and synthesised by MWG Biotech UK Ltd (Milton Keynes, UK) and are presented in Table 2. Dissociation curves were generated to confirm the specificity of the resulting PCR products. The QPCR assay efficiencies were established by plotting the cycling threshold (CT) values derived from fourfold serial dilutions of cDNA against their arbitrary quantities and only assays exhibiting 90–110% efficiency and single products were used in this study. Normalised relative quantities were obtained using the qbase PLUS software (Biogazelle, Ghent, Belgium) from stable reference genes; B2M, ACTB and PPIA. These genes were selected as reference genes based on their M value (< 1.5) generated by the GeNorm algorithm within GeNorm. The genes analysed in the current study are as follows: SLC15A1 (previously known as PEPT1); FABP2; SLC2A1 (previously known as GLUT1); SLC5A1 (previously known as SGLT1); SLC2A2 (previously known as GLUT2); SLC2A5 (previously known as GLUT5); SLC16A; SLC1A4; SLC5A8; CCK; TNF; CXCL8 (previously known as IL8); IL6; IL10; IFNG; MUC2; TGFB1; IL17; CLDN3; MUC1; B2M; ACTB; PPIA.
Microbiological analyses
Microbial DNA extraction
Microbial genomic DNA was extracted using QIAamp PowerFecal Pro DNA Kit (Qiagen, West Sussex, UK) according to the manufacturer’s instructions. The DNA quantity and quality were evaluated using a Nanodrop ND-1000 Spectrophotometer (Thermo Scientific, Wilmington, DE).
Illumina sequencing
Extraction of the bacterial DNA from the caecal digesta samples and high-throughput sequencing of the V3–V4 hypervariable region of the bacterial 16S rRNA gene was performed on an Illumina MiSeq platform according to their standard protocols (Eurofins Genomics, Ebersberg, Germany). Briefly, the V3–V4 region was PCR-amplified using universal primers containing adapter overhang nucleotide sequences for forward and reverse index primers. Amplicons were purified using AMPure XP beads (Beckman Coulter, Indianapolis, IN) and set up for the index PCR with Nextera XT index primers (Illumina, San Diego, CA). The indexed samples were purified using AMPure XP beads, quantified using a fragment analyser (Agilent, Santa Clara, CA), and equal quantities from each sample were pooled. The resulting pooled library was quantified using the Bioanalyzer 7500 DNA kit (Agilent, Santa Clara, CA) and sequenced using the v3 chemistry (2 × 300 bp paired-end reads).
Bioinformatic
The bioinformatic analysis of the resulting sequences was performed by Eurofins Genomics (Ebersberg, Germany) using the open-source software package (version 1.9.1) Quantitative Insights into Microbial Ecology (QIIME)19. All raw reads passing the standard Illumina chastity filter were demultiplexed according to their index sequences (read quality score > 30). The primer sequences were clipped from the starts of the raw forward and reverse reads. If primer sequences were not perfectly matched, read pairs were removed to retain only high-quality reads. Paired-end reads were then merged if possible, to obtain a single, longer read that covers the full target region using the software FLASH 2.2.0020. Pairs were merged with a minimum overlap size of 10 bp to reduce false-positive merges. The forward read was only retained for the subsequent analysis steps when merging was not possible. Merged reads were quality filtered according to the expected length and known length variations of the V3–V4 region (ca. 445 bp). The ends of retained forward reads were clipped to a total read length of 285 bp to remove low quality bases. Merged and retained reads containing ambiguous bases were discarded. The filtered reads (merged and quality clipped retained forward reads) were used for the microbiome profiling. Chimeric reads were identified and removed based on the de-novo algorithm of UCHIME21 as implemented in the VSEARCH package22. The remaining set of high-quality reads were processed using minimum entropy decomposition (MED) to partition reads to operational taxonomic units (OTU)23,24. DC-MEGABLAST alignments of cluster representative sequences to the NCBI nucleotide sequence database were performed for taxonomic assignment (from phylum to species) of each OTU. A sequence identity of 70% across at least 80% of the representative sequence was the minimal requirement for considering reference sequences. Abundances of bacterial taxonomic units were normalized using lineage-specific copy numbers of the relevant marker genes to improve estimates25.
The normalized OTU table combined with the phenotype metadata and phylogenetic tree comprised the data matrix. This matrix was then input into the phyloseq package within R (http://www.r-project.org; version 3.5.0). The dynamics of richness and diversity in the microbiota were computed with the observed, Chao1, ACE, Shannon, Simpson, InvSimpson and Fisher indices. The Simpson and Shannon indices of diversity account for both richness and evenness parameters. The beta diversity measurements are a measure of separation of the phylogenetic structure of the OTU in one sample compared with all other samples. This was estimated by normalising the data so taxonomic feature counts were comparable across samples. Several distance metrics were considered, to calculate the distance matrix of the different multidimensional reduction methods. These included weighted/unweighted UniFrac distance and non-phylogenetic distance metrics (i.e., Bray–Curtis, Jensen–Shannon divergence and Euclidian) using phyloseq in R26,27. Differential abundance testing was performed on tables extracted from the phyloseq object at phylum, family, and genus level. The data was analysed using the PROC Glimmix procedure within Statistical Analysis Software (SAS) 9.4 (SAS Institute, Cary, NC, USA). The model assessed the effect of ‘treatment’, with the piglet (sow pen) as the experimental unit. 10 sows and 10 piglets per treatment group were used for the statistical analysis of the relative bacterial abundances. Results are presented using Benjamini–Hochberg (BH) adjusted P-values.
Feed analysis
The feed samples were milled through a 1 mm screen (Christy and Norris Hammer Mill, Chelmsford, England) and retained for chemical analysis. The gross energy (GE) content was determined using an adiabatic bomb calorimeter (Parr Instruments, Moline, IL USA) as previously described28. The dry matter (DM) content of the feed was determined after drying overnight at 104 °C. Feed samples were analysed for crude ash (AOAC method 942.0529) and nitrogen (N × 6.25; AOAC method 990.0329). All samples were measured in duplicate.
Statistical analysis
The data on performance, intestinal morphology and gene expression were initially checked for normality using the univariate procedure of Statistical Analysis Software30. The data on performance, intestinal morphology, and gene expression were analysed using the GLM procedure within SAS. The model assessed the effect of treatment, with the sow as the experimental unit. For piglet diarrhoea score, the data was analysed by repeated-measures analysis using the PROC MIXED procedure of SAS. All data presented in the tables are expressed as least-square means with their standard errors. The probability level that denoted significance was P < 0.05. Data are presented as least-square means and standard error of the mean.
Ethics statement
All experimental procedures described in the present experiment were approved under the University College Dublin Animal Research Ethics Committee, Ireland (AREC-17-38-Sweeney). The study was conducted in accordance with Irish legislation (SI no. 543/2012) and the EU directive 2010/63/EU for animal experimentation and in compliance with the ARRIVE guidelines. The authors confirm that the ethical policies of the journal, as noted on the journal’s author guidelines page, have been adhered to and the appropriate ethical review committee approval has been received. The authors confirm that they have followed the EU standards for the protection of animals used for scientific purposes and the ARRIVE guidelines.
Results
Gestation length and suckling piglet growth performance
The effect of maternal supplementation on gestation length and piglet body weight and ADG is presented in Table 3. There was no effect of maternal dietary supplementation on gestation length (115.2 vs 115.4 days, P = 0.724) and piglet body weight at birth (1.23 vs 1.15 kg, P = 0.052) and weaning (6.46 vs 6.58, P = 0.721), and ADG (0.19 vs 0.19, P = 0.536) from birth to weaning (P > 0.05).
The effects of maternal dietary supplementation on the sow faecal and piglet caecal and colonic microbiota
Bacterial richness and diversity
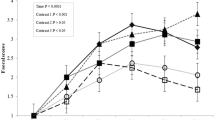
The effects of maternal supplementation on the measures of beta diversity and alpha diversity are presented in Fig. 1 and Table 4, respectively. Beta diversity in the piglets caecal and colonic microbiome differed to that of the sow (P < 0.05) based on Permanova analysis and through visualisation using the Bray Curtis distance matrix and multi-dimensional scaling. Maternal dietary supplementation had no effect on the Observed, Chao1, ACE, Shannon, Simpson, InvSimpson and Fisher index measures of alpha diversity (P > 0.05) in either the sows faeces or the piglets caecal and colonic digesta.
Differential bacterial abundance analysis
All data on bacterial abundances at phylum, family and genus level are provided in Tables 5 and 6.
Sow faeces at farrowing
Phylum
There were six bacterial phyla identified with Firmicutes being the dominant phyla (~ 87.69%) followed by Proteobacteria (~ 6.52%), Spirochaetes (~ 3.50%), Actinobacteria (~ 1.38%), Tenericutes (~ 0.47%), and Synergistetes (~ 0.10%). Firmicutes were increased (P < 0.05) and Proteobacteria were decreased (P < 0.05) in the supplemented sows compared to the control sows.
Family
At family level, dietary supplementation increased the relative abundance of Lactobacillaceae (P < 0.0001) and Lachnospiraceae (P < 0.05) compared with the control group. Dietary supplementation decreased the relative abundance of Christensenellaceae (P < 0.05), Planococcaceae (P < 0.05) and Moraxellaceae (P < 0.0001) compared to the control.
Genus
At genus level, dietary supplementation increased the relative abundance of Lactobacillus (P < 0.001), Metabacterium (P < 0.01), Niameybacter (P < 0.01) and Gemmiger (P < 0.05) compared to the control. Dietary supplementation decreased the relative abundance of Christensenella (P < 0.05), Kineothrix (P < 0.05) and Acinetobacter (P < 0.0001) compared to the control.
Piglets caecal and colonic microbiota
Phylum
In the caecum and colon, there were six bacterial phyla identified with Firmicutes being the dominant phyla followed by Actinobacteria, Proteobacteria, Verrucomicrobia, Fusobacteria, and Bacteroidetes. The relative abundance of Actinobacteria was decreased (P < 0.05) in the caecum and colon of piglets from supplemented sows compared to piglets from control sows.
Family
In the caecum, the relative abundance of Lactobacillacea and Oscillospiraceae were increased (P < 0.05), while the relative abundance of Coriobacteriaceae, Ruminococcaceae and Akkermansiaceae decreased (P < 0.05) in piglets from supplemented sows compared to piglets from control sows. In the caecum and colon, the relative abundance of Erysipelotrichaceae and Christensenellaceae were increased (P < 0.05), while the relative abundance of Atopobiaceae was decreased (P < 0.05) in piglets from supplemented sows compared to piglets from control sows.
Gene expression in the small intestine
The intestinal expression of genes related to nutrient digestion and absorption, mucosal barrier function, and immunity in the duodenum, jejunum and ileum of piglets are presented in Table 7.
Nutrient transporter gene expression
The gene expression of FABP2 (fatty acid binding protein 2, P < 0.01), SLC15A1 (peptide transporter 1, P < 0.01), SLC2A2 (glucose transporter 2, P < 0.05), SLC2A5 (glucose transporter 5, P < 0.05) and SLC1A4 (solute carrier family 1 member 4, P < 0.05) was upregulated in the duodenum and jejunum of piglets suckling the supplemented sows compared with those suckling the control sows. In the jejunum, the gene expression of SLC5A1 (sodium transporter 1, P < 0.01) was upregulated and there was a numerical tendency for SLC16A (solute carrier family 16 member 1, P < 0.07) and SLC5A8 (solute carrier family 5 member 8, P < 0.07) to be upregulated in piglets suckling the supplemented sows compared with those suckling the control sows.
Expression of genes involved in inflammation and the epithelial barrier
The expression of IL6 (interleukin-6, P < 0.05), TNF (tumor necrosis factor, P < 0.05) and TGFB (transforming growth factor beta-1, P < 0.05) were down-regulated in the duodenum and jejunum of piglets suckling the supplemented sows compared with those suckling the control sows. The expression of CLDN3 (claudin-3, P < 0.01) was up-regulated in the duodenum and jejunum of piglets suckling the supplemented sows compared with those suckling the control sows. The expression of IL10 (interleukin-10, P < 0.01) was down-regulated in the jejunum of piglets suckling the supplemented sows compared with those suckling the control sows. The expression of CXCL8 (interleukin 8, P < 0.01) and CLDN3 (P < 0.0001) were upregulated in the jejunum of piglets suckling the supplemented sows compared with those suckling the control sows.
Ileum
The expression of IFN (interferon gamma, P < 0.05) was downregulated in the ileum of piglets suckling the supplemented sows compared with those suckling the control sows.
Piglet small-intestinal morphology
The effect of maternal supplementation on piglet small intestinal morphology is presented in Table 8.
In the duodenum, piglets suckling the supplemented sows had larger VH (P < 0.05) compared with those suckling the control sows. In the ileum, piglets suckling supplemented sows had a larger VH:CD ratio (P < 0.05) compared with those suckling the control sows.
Discussion
In the present study, it was hypothesised that maternal supplementation with a casein hydrolysate and yeast β-glucan (CH-YBG) from day 83 of gestation would alter the GIT microbiota of the sow and modulate selected intestinal microbial populations, the inflammatory response, and aspects of intestinal health of her progeny at weaning. Maternal supplementation with CH-YBG altered the sow’s faecal microbiota by increasing the relative abundance of bacterial members of the Firmicutes phylum, including Lactobacillus. The positive response observed in piglets suckling the CH-YBG supplemented sows, such as increased abundance of numerous Firmicutes, including Lactobacillus and Christensenella in the caecal microbiota, improved intestinal morphology, decreased expression of proinflammatory cytokine genes, and increased expression of nutrient transporter, tight junction, and mucin genes at the time of weaning supports this hypothesis.
The early colonization of the piglets intestinal microbiota plays an essential role in the development of the piglets intestinal immune system, barrier function and nutrient absorption31,32. The major microbial changes in this experiment were observed within the Firmicutes, with supplemented sows having a higher abundance of beneficial bacteria such as Lactobacillus compared to the control sows. The abundance of Lactobacillus, Christensenella and Erysipelotrichaceae was also increased in the progeny of supplemented sows at 28 days of age. These genera are positively correlated with a healthy gut, with low numbers associated with inflammatory disorders such as inflammatory bowel disease33,34 and Crohns disease35,36. The increase in Lactobacillus in the sow is likely to be attributable to the casein hydrolysate’s role as a substrate for the growth of Lactobacilli, as casein hydrolysate supplementation increased the growth of Lactobacillus in rats and pigs37,38. Both Lactobacillus and Christensenella species have strong anti-inflammatory properties and play an important role in maintaining microbial symbiosis, thereby contributing to the establishment of an “optimal” microbiota in piglets at weaning. These results indicate that maternal supplementation positively influenced the microbial composition in the faeces of the sow and in the large intestine of her progeny and thus, may play a role in improving the health of piglets at weaning.
The small intestine plays an essential role in the digestion and absorption of nutrients and serves as a key regulator of growth. Under conventional practice, abrupt weaning is often associated with undesirable morphological changes, such as villous atrophy and crypt hyperplasia, leading to functional changes such as a decrease in nutrient digestion and absorption2,39. Thus, supplements that positively influence intestinal health should aid in the amelioration of the intestinal dysfunction associated with weaning stress. Supplementation with bacterial members of the genus Lactobacillus has been shown to improve intestinal health in broilers through the improvement of small intestinal villous height and crypt depth40,41. In the present study, pigs weaned from supplemented sows had improved VH in the duodenum and VH to CD ratio in the ileum.
The improvement in intestinal health in response to maternal supplementation, consequently enhanced the digestive and absorptive capacity of the intestinal epithelium resulting in an upregulation of nutrient transporters in the small intestine. Maternal dietary supplementation with CH-YBG increased the expression of fatty acid transporter FABP2, peptide transporter SLC15A1, glucose transporters SLC2A1, SLC2A2, SLC2A5, and SGLT1, amino acid transporter SLC1A4, and sodium transporter SLC5A8 and had a tendency to increase solute carrier SLC16A. This suggests there is increased availability and transport of fatty acids, sugars, sodium, peptides and amino acids in the duodenum and jejunum of pigs suckling supplemented sows. The upregulation of these nutrient transporters indicates a positive functional adaptation in the pigs, and potentially improving their ability to digest a diet primarily comprised of carbohydrates and protein post-weaning.
In the present study, maternal supplementation increased the expression of the tight junction gene, CLDN3 and the mucin gene MUC2. Claudin 3 is a tight junction integral protein that decreases paracellular permeability, thus preventing the translocation of luminal antigens42. Mucins provide a barrier against potential pathogens while also modulating the expression of inflammatory cytokines43. It is possible that the increased expression of CLDN3 reduced the translocation of bacteria across the intestinal barrier, while the increased expression of MUC2 reduced the adherence of bacteria to the intestinal cells, thereby reducing the risk of pigs to infection and diarrhoea. The increase in genes associated with intestinal barrier function was concomitant with a decrease in the expression of pro-inflammatory cytokine genes. Overall, these effects indicate a better host immune response with regard to potential environmental or pathogenic challenge at weaning time. Maternal supplementation from day 83 of gestation and throughout the lactation period decreased expression of the pro-inflammatory cytokine genes IL6, TGFB, TNF and IFNG. IL-6 and TNF are potent mediators of inflammation and play an important role in the pathogenesis of E. coli caused diarrhoea44,45. It is likely that the increase in the expression of the tight junction genes strengthened the intestinal barrier, thus limiting the ability of antigens to enter the body and prevent adverse immune responses. When the immune system is activated, and inflammation occurs, maintenance energy and amino acid requirements are increased, thereby reducing the proportion of daily nutrient intake directed towards growth46. Thus, the potential decrease in immune system activity in maternally supplemented pigs may result in an improvement in growth performance during the post-weaning period.
While it was not within the scope of this study to identify what effects each bioactive had on the gastrointestinal health parameters measured, it is worthy to note that the 5 kDa retentate (5kDaR) of a CH has established microbial activity and anti-inflammatory effects in vitro9,47,48. Furthermore, in a previous study the combination of the 5kDaR + YBG positively enhanced gastrointestinal parameters in pigs post-weaning, whereas the individual inclusion of either 5kDaR or YBG were not effective10. It is evident in the present study that maternal supplementation with a CH-YBG improved several gastrointestinal health parameters that influence performance. Thus, it may be anticipated that this would translate into an improvement in growth performance at weaning, however piglet weights at weaning were not influenced by sow diet. A similar effect was observed following maternal supplementation with seaweed-derived polysaccharides, containing laminarin and fucoidan49. Furthermore, maternal supplementation with Bacillus altitudinis50 did not influence the growth performance of piglets at weaning, however maternal supplementation had an impact on the progeny’s performance post-weaning. A further study to explore the effects of maternal supplementation with a CH-YBG on piglet growth performance post-weaning is warranted.
Conclusion
Maternal dietary supplementation with a combination of a casein hydrolysate + yeast β-glucan from day 83 of gestation to weaning had a positive effect on the microbial population of sows and their piglets, represented by a higher abundance of several bacterial members of the phylum Firmicutes, including Lactobacillus and Christensenella. Maternal supplementation enhanced piglet gastrointestinal health by improving intestinal morphology, reducing the expression of proinflammatory cytokine genes, and increasing the expression of nutrient transporter, tight junction, and mucin genes at weaning. These findings indicate that maternal casein hydrolysate + yeast β-glucan supplementation may be a promising strategy for improving the gastrointestinal health and function of piglets at weaning, making piglets more resilient to post-weaning challenges.
Data availability
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
References
Spreeuwenberg, M. A., Verdonk, J. M., Gaskins, H. R. & Verstegen, M. W. Small intestine epithelial barrier function is compromised in pigs with low feed intake at weaning. J. Nutr. 131, 1520–1527. https://doi.org/10.1093/jn/131.5.1520 (2001).
Pluske, J. R. Feed- and feed additives-related aspects of gut health and development in weanling pigs. J. Anim. Sci. Biotechnol. 4, 1–1. https://doi.org/10.1186/2049-1891-4-1 (2013).
Luppi, A. et al. Prevalence of virulence factors in enterotoxigenic Escherichia coli isolated from pigs with post-weaning diarrhoea in Europe. Porc. Health Manag. 2, 1–6 (2016).
Sweeney, T. & Odoherty, J. V. Marine macroalgal extracts to maintain gut homeostasis in the weaning piglet. Domest. Anim. Endocrinol. 56, S84–S89 (2016).
O’Doherty, J., Bouwhuis, M. & Sweeney, T. Novel marine polysaccharides and maternal nutrition to stimulate gut health and performance in post-weaned pigs. Anim. Prod. Sci. 57, 2376 (2017).
de Agüero, M. G. et al. The maternal microbiota drives early postnatal innate immune development. Science 351, 1296–1302 (2016).
Marcone, S., Belton, O. & Fitzgerald, D. J. Milk derived bioactive peptides and their health promoting effects: A potential role in atherosclerosis. Br. J. Clin. Pharmacol. 83, 152–162. https://doi.org/10.1111/bcp.13002 (2017).
Mukhopadhya, A. & Sweeney, T. Milk proteins: Processing of bioactive fractions and effects on gut health. Milk Proteins. 83 (2016).
Mukhopadhya, A. et al. Anti-inflammatory effects of a casein hydrolysate and its peptide-enriched fractions on TNFα-challenged Caco-2 cells and LPS-challenged porcine colonic explants. Food Sci. Nutr. 2, 712–723. https://doi.org/10.1002/fsn3.153 (2014).
Mukhopadhya, A., O’Doherty, J. V. & Sweeney, T. A combination of yeast beta-glucan and milk hydrolysate is a suitable alternative to zinc oxide in the race to alleviate post-weaning diarrhoea in piglets. Sci. Rep. 9, 616. https://doi.org/10.1038/s41598-018-37004-9 (2019).
Du, B., Lin, C., Bian, Z. & Xu, B. An insight into anti-inflammatory effects of fungal beta-glucans. Trends Food Sci. Tech. 41, 49–59 (2015).
Suchecka, D. et al. Antioxidative and anti-inflammatory effects of high beta-glucan concentration purified aqueous extract from oat in experimental model of LPS-induced chronic enteritis. J. Funct. Foods. 14, 244–254 (2015).
Cao, H. et al. Effects of dietary supplementation with β-glucan and Bacillus subtilis on growth, fillet quality, immune capacity, and antioxidant status of Pengze crucian carp (Carassius auratus var. Pengze). Aquaculture 508, 106–112. https://doi.org/10.1016/j.aquaculture.2019.04.064 (2019).
Sweeney, T. et al. Effect of purified β-glucans derived from Laminaria digitata, Laminaria hyperborea and Saccharomyces cerevisiae on piglet performance, selected bacterial populations, volatile fatty acids and pro-inflammatory cytokines in the gastrointestinal tract of pigs. Br. J. Nutr. 108, 1226–1234. https://doi.org/10.1017/s0007114511006751 (2012).
Mukhopadhya, A., Sweeney, T., O’Shea, C. & O’Doherty, J. A comparative study of alternatives to pharmacological doses of zinc for improving gut health parameters in weaning piglets. J. Anim. Sci. 94, 472–475 (2016).
Council, N. R. Nutrient Requirements of Swine (National Academies Press, 2012).
Pérez, J. M. & Sauvant, D. Tables of Composition and Nutritional Value of Feed Materials (No. 636.085 Sa89t Ej. 1 021121). INRA (2004).
Rattigan, R. et al. Laminarin-rich extract improves growth performance, small intestinal morphology, gene expression of nutrient transporters and the large intestinal microbial composition of piglets during the critical post-weaning period. Br. J. Nutr. 123, 255–263. https://doi.org/10.1017/S0007114519002678 (2020).
Caporaso, J. G. et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. https://doi.org/10.1038/nmeth.f.303 (2010).
Magoč, T. & Salzberg, S. L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27, 2957–2963. https://doi.org/10.1093/bioinformatics/btr507 (2011).
Edgar, R. C., Haas, B. J., Clemente, J. C., Quince, C. & Knight, R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27, 2194–2200. https://doi.org/10.1093/bioinformatics/btr381 (2011).
Rognes, T., Flouri, T., Nichols, B., Quince, C. & Mahé, F. VSEARCH: A versatile open source tool for metagenomics. PeerJ 4, e2584 (2016).
Eren, A. M. et al. Oligotyping: Differentiating between closely related microbial taxa using 16S rRNA gene data. Methods Ecol. Evol. 4, 1111–1119 (2013).
Eren, A. M. et al. Minimum entropy decomposition: unsupervised oligotyping for sensitive partitioning of high-throughput marker gene sequences. ISME J. 9, 968–979 (2015).
Angly, F. E. et al. CopyRighter: A rapid tool for improving the accuracy of microbial community profiles through lineage-specific gene copy number correction. Microbiome 2, 1–13 (2014).
McMurdie, P. J. & Holmes, S. phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 8, e61217 (2013).
Hamady, M., Lozupone, C. & Knight, R. Fast UniFrac: Facilitating high-throughput phylogenetic analyses of microbial communities including analysis of pyrosequencing and PhyloChip data. ISME J. 4, 17–27 (2010).
O’shea, C., McAlpine, P., Sweeney, T., Varley, P. & O’doherty, J. Effect of the interaction of seaweed extracts containing laminarin and fucoidan with zinc oxide on the growth performance, digestibility and faecal characteristics of growing piglets. Br. J. Nutr. 111, 798–807 (2014).
AOAC. Official Methods of Analysis 18th edn. (AOAC International, 2005).
Littell, R. C., Milliken, G. A., Stroup, W. W., Wolfinger, R. D. & Schabenberger, O. SAS System for Mixed Models Vol. 633 (SAS Institute Cary, 1996).
Matamoros, S., Gras-Leguen, C., Le Vacon, F., Potel, G. & de La Cochetiere, M.-F. Development of intestinal microbiota in infants and its impact on health. Trends Microbiol. 21, 167–173. https://doi.org/10.1016/j.tim.2012.12.001 (2013).
Choudhury, R. et al. Impact of early-life feeding on local intestinal microbiota and digestive system development in piglets. Sci. Rep. 11, 4213. https://doi.org/10.1038/s41598-021-83756-2 (2021).
Waters, J. L. & Ley, R. E. The human gut bacteria Christensenellaceae are widespread, heritable, and associated with health. BMC Biol. 17, 83–83. https://doi.org/10.1186/s12915-019-0699-4 (2019).
Kropp, C. et al. The Keystone commensal bacterium Christensenella minuta DSM 22607 displays anti-inflammatory properties both in vitro and in vivo. Sci. Rep. 11, 11494. https://doi.org/10.1038/s41598-021-90885-1 (2021).
Dey, N., Soergel, D. A. W., Repo, S. & Brenner, S. E. Association of gut microbiota with post-operative clinical course in Crohn’s disease. BMC Gastroenterol. 13, 131. https://doi.org/10.1186/1471-230X-13-131 (2013).
Gevers, D. et al. The treatment naive microbiome in new onset Crohn’s disease. Cell Host. Microbe. 15, 382–392. https://doi.org/10.1016/j.chom.2014.02.005 (2014).
Wang, H., Shen, J., Pi, Y., Gao, K. & Zhu, W. Low protein diets supplemented with casein hydrolysate favor the microbiota and enhance the mucosal humoral immunity in the colon of pigs. J. Anim. Sci. Biotechnol. 10, 79. https://doi.org/10.1186/s40104-019-0387-9 (2019).
Visser, J. et al. Potential mechanisms explaining why hydrolyzed casein-based diets outclass single amino acid-based diets in the prevention of autoimmune diabetes in diabetes prone BB rats. Diabetes Metab. Res. Rev. 28, 505–513 (2012).
Wijtten, P. J. A., Meulen, J. V. D. & Verstegen, M. W. A. Intestinal barrier function and absorption in pigs after weaning: A review. Br. J. Nutr. 105, 967–981. https://doi.org/10.1017/S0007114510005660 (2011).
Nii, T., Jirapat, J., Isobe, N. & Yoshimura, Y. Effects of oral administration of Lactobacillus reuteri on mucosal barrier function in the digestive tract of broiler chicks. J. Poult. Sci. 57, 67–76. https://doi.org/10.2141/jpsa.0190035 (2020).
Awad, W., Ghareeb, K. & Böhm, J. Effect of addition of a probiotic micro-organism to broiler diet on intestinal mucosal architecture and electrophysiological parameters. J. Anim. Physiol. Anim. Nutr. 94, 486–494 (2010).
Garcia-Hernandez, V., Quiros, M. & Nusrat, A. Intestinal epithelial claudins: Expression and regulation in homeostasis and inflammation. Ann. N. Y. Acad. Sci. 1397, 66–79. https://doi.org/10.1111/nyas.13360 (2017).
McGuckin, M. A., Lindén, S. K., Sutton, P. & Florin, T. H. Mucin dynamics and enteric pathogens. Nat. Rev. Microbiol. 9, 265–278. https://doi.org/10.1038/nrmicro2538 (2011).
McLamb, B. L., Gibson, A. J., Overman, E. L., Stahl, C. & Moeser, A. J. Early weaning stress in pigs impairs innate mucosal immune responses to enterotoxigenic E. coli challenge and exacerbates intestinal injury and clinical disease. PLoS ONE 8, e59838. https://doi.org/10.1371/journal.pone.0059838 (2013).
Belkaid, Y. & Hand, T. W. Role of the microbiota in immunity and inflammation. Cell 157, 121–141. https://doi.org/10.1016/j.cell.2014.03.011 (2014).
Williams, N., Stahly, T. & Zimmerman, D. Effect of chronic immune system activation on the rate, efficiency, and composition of growth and lysine needs of pigs fed from 6 to 27 kg. J. Anim. Sci. 75, 2463–2471 (1997).
Mukhopadhya, A. et al. The anti-inflammatory potential of a moderately hydrolysed casein and its 5 kDa fraction in in vitro and ex vivo models of the gastrointestinal tract. Food Funct. 6, 612–621 (2015).
Venardou, B. et al. Evaluation of the in vitro effects of the increasing inclusion levels of yeast β-glucan, a casein hydrolysate and its 5 kDa retentate on selected bacterial populations and strains commonly found in the gastrointestinal tract of pigs. Food Funct. 12, 2189–2200 (2021).
Heim, G., Sweeney, T., O’Shea, C. J., Doyle, D. N. & O’Doherty, J. V. Effect of maternal supplementation with seaweed extracts on growth performance and aspects of gastrointestinal health of newly weaned piglets after challenge with enterotoxigenic Escherichia coli K88. Br. J. Nutr. 112, 1955–1965. https://doi.org/10.1017/S0007114514003171 (2014).
Crespo-Piazuelo, D. et al. Maternal supplementation with Bacillus altitudinis spores improves porcine offspring growth performance and carcass weight. Br. J. Nutr. 1–18 (2021).
Acknowledgements
This research was a part of Food for Health Ireland (www.fhi.ie) project funded by Enterprise Ireland (Grant number TC2013-0001).
Author information
Authors and Affiliations
Contributions
The author’s contributions were as follows: T.S. and J.V.O.D. designed the experiment and supervised data collection; A.M. and S.M. performed the experiment and collected the samples; A.D. carried out the laboratory analyses and wrote the manuscript; E.C., and M.T.R. contributed to laboratory analyses. T.S., J.V.O.D., and S.V. performed the statistical analyses and corrected the manuscript; all authors approved the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Dowley, A., O’Doherty, J.V., Mukhopadhya, A. et al. Maternal supplementation with a casein hydrolysate and yeast beta-glucan from late gestation through lactation improves gastrointestinal health of piglets at weaning. Sci Rep 12, 17407 (2022). https://doi.org/10.1038/s41598-022-20723-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-022-20723-5
- Springer Nature Limited