Abstract
Leptonetidae are tiny, rarely encountered spiders that mainly inhabit moist environments, such as caves, leaf litter, and rock piles. Because they are microhabitat specialists, most leptonetid species have short-range endemism, and rarely occur in sympatry. Their small size, relatively simple habitus features and reproductive organ structure increase the difficulty of identification. The identification of leptonetids and other spiders may also be time-consuming due to their sexual dimorphism, polymorphism, and lack of diagnostic characteristics in juveniles. DNA barcoding has been used as an effective tool for species identification to overcome these obstacles. Herein, we conducted a test of DNA barcoding based on 424 specimens of Korean Leptonetidae representing 76 morphospecies. A threshold of 4.2% based on maximum intraspecific genetic divergence was estimated to efficiently differentiate the morphospecies. The species assignments tested by five species delimitation methods (ABGD, ASAP, GMYC, PTP, and bPTP) were consistent with the morphological identifications for only 47 morphospecies (61.8%), indicating many cases of cryptic diversity among the remaining morphospecies. Furthermore, sympatry in leptonetids, which are known to be rare, was revealed to be common in South Korea, especially in epigean species. Our results showed that sympatries within families, congeners, and intraclades potentially occur throughout the entire region of Korea.
Similar content being viewed by others
Introduction
Spiders of the family Leptonetidae Simon, 1890 currently include 21 genera and 368 species that mainly inhabit the Holarctic region. To date, 57 species in seven genera have been recognized in the Nearctic region, 74 species in eight genera have been identified in the Mediterranean region, and 237 species in eight genera have been identified in Asia1.
Leptonetids are small (1–3 mm) spiders that build sheet webs, on which they hang below. They are known to prefer dark and moist microhabitats, such as leaf litter, layered rock piles, mines, and caves2,3,4,5,6,7. This family is well-known to have a strong cave association because more than 50% of the species have only been identified in cave habitats. Many of these cave species have highly troglomorphic morphologies, such as eye reduction, depigmentation, and elongation of the appendages8. In contrast, in South Korea, only 17% (9 species out of 52 species) of the species have only been identified in caves, and the remainder have been identified in epigean habitats2,3,7,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23. Subterranean habitats and islands are known as one of the best barriers to geographic isolation, resulting in high endemism in arthropod taxa, including spiders24,25,26,27,28,29,30. However, within leptonetids, especially within epigean species, the speciation barrier is ambiguous and poorly understood. Because they are rarely seen, the general biology and life history of leptonetid spiders are poorly understood, and only some of the reproductive biology has been reported thus far4,30,31. Due to their habitat preferences, most species have a limited distribution range and rarely occur in sympatry, with this characteristic only observed in a few epigean populations4,5.
In studies of North American leptonetids, Gertsch suggested intraspecific polymorphism in troglomorphic characters, such as the reduction of eyes and depigmentation in Neoleptoneta capilla32. Additionally, Tayshaneta myopica and Tayshaneta paraconcinna presented more complex polymorphisms and genetic diversity that was dependent on the population and habitat type6,33. Similar patterns were also found in South Korea, but only between epigean populations. Compared with research performed in North America, studies performed on Korean leptonetids have focused on α-level diversity and treated cases of polymorphism as a separate species2,3,7,22,23.
Sexual dimorphism, polymorphisms, and limited morphological information on juveniles or larvae have been obstacles for morphological identification34,35. To overcome these obstacles, DNA barcoding has been developed, and it represents an efficient and popular tool for taxonomic investigations for the accurate species identification, cryptic species discovery and biodiversity estimates of animal taxa36,37,38,39,40,41,42,43,44, including spiders45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60,61,62. DNA barcoding of leptonetids has also been performed based on genetic distance and the Automatic Barcode Gap Discovery (ABGD) method in Leptonetela taxa62. Their study included numerous species from over a hundred cave populations throughout China and Europe. However, their sampling was mainly based on populations from caves and did not include populations from epigean habitats.
In this study, we (i) tested the utility of DNA barcodes for species identification, delimitation, and new species discovery, (ii) compared morphology-based species delimitation and barcode-based approaches, (iii) searched for types of barriers that lead to genetic diversity and speciation, including epigean leptonetids, by determining the range of species distribution, (iv) provided morphological characters that support results of delimitation methods, (v) detected cases of cryptic species, and (vi) generated a DNA barcode reference library of described species and putative new leptonetid species.
Material and methods
Taxon sampling and morphological identification
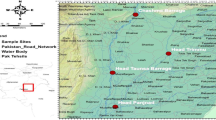
As part of an ongoing revision of the Korean Leptonetidae, we examined approximately 900 specimens (including more than 600 adult specimens) collected from approximately 150 sites in 15 (out of 17) administrative districts, including 22 caves/mines and 11 islands of South Korea (Fig. 1, Supplementary Table S1 online). Samples were mainly collected by net sifting, pit-fall traps, and exploration in caves or mines. Between 1 and 26 individuals were sampled per site, placed directly into 98% ethanol, and stored at − 20 °C. Before DNA extraction, the habitus and the male palp of the specimens were examined under Leica Z16 APO stereomicroscope. Female genitalia were separated from opisthosoma using a microsurgery stiletto knife. Separated genitalia were cleared by heating in 5 ml tubes of lactic acid or 10% KOH solution for 1 h to dissolve extraneous tissue, and then examined and photographed with Olympus BX53 compound microscope. All specimens with vouchers are deposited in the College of Agriculture and Life Sciences, Seoul National University (CALS, SNU, Seoul), National Institute of Biological Resources (NIBR, Incheon), and Yangpyeong Insect Museum (YIM, Yangpyeong), Republic of Korea. The sampling map was generated with QGIS 3.22.1 (https://www.qgis.org/ko/site/). Digital editing of the figures, microscopic photographs, and maps was prepared using Adobe Photoshop CC 2018 (Adobe Systems Incorporated, https://www.adobe.com/products/photoshop.html).
Map with sampling sites for collected specimens used in the present study. Pie charts indicate the proportion of each genus sampled from respective administrative localities. The abbreviations of each administrative district follow: GG, Gyeonggi-do; GW, Gangwon-do; CB, Chungcheongbuk-do; CN, Chungcheongnam-do; JB, Jeollabuk-do; JN, Jeollanam-do; GB: Gyeongsangbuk-do; GN, Gyeongsangnam-do; JJ, Jeju-do. The map was prepared using QGIS 3.22.1 (https://www.qgis.org/ko/site/).
DNA extraction, PCR amplification, and Sequencing
Genomic DNA was extracted from muscle tissue by grinding usually 2–4 legs, or whole body except the abdomen using DNeasy Blood and Tissue kit (QIAGEN, Hilden, Germany) following the manufacturer’s protocols. For PCR amplification for mitochondrial cytochrome oxidase subunit I (mtCOI, ~ 901 bp), we used universal primers or primers developed and used in arachnid taxa. Primer combinations we used in this study were forward LCO1490 (5’-GGTCAACAAATCATCATAAAGATATTGG-3’)63 with reverse HCO2568 (5’-GCTACAACATAATAAGTATCATG-3’)64 or HCOoutout (5’-GTAAATATATGRTGDGCTC-3’)65. Amplification was performed using AccuPower PCR Premix (Bioneer, Daejeon, Republic of Korea) following the standard protocols. The PCR condition consisted to initial denaturation at 95 °C for 2 min, followed by 35 cycles of denaturation at 95 °C for 30 s, annealing at 45–50 °C for 30 s, extension at 72 °C for 45 s, and a final extension at 72 °C for 10 min. Successfully amplified PCR products were checked in 1.2% agarose gels and were purified and sequenced at BIONICS, Inc. (Seongdong-gu, Seoul, Republic of Korea).
Sequence analysis, genetic distance, phylogenetic analysis
Raw sequences of the COI region were assembled and edited using SeqManTMII (version 5.01, 2001; DNA-star™). We eliminated poor quality and short DNA sequences in order to prevent any risk of confusion or errors, and ended up with 411 COI sequences. 13 additional sequences were downloaded from NCBI (see Supplementary Table S1 for accession numbers). Therefore, a total of 424 sequences were carried out for alignment using MAFFT version 766 through the EMBL-EBI online portal using the L-INS-i method. The sequences were deposited in GenBank (ON041801–ON042211, Supplementary Table S1). The sequence data were then combined using SequenceMatrix windows ver. 1.867. To calculate intra/interspecific pairwise genetic distances, we implemented the Kimura-2 parameter (K2P) model using MEGA 7.068. Finally, haplotype data were generated in DnaSP6.1269 to identify the distinct haplotypes.
For phylogenetic analysis, Maximum likelihood (ML), and Bayesian Inference (BI) were generated in order to test monophyletic criteria of species delimitation. Aligned sequences were carried out to PartitionFinder v2.1.170 to choose the best fit model for the analysis. RaxML GUI v2.0 was used under GTR + GAMMA model for ML tree analysis71,72. IQ-tree was also conducted as ML tree analysis using IQ-TREE version 1.6.7.173. BI tree was constructed using MrBayes v3.2.774. The Markov chain Monte Carlo search for the data matrix ran four chains for 10,000,000 generations with 25% burn-in, and sampling every 100 generations. Finally, the phylogenetic tree was visualized in FigTree v1.4.475.
Barcoding, species delimitation, and MOTUs estimation
To estimate the number of Molecular Operational Taxonomic Units (MOTUs), we used five species delimitation methods: Automatic Barcode Gap Discovery (ABGD)76, Assemble Species by Automatic Partitioning (ASAP)77, Generalized Mixed Yule-coalescent (GMYC)78, Poisson-Tree-Processes (PTP), and Bayesian implementation of the PTP (bPTP)79.
ABGD method delimit hypothetical species by calculating the pairwise distance based on the barcode gap. The ABGD analysis was performed online (https://bioinfo.mnhn.fr/abi/public/abgd/) under Jukes-Cantor (JC69), Kimura 2-parameter (K2P), and uncorrected distance (p-distance) model with relative gap width (X = 1.5).
ASAP is a recently developed method designed to propose species partitions using a hierarchical clustering algorithm based on pairwise genetic distances. The aligned sequences were submitted online (https://bioinfo.mnhn.fr/abi/public/asap/) under models the same as the ABGD method in default settings.
In the GMYC analysis, we used BEAST v2.6.680 to obtain an ultrametric tree, under a strict molecular clock model. In the prior, we used the Yule speciation model running 20 million generations, sampling every 1000 generations. We further checked for stationarity and determined burn-in using TRACER v1.7.281, and then discarded as 15% burn-in, 0.5 posterior probability using TreeAnnotator80. For the GMYC analysis, the ultrametric tree was then carried out in RStudio (https://www.r-project.org/) using the “splits” package82.
PTP is a coalescent-based delimitation method that requires a phylogenetic input tree, and bPTP is an updated version of the original PTP, adding Bayesian support (BS) values to delimited species on the input tree. We used the BI tree as the input tree for PTP and bPTP analyses, implemented online (https://species.h-its.org/ptp), running 100,000 MCMC generations, with a thinning of 100, burn-in of 0.1, and removing the outgroups for improved results.
Results
Morphological identification
Based on the morphological examination, 75 morphospecies in four genera (Falcileptoneta, Leptoneta, Longileptoneta, and Masirana) of leptonetids were identified from 409 specimens (Supplementary Table S2 online). Additionally, two individuals of Telemidae from South Korea, were also included as an outgroup.
Although leptonetids are known as microhabitat specialists and highly localized, many species, especially epigean leptonetids, were sampled from various distant populations, indicating that they are more widely distributed than expected around the Korean Peninsula (Falcileptoneta odaesanensis, Leptoneta taeguensis, Leptoneta chilbosanensis, Leptoneta spinipalpus, Longileptoneta weolakensis, Longileptoneta songniensis, and Longileptoneta sp3). In some cases, the same species were distributed across the islands (Falcileptoneta sp17, Falcileptoneta sp21, Leptoneta namhensis, and Leptoneta paikmyeonggulensis).
Genetic distance divergence of species identification
A haplotype data analysis revealed 200 distinct haplotypes (Supplementary Data S1 online). The average K2P genetic divergence was 20.74% across all specimens of the dataset. The mean of intraspecific K2P genetic divergence was 1.27% (ranging from 0 to 18.41%), with an increase in divergence between congeners of approximately 13 times to a value of 16.64% (ranging from 14.49 to 21.22%) on average (Table 1). However, the interspecies divergence within genera varied from 10.46% to 21.22%: Falcileptoneta (mean = 20.40%), Longileptoneta (mean = 10.46%), Leptoneta (mean = 21.22%), and Masirana (mean = 14.49%). Detailed values of each genus are provided in Table 2. The number of sampled sites, sampled specimens, and intraspecific genetic divergences of each morphospecies are listed in Supplementary Data S2.
A threshold was determined to evaluate the number of MOTUs in Leptonetidae, and the maximum value of intraspecific K2P genetic divergence was less than 4.2%, with 11 morphospecies presenting a maximum intraspecific divergence over 4.2% (Fig. 3).
MOTU estimation
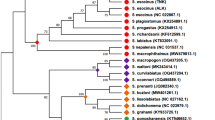
The species delimitation methods of ABGD, ASAP, GMYC, PTP, and bPTP yielded 92, 98, 112, 117, and 120 MOTUs, respectively. Color bars in the ML tree indicate the results of MOTUs delineated by different methods (Fig. 2; IQ-tree in Supplementary Fig. S1). Compared with the morphological identification method, species delimitation methods are mostly focused on the population level rather than morphological appearance (see Supplementary Table S3 online). Phylogenetic tree-based methods generally exhibit greater oversplitting compared with genetic distance-based methods. As a result, 47 MOTUs, which accounted for 61.8% of the total, matched the morphological delimitation of the species.
Maximum Likelihood (ML) tree of 424 COI sequences, including morphological analysis and results of five different species delimitation methods. Color bars indicate the results of each delimitation method. In the ML tree, blue panels indicate that all species delimitation methods match with morphological identification, while red panels show one or more methods disagree.
An ABGD analysis of each JC69, K2P, and p-distance substitution model produced nonidentical MOTUs, resulting in 82, 92, and 91 MOTUs that with 85.5%, 85.5%, and 82.9% consistency with the morphology, respectively (all P = 0.035938 (for more details, see Supplementary Table S4 online)). Considering the morphological identifications and comparisons with other species delimitations used in this study, we display the ABGD results based on the K2P substitution model. Six morphospecies, Falcileptoneta secula, Falcileptoneta maewhaensis, Falcileptoneta sp2, Falcileptoneta odaesanensis, Falcileptoneta sp17, and Leptoneta chilbosanensis were split into two MOTUs, Falcileptoneta chiakensis was split into five MOTUs, and Leptoneta taeguensis was split into six MOTUs by ABGD. On the other hand, Longileptoneta weolakensis and Leptoneta spinipalpus were merged into a single MOTU.
Compared with the ABGD method, the ASAP analysis produced identical MOTU results on each JC69, K2P, and p-distance substitution model, which all yielded 98 MOTUs with ASAP scores of 7.00, 9.50, and 6.50, respectively (for more details, see Supplementary Table S5 online). Compared with ABGD, additional MOTUs were observed in seven morphospecies: Falcileptoneta chiakensis, Falcileptoneta hansanensis, Longileptoneta gayaensis, Longileptoneta weolakensis, Leptoneta spinipalpus, Leptoneta namhensis, and Masirana ilweolensis.
The GMYC analysis produced more MOTUs, and it likely oversplit them into 112 MOTUs. Compared with the ABGD and ASAP methods, 12 species were split into additional MOTUs by the GMYC method: Falcileptoneta sp2, Falcileptoneta sp3, Falcileptoneta sp4, Falcileptoneta juwangensis, Falcileptoneta geumsanensis, Falcileptoneta unmunensis, Longileptoneta byeonsanbando, Longileptoneta sp3, Longileptoneta sp4, Longileptoneta sp6, Leptoneta taeguensis, and Leptoneta paikmyeonggulensis. Conversely, Falcileptoneta hansanensis was recovered as a single MOTU in the GMYC analysis.
The PTP analysis yielded 117 MOTUs, which was similar to the GMYC delineation. However, the MOTUs of the five morphospecies were split by the PTP method from GMYC: Falcileptoneta geumsanensis, Falcileptoneta hansanensis, Longileptoneta sp4, Leptoneta sp1, and Leptoneta paikmyeonggulensis. Additionally, Falcileptoneta juwangensis was recovered as a single MOTU from GMYC.
The bPTP produced the most MOTUs among the methods we used in this study, and it yielded 120 MOTUs. Although bPTP recovered Leptoneta sp1 into a single MOTU, which was split by the PTP method, it split the other morphospecies, e.g., Falcileptoneta unmunensis was split into three MOTUs, Leptoneta paikmyeonggulensis was split into four MOTUs, and Masirana ilweolensis was split into five MOTUs.
Population-dependent cryptic diversity
The threshold based on the maximum intraspecific K2P genetic divergence was estimated at 4.2% from the dataset, with 11 morphospecies showing divergence over the maximum estimated threshold (5.8–18.4%) (Fig. 3). In particular, nine morphospecies were split into multiple MOTUs by all five species delimitation methods. The morphospecies had a maximum divergence of over 4.2% and their delimitation results were mainly population dependent, especially the epigean leptonetids.
First, among the epigean leptonetids (Fig. 4), the morphospecies Leptoneta taeguensis was split into six to seven MOTUs (Fig. 4a), Falcileptoneta odaesanensis was into two MOTUs (Fig. 4b), and Falcileptoneta chiakensis was split into five to six MOTUs (Fig. 4c) depending on each populational locality by the species delimitation methods used in this study (morphological details on Fig. 5, see the original description of the morphospecies Falcileptoneta chiakensis18).
Cryptic species between various populations of epigean leptonetids with a map of sampled localities. All maps were prepared using QGIS 3.22.1 (https://www.qgis.org/ko/site/). (a) Leptoneta taeguensis from (1) Hapcheon-gun, (2) Miryang-si, (3) Yangsan-si, (4) Daegu, (5) Cheongdo-gun, and (6) Ulju-gun; (b) Falcileptoneta odaesanensis from (7) Goseong-gun, (8) Inje-gun, Yangyang-gun, Gangneung-si, Pyeongchang gun, and Hongcheon-gun, (c) Falcileptoneta chiakensis from (9) Danyang-gun, (10) Wonju-si, (11) Pyeongchang-gun, (12) Yeongju-si, and (13) Jaecheon-si. Asterisks represent the individuals collected from type localities.
Morphological photographs of each cryptic species from Fig. 4. upper panels: habitus (dorsal and ventral view); lower panels: left palp (prolateral and retrolateral view), (6) for right palp (retrolateral and prolateral view). The numbers of each panel match the numbers from Fig. 4. Scale bars: upper panels = 0.5 mm, lower panels = 0.1 mm.
Second, in troglophilic leptonetids (Fig. 6), separated MOTUs were detected between the cave population and epigean population, although those populations were not readily diagnosable (Falcileptoneta secula and Falcileptoneta maewhaensis) (Fig. 6a,b, morphological details on Fig. 7, see the original description of Falcileptoneta maewhaensis21). As an exception, the cave population and epigean population of the morphospecies Falcileptoneta simboggulensis resulted in merged MOTU.
Cryptic species between cave populations vs. epigean populations, and mainland populations versus island populations with a map of sampled localities. All maps were prepared using QGIS 3.22.1 (https://www.qgis.org/ko/site/). (a) Falcileptoneta secula from (14) Jumunjin (Gangneung-si) and (15) Magwihalmigul cave (Gangneung-si), (b) Falcileptoneta maewhaensis from (16) Gaya-myeon (Hapcheon-gun), and (17) Betishale cave (Hapcheon-gun), (c) Falcileptoneta sp17 from (18) Geogumdo island (Goheung-gun), and (19) Mt. Duryunsan (Haenam-gun), Wando island (Wando-gun). Asterisks represent the individuals collected from type localities.
Finally, species that were geographically isolated by islands showed both single and double MOTUs by all five species delimitation methods (Fig. 6c, morphological details in Fig. 7). The population from the mainland of South Korea (Haenam-gun, Jeollanam-do), resulted in a MOTU merged with the population from the island nearby (Wando Island, Wando-gun, Jeollanam-do), while the MOTU that split from the population were distributed relatively distant from the island (Geogumdo Island, Goheung-gun, Jeollanam-do).
Widely distributed species, and congeneric sympatry in leptonetids
First, in many cases, particularly for members of the genus Longileptoneta, a single MOTU was sampled between distant populations, indicating that many of the species are distributed in a large range and may present sympatry within congeners (Fig. 8a): (i) Longileptoneta sp3, which is a potentially new species distributed nationwide in the western part of South Korea (Pocheon-si, Jaecheon-si, Daejeon, Haenam-gun); (ii) Longileptoneta songniensis, which was previously only found at Mt. Songnisan (Boeun-gun, Chungcheongbuk-do) but has also been found on Ganghwado Island (Incheon); (iii) Longileptoneta weolakensis, which was previously only found at Mt. Weolaksan (Jecheon-si) but has also been found on Mt. Baegunsan (Pocheon-si, Gyeonggi-do).
Maps showing sympatric localities between congeneric species. The maps were prepared using QGIS 3.22.1 (https://www.qgis.org/ko/site/). Members of Longileptoneta are mainly wider distributed compared to species of Falcileptoneta, suggesting a possibility that this genus may have potentially more sympatric distributions.
Second, some cases revealed that different populations across the islands were merged into a single MOTU: (i) Falcileptoneta sp17 from Mt. Duryunsan (Haenam-gun, Jeollanam-do) and Wando Island (Wando-gun, Jeollanam-do) was different from that on Geogumdo Island (Goheung-gun, Jeollanam-do) (Fig. 6c); (ii) Falcileptoneta sp21 from Namsan Park (Goseong-gun, Gyeongsangnam-do) and Namhae Island (Namhae-gun, Gyeongsangnam-do); (iii) Longileptoneta songniensis from Mt. Songnisan (Boeun-gun, Chungcheongbuk-do) and Ganghwado Island (Incheon); and (iv) Leptoneta paikmyeonggulensis from Namhae Island (Namhae-gun, Gyeongsangnam-do) and Dolsan Island (Yeosu-si, Jeollanam-do) resulted in single MOTUs.
Finally, two or more species of epigean populations belonging to the genus Falcileptoneta found at the same locality were detected (Fig. 8b): (i) F. chiakensis and Falcileptoneta sp2 from Mt. Chiaksan (Wonju-si, Gangwon-do); (ii) Falcileptoneta umyeonsanensis and Leptoneta chilbosanensis on Mt. Kwanaksan (Seoul); (iii) Falcileptoneta odaesanensis, Falcileptoneta sp4 and Leptoneta chilbosanensis on Mt. Seoraksan (Inje-gun, Gangwon-do); (iv) Falcileptoneta hansanensis and Falcileptoneta sp23 from Mt. Mireuksan (Tongyeong-si, Gyeongsangnam-do); (v) Falcileptoneta geumsanensis, Falcileptoneta sp21 and Falcileptoneta sp22 in Namhae Island (Namhae-gun, Gyeongsangnam-do); and (vi) Falcileptoneta sp8 and Falcileptoneta sp17 from Wando Island (Wando-gun, Jeollanam-do). However, in cave habitats, no sympatry was found.
Sympatry in intraclade species
The clade that includes Falcileptoneta sp2, Falcileptoneta sp3, Falcileptoneta umyeonsanensis, Falcileptoneta sp4, Falcileptoneta sp5, Leptoneta kwangreungensis, and Leptoneta chilbosanensis has sharp tooth-like tibial apophyses in common. The identification between those species is quite challenging, although they can be diagnosed by the thickness and length of the tibial apophysis of the male palp, spotted patterns of the abdomen, and length of the body. Most of the species appear to present short-range endemism and have only been found in a small range of habitats (one or two mountain ranges). However, clade 8 (Fig. 9) was detected as a cryptic species of Leptoneta chilbosanensis, had a large-range distribution and was collected from three administrative districts. Unlike other morphospecies in which cryptic species were detected depending on the population-level, cryptic species of this group were found to be nonpopulation dependent. Moreover, the population from Mt. Surisan (Anyang-si, Gyeonggi-do), one individual among the population of Falcileptoneta umyeonsanensis from Mt. Gwanaksan (Seoul), and one individual among the population of Falcileptoneta sp4 from Mt. Seoraksan (Inje-gun, Gangwon-do) were all merged to a single MOTU and shared habitat with numerous intraclade species populations.
Intraclade sympatry occurring in central region of Korean peninsula. The map was prepared using QGIS 3.22.1 (https://www.qgis.org/ko/site/).
Discussion
Accurate identification of leptonetid species can be challenging due to their small body size, similar habitus among different species, and limited morphological information, such as the structure of the male palp. In particular, morphological identification using female and juvenile specimens is problematic due to the lack of diagnosable morphological characteristics, including female genitalia4,5,33. Therefore, we tested the utility of DNA barcodes in species identification based on intraspecific divergences and five species delimitation methods (ABGD, ASAP, GMYC, PTP, and bPTP) using Korean leptonetids. Consequently, our results showed some cases of population-dependent cryptic diversity, both observed in short-range (one or two mountain ranges) and large-range distributions (at least including a range of three administrative districts), and at the same time, various types of sympatry, which are rare in this family.
In some spider lineages, especially in several haplogyne spiders such as Hypochilidae, the intraspecific genetic divergence value is extremely high and can exceed 15%83,84. Likewise, our results also revealed an extreme divergence of intraspecific genetic distances that exceed 18% (in Leptoneta taeguensis), with variations mainly observed at the population level. However, by using species delimitation methods, morphospecies with high intraspecific divergence were split into multiple MOTUs, resulting in cryptic species detection. These results may suggest that cross-validation between intraspecific divergence and species delimitation methods is needed to delineate species boundaries using DNA barcodes. In our study, Korean leptonetids were delineated as a threshold value of 4.2%, which was estimated based on the maximum intraspecific divergences, could serve as an efficient standard for preliminary species delimitation. In particular, the Korean Peninsula includes unique and diverse geographical structures, such as having thousands of caves85, presenting a land area that consists of over 70% mountains, and having over 3500 islands off the coast86, which may cause frequent geographic isolations. Additionally, it has relatively strong seasonal fluctuations and rich environmental diversity and biodiversity, with over 56,000 biological species87, including approximately 2200 endemic species88. Therefore, given the different environments around the world, and differences in intraspecific genetic divergences in other spider taxa, this value can be specifically applied to Korean Leptonetidae while other fauna should be further discussed.
In our study, we used various species delimitation methods that are considered the most reliable. ABGD is a genetic distance-based method that is one of the most popular barcode-gap methods43,44,45,46,49,62, while GMYC, PTP, and bPTP are tree-based delimitation methods used for spider lineages45,46,49,61,89 and other arthropod members43,44. In most studies of species delimitation methods, phylogenetic tree-based delimitation methods tend to be more sensitive and present more splits into multiple MOTUs compared to barcode gap-based methods, and similar results were obtained in our study (Fig. 2, Supplementary Table S3 online). Tree-based species delimitation methods, especially coalescent-based methods, such as GMYC or BP&P, have been constantly introduced because they are affected by the population structure in the dataset and oversplit taxa, especially when a single gene is applied45,46,50,89,90. Because a single gene was applied here, we concede at least four congruent MOTU results among the five different species delimitation methods used in this study as a hypothetical cryptic species (Supplementary Data S3). These results may be important for increasing our understanding of biodiversity and evolution and connecting taxonomy and phylogenetic studies91. In further studies, however, a combination of effective markers, such as ribosomal or nuclear genes, would be sufficient for species-level delimitation, as like other arthropods45,50,59,92,93,94.
Morphospecies that resulted in MOTU splits by species delimitation methods and presented high genetic divergence depending on the population level (Falcileptoneta secula, Falcileptoneta odaesanensis, Falcileptoneta maewhaensis, Falcileptoneta sp17, and Leptoneta taeguensis) were shown to belong to the genus Falcileptoneta (Figs. 4, 5, 6, and 7). Compared to other Korean leptonetid genera, especially Longileptoneta, species in Falcileptoneta tended to have more dispersal-limited endemism. Our sampling data around South Korea indicate that in most cases, many individuals of Falcileptoneta species were sampled but in a small range, while few individuals of Longileptoneta species were sampled but in larger ranges than expected (Fig. 8a). Based on the analysis of the sampling data, we identified the phenomenon that species in the genus Longileptoneta mainly tend to spread out while Falcileptoneta gathers.
Leptonetids are known to be dispersal-limited spiders and prefer specific types of microhabitats, such as leaf litter, caves, and mines, creating distributional patterns as ‘narrow endemism’5,33. As demonstrated in many other arachnid lineages, the biological traits of restricted dispersion ability and high microhabitat specialization generally lead to biogeographic histories dominated by vicariance, with few dispersal events95,96,97. Additionally, islands and caves play a role in vicariance, thus causing high endemism in arthropod lineages98,99,100,101,102. Throughout this study, we found that a considerable number of leptonetid species have expanded their distributions and are being found beyond restricted zones, such as caves and islands, and they also occur in sympatries between different genera, species, and intraspecific clades (Figs. 8, 9). The details of the biogeographical mechanisms leading to these sympatries and a discussion of the phylogenetic relationships will be outlined in a future study (under preparation).
Single morphospecies between the cave population and epigean population near the cave mainly resulted in a split of MOTUs in the species delimitation methods we used in this study (Fig. 5a, b). Ledford and colleagues discussed similar cases in Tashaneta species based on multigene phylogeny, and they treated these cases as intraspecific polymorphisms despite furcation on phylogenetic tree33. Similarly, our results also showed that populations between the epigean habitat and cave habitats had a high intraspecific genetic divergence, ranging up to a maximum of 11.5%, with a split of MOTUs. However, an exceptional case in this study showed that, Falcileptoneta simboggulensis, which is known as a troglobitic spider that has only been found in Simboggul Cave was merged to a single MOTU with epigean individuals which was sampled near the cave (with 0.2% of maximum intraspecific divergence), indicating that this cave does not function as a barrier of gene flow between the cave and epigean populations. Because we lack morphological data for epigean F. simboggulensis, additional sampling, especially for male specimens, is needed in future studies.
In the traditional taxonomy of spiders, structures of the female genitalia and the male palp have been key features for species identification. However, many cases in our results showed that species that are indistinguishable based on the male palp resulted in a split of MOTUs depending on the population level. Rather, the shape of the sternum, patterns in the abdomen, and the ratio between the length of the body and leg were somewhat diagnosable from dependent MOTUs (Figs. 5, 7).
In our study, we included several species in the genus Leptoneta. However, the phylogenomic and biogeographic study of the family Leptonetidae shows that Leptoneta species are restricted in Mediterranean Europe, and all of the species in Leptoneta are morphologically, and geographically misplaced5. Although Seo transferred many species of Leptoneta to Falcileptoneta (2015)18, more than ten species of Korean leptonetids remain in this genus. Thus, accurate identification based on morphological and phylogenetic studies of misplaced species, especially Leptoneta, would be important to better understanding the systematics, biogeography, and evolution of the family Leptonetidae (under preparation).
Data availability
Accession Codes: The COI sequences generated and analyzed during the current study are available in the Genbank repository (https://www.ncbi.nlm.nih.gov/genbank/), from ON041801 to ON042211.
References
World Spider Catalog. Version 23.0. Natural History Museum Bern, online at http://wsc.nmbe.ch Accessed from 26 May 2022.
Kim, S. T., Yoo, J. S. & Lee, S. Y. Two new species of the genus Leptoneta Simon, 1872 (Araneae: Leptonetidae) from Korea. J. Spec. Res. 5, 596–600 (2016).
Lan, T. et al. Five new species of the genera Falcileptoneta and Longileptoneta (Araneae, Leptonetidae) from South Korea. ZooKeys 1010, 97–116 (2021).
Ledford, J. M. A revision of the spider genus Calileptoneta Platnick (Araneae, Leptonetidae), with notes on morphology, natural history and biogeography. J. Arachnol. 32, 231–269 (2004).
Ledford, J. et al. Phylogenomics and biogeography of leptonetid spiders (Araneae: Leptonetidae). Invert. Syst. 35, 332–349 (2021).
Ledford, J., Paquin, P., Cokendolpher, J., Campbell, J. & Griswold, C. Systematics of the spider genus Neoleptoneta Brignoli, 1972 (Araneae: Leptonetidae) with a discussion of the morphology and relationships of the North American Leptonetidae. Invert. Syst. 35, 334–388 (2011).
Xu, M., Kim, S. T., Yoo, J. S., Nam, E. J. & Li, S. Three new species of the genus Falcileptoneta Komatsu, 1970 (Araneae: Leptonetidae) from Korea. ZooKeys 872, 1–12 (2019).
Mammola, S. & Isaia, M. Spiders in caves. Proc. R. Soc. Lond. B Biol. Sci. 284, 20170193 (2017).
Paik, K. Y. Cave-dwelling spiders from southern part of Korea. II. Spiders of Simbog-gul cave and Sungyoo-gul cave. In: Thes. Coll. Comm. 60th Birth. Prof. Yung Ho Choi 299–313 (1971).
Paik, K. Y. The spider fauna of Dae Heuksan-do Isl., So Heuksan-do Isl., Jeunlanam-do, Korea. Kyungpook Educ. Forum Kyungpook Natl. Univ. 22, 153–173 (1980).
Paik, K. Y. A new spider of the genus Leptoneta (Araneae: Leptonetidae) from Korea. J. Inst. Nat. Sci. 4, 113–117 (1985).
Paik, K. Y., Yaginuma, T. & Namkung, J. Results of the speleological survey in South Korea 1966 XIX. Cave-dwelling spiders from the southern part of Korea. Bull. Nat. Sci. Mus. Tokyo 12, 836–843 (1969).
Paik, K. Y. & Seo, B. K. A new spider of the genus Leptoneta (Araneae: Leptonetidae) from Korea. Acta Arachnol. 30, 65–70 (1982).
Paik, K. Y. & Seo, B. K. Two new species of Korean spiders (Araneae: Leptonetidae and Linyphiidae). J. Inst. Nat. Sci. 3, 97–103 (1984).
Namkung, J. Two new cave spiders of the genus Leptoneta (Araneae: Leptonetidae) from Korea. Korean Arachnol. 3, 83–90 (1987).
Namkung, J. The Spider of Korea 2nd edn, 35–42 (Kyo-Hak Publ, Seoul, 2003).
Kim, J. P., Ye, S. H. & Kim, T. W. A new species of the Genus Leptoneta Simon, 1872 (Araneae: Leptonetidae) from Korea. Korean Arachnol. 31, 31–36 (2015).
Ten Seo, B. K. new species of the genus Falcileptoneta (Araneae, Leptonetidae) from Korea. Korean J. Environ. Biol. 33, 290–305 (2015).
Seo, B. K. Four new species of the genera Masirana and Longileptoneta (Araneae, Leptonetidae) from Korea. Korean J. Environ. Biol. 33, 306–313 (2015).
Seo, B. K. Four new species of the genus Longileptoneta (Araneae, Leptonetidae) from Korea. J. Spec. Res. 5, 584–589 (2016).
Seo, B. K. Four new species of the genus Falcileptoneta (Araneae, Leptonetidae) from Korea. J. Spec. Res. 5, 590–595 (2016).
Kim, S. T., Lee, J. H. & Namkung, J. Two New Ground-inhabiting Leptoneta Spiders (Araneae: Leptonetidae) from Korea. J. Asia-Pac. Entomol. 7, 257–261 (2004).
Kim, S. T., Jung, M. P., Kim, H. S., Lee, J. H. & Koo, H. M. Two new species of litter-inhabiting spiders of the genus Leptoneta from Korea (Araneae: Leptonetidae). Can. Entomol. 136, 639–644 (2004).
Barr, T. C. Jr. & Holsinger, J. R. Speciation in cave faunas. Ann. Rev. Ecol. Syst. 16, 313–337 (1985).
Caccone, A. Gene flow in cave arthropods: A qualitative and quantitative approach. Evolution 39, 1223–1235 (1985).
Chapman, P. The origin of troglobites. Proc. Univ. Bristol. Spelaeol. Soc. 16, 133–141 (1982).
Culver, D. C., Master, L. L., Christman, M. C. & Hobbs, H. H. III. Obligate cave fauna of the 48 contiguous United States. Conserv. Biol. 14, 386–401 (2000).
Waterman, T. H. Evolutionary challenges of extreme environments (part 2). J. Exp. Zool. 291, 130–168 (2001).
Yao, Z., Dong, T., Zheng, G., Fu, J. & Li, S. High endemism at cave entrances: A case study of spiders of the genus Uthina. Sci. Rep. 6, 1–9 (2016).
Cokendolpher, J. C. A new Neoleptoneta spider from a cave in Camp Bullis, Bexar County, Texas (Araneae: Leptonetidae). Texas Mem. Mus. Speleol. Monogr. 6, 63–69 (2004).
Ledford, J. M. & Griswold, C. E. A study of the subfamily Archoleptonetinae (Araneae: Leptonetidae) with a review of the morphology and relationships for the Leptonetidae. Zootaxa 2391, 1–32 (2010).
Gertsch, W. J. The spider family Leptonetidae in North America. J. Arachnol. 1, 145–203 (1974).
Ledford, J. M., Paquin, P., Cokendolpher, J. C., Campbell, J. & Griswold, C. Systematics, conservation and morphology of the spider genus Tayshaneta (Araneae, Leptonetidae) in central Texas Caves. ZooKeys 167, 1–102 (2012).
Coddington, J. A. & Levi, H. W. Systematics and evolution of spiders Araneae. Ann. Rev. Ecol. Syst. 22, 565–592 (1991).
Magalhaes, I. L., Martins, P. H., Nogueira, A. A. & Santos, A. J. Finding hot singles: Matching males to females in dimorphic spiders (Araneidae: Micrathena) using phylogenetic placement and DNA barcoding. Invert. Syst. 31, 8–36 (2017).
Will, K. W. & Rubinoff, D. Myth of the molecule: DNA barcodes for species cannot replace morphology for identification and classification. Cladistics 20, 47–55 (2004).
Hebert, P. D., Cywinska, A., Ball, S. L. & DeWaard, J. R. Biological identifications through DNA barcodes. Proc. R. Soc. Lond. B Biol. Sci. 270, 313–321 (2003).
Hebert, P. D. N., Ratnasingham, S. & DeWaard, J. R. Barcoding animal life: Cytochrome c oxidase subunit 1 divergences among closely related species. Proc. R. Soc. Lond. B Biol. Sci. 270, S96–S99 (2003).
Hebert, P. D. N., DeWaard, J. R. & Landry, J. F. DNA barcodes for 1/1000 of the animal kingdom. Biol. Lett. 6, 359–362 (2010).
Ward, R., Hanner, R. & Hebert, P. D. N. The campaign to DNA barcode all fishes. J. Fish Biol. 74, 329–356 (2009).
Radulovici, A. E., Sainte-Marie, B. & Dufresne, F. DNA barcoding of marine crustaceans from the Estuary and Gulf of St Lawrence: A regional-scale approach. Mol. Ecol. Res. 9, 181–187 (2009).
Tavares, E. S. & Baker, A. J. Single mitochondrial gene barcodes reliably identify sister-species in diverse clades of birds. BMC Evol. Biol. 8, 81 (2008).
Tyagi, K. et al. DNA Barcoding studies on Trips in India: Cryptic species, Species complexes. Sci. Rep. 7, 4898 (2017).
Kim, S., Lee, Y., Mutanen, M., Seung, J. & Lee, S. High functionality of DNA barcodes and revealed cases of cryptic diversity in Korean curved-horn moths (Lepidoptera: Gelechioidea). Sci. Rep. 10, 1–12 (2020).
Cao, X. et al. Rapid dissemination of taxonomic discoveries based on DNA barcoding and morphology. Sci. Rep. 6, 1–13 (2016).
Tyagi, K. et al. Identification of Indian spiders through DNA barcoding: Cryptic species and species complex. Sci. Rep. 9, 1–13 (2019).
Thomas, S. M. & Hedin, M. Multigenic phylogeographic divergence in the paleoendemic southern Appalachian opilionid Fumontana deprehendor Shear (Opiliones, Laniatores, Triaenonychidae). Mol. Phylogenet. Evol. 46, 645–658 (2008).
Zhang, Y. & Li, S. A spider species complex revealed high cryptic diversity in South China caves. Mol. Phylogenet. Evol. 79, 353–358 (2014).
Ortiz, D., Pekár, S., Bilat, J. & Alvarez, N. Poor performance of DNA barcoding and the impact of RAD loci filtering on the species delimitation of an Iberian ant-eating spider. Mol. Phylogenet. Evol. 154, 106997 (2021).
Hamilton, C. A., Formanowicz, D. R. & Bond, J. E. Species delimitation and phylogeography of Aphonopelma hentzi (Araneae, Mygalomorphae, Theraphosidae): Cryptic diversity in North American tarantulas. PLoS ONE 6, e26207 (2011).
Ashfaq, M. et al. Assembling a DNA barcode reference library for the spiders (Arachnida: Araneae) of Pakistan. PLoS ONE 14, e0217086 (2019).
Coddington, J. A. et al. DNA barcode data accurately assign higher spider taxa. PeerJ 4, e2201 (2016).
Barrett, R. D. H. & Hebert, P. D. N. Identifying spiders through DNA barcodes. Can. J. Zool. 83, 481–491 (2005).
Blagoev, G. A., deWaard, J. & Hebert, P. D. N. Building a DNA barcode reference library for Canadian spiders (Araneae). Genome 58, 197–197 (2015).
Gergin, A. et al. Untangling taxonomy: A DNA barcode reference library for Canadian spiders. Mol. Ecol. Resour. 16, 325–341 (2016).
Ivanov, V., Lee, K. M. & Mutanen, M. Mitonuclear discordance in wolf spiders: Genomic evidence for species integrity and introgression. Mol. Ecol. 27, 1681–1695 (2018).
Newton, L. G., Starrett, J., Hendrixson, B. E., Derkarabetian, S. & Bond, J. E. Integrative species delimitation reveals cryptic diversity in the southern Appalachian Antrodiaetus unicolor (Araneae: Antrodiaetidae) species complex. Mol. Ecol. 29, 2269–2287 (2020).
Gaikwad, S., Warudkar, A. & Shouche, Y. Efficacy of DNA barcoding for the species identification of spiders from Western Ghats of India. Mitochondrial DNA Part A 28, 638–644 (2017).
Wang, Z. L., Yang, X. Q., Wang, T. Z. & Yu, X. Assessing the effectiveness of mitochondrial COI and 16S rRNA genes for DNA barcoding of farmland spiders in China. Mitochondrial DNA Part A 29, 695–702 (2018).
Macrini, C. M. T., Peres, E. A. & Solferini, V. N. Cryptic diversity of Aglaoctenus lagotis (Araneae, Lycosidae) in the Brazilian Atlantic Rainforest: Evidence from microsatellite and mitochondrial DNA sequence data. J. App. Biol. Biotech. 3, 9–14 (2015).
Muster, C. & Michalik, P. Cryptic diversity in ant-mimic Micaria spiders (Araneae, Gnaphosidae) and a tribute to early naturalists. Zool. Scr. 49, 197–209 (2020).
Wang, C. C., Xu, X. & Li, S. Q. Integrative taxonomy of Leptonetela spiders (Araneae: Leptonetidae), with descriptions of 46 new species. Zool. Res. 38, 321–448 (2017).
Folmer, O., Black, M., Hoeh, W., Lutz, R. & Vrijenhoek, R. DNA primers for amplification of mitochontrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotech. 3, 264–299 (1994).
Hedin, M. C. & Maddison, W. P. A combined molecular approach to phylogeny of the jumping spider subfamily Dendryphantinae (Araneae: Salticidae). Mol. Phylogenet. Evol. 18, 386–403 (2001).
Schulmeister, S., Wheeler, W. C. & Carpenter, J. M. Simultaneous analysis of the basal lineages of Hymenoptera (Insecta) using sensitivity analysis. Cladistics 18, 455–484 (2002).
Katoh, K. & Standley, D. M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 34, 772–780 (2013).
Vaidya, G., Lohman, D. J. & Meier, R. SequenceMatrix: Concatenation sofware for the fast assembly of multi-gene datasets with character set and codon information. Cladistics 27, 171–180 (2010).
Kumar, S., Stecher, G. & Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0. for bigger datasets. Mol. Biol. Evol. 33(7), 1870–1874 (2016).
Rozas, J. et al. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 34, 3299–3302 (2017).
Lanfear, R., Frandsen, P. B., Wright, A. M., Senfeld, T. & Calcott, B. PartitionFinder 2: New methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol. Biol. Evol. 34, 772–773 (2017).
Edler, D., Klein, J., Antonelli, A. & Silvestro, D. raxmlGUI 2.0: A graphical interface and toolkit for phylogenetic analyses using RAxML. Methods Ecol. Evol. 12, 373–377 (2021).
Stamatakis, A. RAxML Version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313 (2014).
Nguyen, L-T., Schmidt, H. A., von Haeseler, A. & Minh, B. Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum likelihood phylogenies. Mol. Biol. Evol. 32, 268–274
Ronquist, F. & Huelsenbeck, J. P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574 (2003).
Rambaut, A. FigTree, tree figure drawing tool. V. 1.4.2. Institute of Evolutionary Biology, University of Edinburgh, Edinburgh. http://tree.bio.ed.ac.uk.software/figtree/ Accessed from 06 Dec 2021.
Puillandre, N., Lambert, A., Brouillet, S. & Achaz, G. ABGD, automatic barcode gap discovery for primary species delimitation. Mol. Ecol. 21, 1864–1877 (2012).
Puillandre, N., Brouillet, S. & Achaz, G. ASAP: Assemble species by automatic partitioning. Mol. Ecol. Resour. 21, 609–620 (2021).
Fujisawa, T. & Barraclough, T. G. Delimiting species using single-locus data and the Generalized Mixed Yule Coalescent approach: A revised method and evaluation on simulated data sets. Syst. Biol. 62, 707–724 (2013).
Zhang, J., Kapli, P., Pavlidis, P. & Stamatakis, A. A general species delimitation method with applications to phylogenetic placements. Bioinformatics 29, 2869–2876 (2013).
Bouckaert, R. et al. BEAST 2.5: An advanced software platform for Bayesian evolutionary analysis. PLoS Comput. Biol. 15, e1006650 (2019).
Rambaut, A., Drummond, A. J., Xie, D., Baele, G. & Suchard, M. A. Posterior summarization in Bayesian phylogentics using Tracer 1.7. Syst. Biol. 67, 901–904 (2018).
Ezard, T., Fujisawa, T. & Barraclough, T. G. Splits: SPecies’ LImits by Treshold Statistics. R package version 1.0–14/r31, http://R-Forge.R-project.org/projects/splits/ (2009).
Ciaccio, E., Debray, A. & Hedin, M. Phylogenomics of paleoendemic lampshade spider (Araneae, Hypochilidae, Hypochilus), with the description of a new species from montane California. ZooKeys 1086, 163 (2022).
Hedin, M. C. Molecular insights into species phylogeny, biogeography, and morphological stasis in the ancient spider genus Hypochilus (Araneae: Hypochilidae). Mol. Phylogenet. Evol. 18, 238–251 (2001).
Cultural Heritage Administration. A report on the fundamental survey of caves in Gangwon-do X (2020) (translated by author). Retrived from http://116.67.83.213:18080/streamdocs/view/sd;streamdocsId=72059246730938970
Toughtco. Geography of the Korean Peninsula, https://www.thoughtco.com/the-korean-peninsula-1435252. Retrieved from 26 May 2022.
National list of species of Korea (2021). National Institute of Biological Resources, online at https://kbr.go.kr Accessed from 26 May
National Institute of Biological Resources. The Inventory of Endemic Species on the Korean Peninsula (2020). Retrieved from https://www.nibr.go.kr/aiibook/ecatalog5.jsp?Dir=1059&catimage=&callmode=admin
Hamilton, C. A., Hendrixon, B. E., Brewer, M. S. & Bond, J. E. An evaluation of sampling effects on multiple DNA barcoding methods leads to an integrative approach for delimiting species: A case study of the North American tarantula genus Aphonopelma (Araneae, Mygalomorphae, Theraphosidae). Mol. Phylogenet. Evol. 71, 79–93 (2014).
Sukumaran, J. & Knowles, L. L. Multispecies coalescent delimits structure, not species. Proc. Natl. Acad. Sci. U.S.A. 114, 1607–1612 (2017).
Struck, T. H. et al. Finding evolutionary processes hidden in cryptic species. Trends Ecol. Evol. 33, 153–163 (2018).
Õunap, E. & Viidalepp, J. Description of Crypsiphona tasmanica sp. Nov.(Lepidoptera: Geometridae: Geometrinae), with notes on limitations in using DNA barcodes for delimiting species. Aust. J. Entomol. 48, 113–124 (2009).
Nieukerken, E. J. V., Doorenweerd, C., Stokvis, F. R. & Groenenberg, D. S. DNA barcoding of the leaf-mining moth subgenus Ectoedemia s. str.(Lepidoptera: Nepticulidae) with COI and EF1-α: Two are better than one in recognising cryptic species. Contrib. Zoo. 81, 1–24 (2012).
Kim, S., Lee, W. & Lee, S. Estimation of a new molecular marker of the genus Stathmopoda (Lepidoptera: Stathmopodidae): Comparing EF1a and COI sequences. J. Asia-Pacific. Entomol. 20, 269–280 (2017).
Harrison, S. E. et al. An African mygalomorph lineage in temperate Australia: The trapdoor spider genus Moggridgea (Araneae: Migidae) on Kangaroo Island, South Australia. Austral Entomol. 55, 208–216 (2016).
Hedin, M. & McCormack, M. Biogeographical evidence for common vicariance and rare dispersal in a southern Appalachian harvestman (Sabaconidae, Sabacon cavicolens). J. Biogeogr. 44, 1665–1678 (2017).
Baker, C. M. et al. Molecular phylogeny and biogeography of the temperate Gondwanan family Triaenonychidae (Opiliones: Laniatores) reveals pre-Gondwanan regionalization, common vicariance, and rare dispersal. Invert. Syst. 34, 637–660 (2020).
Crews, S. C. & Esposito, L. A. Towards a synthesis of the Caribbean biogeography of terrestrial arthropods BMC. Evol. Biol. 20, 1–27 (2020).
Esposito, L. A. & Prendini, L. Island ancestors and new world biogeography: A case study from the scorpions (Buthidae: Centruroidinae). Sci. Rep. 9, 1–11 (2019).
Pfingstl, T. et al. Phylogeographic patterns of intertidal arthropods (Acari, Oribatida) from southern Japanese islands reflect paleoclimatic events. Sci. Rep. 9, 1–20 (2019).
Čandek, K., Agnarsson, I., Binford, G. J. & Kuntner, M. Biogeography of the Caribbean Cyrtognatha spiders. Sci. Rep. 9, 1–14 (2019).
McHugh, A., Yablonsky, C., Binford, G. & Agnarsson, I. Molecular phylogenetics of Caribbean Micrathena (Araneae: Araneidae) suggests multiple colonization events and single island endemism. Invert. Syst. 28, 337–349 (2014).
Acknowledgements
We thank Minho Lee, Duk-Young Park, Minseok Oh & Jaeseok Oh (Department of Agricultural Biotechnology, Seoul National University, Seoul, Korea), Sungwon Yoon (Department of Forest Landscape Architecture, Sangji University, Wonju, Korea), Jeong-Hoon Ahn (Goseong Middle School), Jun-Gi Lee, Jun-Ho Lee (College of Agriculture & Life Sciences, Kangwon National University, Chuncheon, Korea), Wonoong Kim (College of Literature, Science, and the Arts, University of Michigan, Ann Arbor, Michigan, USA), Hyeonjun Shin (Department of Biology, Soonchunhyang University, Asan, Korea) and Jangwon Seo (Department of Biological Science and Biotechnology, Chungbuk National University, Chungju, Korea) for their help collecting specimens. We also thank Seung-Ha Hwang (Department of Game, Chungkang University, Icheon, Korea) for assistance in preparing the figures. The first author is especially grateful to Yong-Gun Choi (The Korean Institute of Biospeleology, Daejeon, Korea), Dr. Jung Sun Yoo (National Institute of Biological Resources, Incheon, Korea), Jae-Won Kim (Yangpyeong Insectarium, Yangpyeong-gun, Korea), and Dongyoung Kim (Department of Biological Sciences, Ajou University) for providing specimens with sharing valuable comments during this study. Finally, we are grateful to Dr. Joel Ledford (Department of Plant Biology, University of California–Davis, Davis, California, USA) for advice at the beginning of this study, and Dr. Dimitar Dimitrov (Department of Natural History, University of Bergen, Bergen, Norway) for valuable and helpful comments on the draft version of the manuscript. This work was supported by a grant from the National Institute of Biological Resources (NIBR), funded by the Ministry of Environment (MOE) of the Republic of Korea (NIBR202002204, NIBR202102203), and Korea Institute of Planning and Evaluation for Technology in Food, Agriculture and Forestry (IPET) through Agriculture, Food and Rural Affairs Convergence Technologies Program for Educating Creative Global Leader Program (or Project), funded by Ministry of Agriculture, Food and Rural Affairs (MAFRA)(no.321001-03), and the Basic Science Research Program through the National Research Foundation of Korea (NRF), funded by the Ministry of Education (NRF2020R1I1A2069484).
Author information
Authors and Affiliations
Contributions
All authors conceived and designed the study; S.K and S.L supported funding acquisition; J.H.O conducted the sampling, morphological identification, DNA extraction, and PCR experiments, performed sequence data analyses, MOTU analyses, prepared figures and tables; J.H.O and S.K wrote and revised the manuscript; S.K. edited the manuscript; All authors confirmed and approved the final version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Oh, JH., Kim, S. & Lee, S. DNA barcodes reveal population-dependent cryptic diversity and various cases of sympatry of Korean leptonetid spiders (Araneae: Leptonetidae). Sci Rep 12, 15528 (2022). https://doi.org/10.1038/s41598-022-18666-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-022-18666-y
- Springer Nature Limited