Abstract
Nitrogen transport is one of the most important processes in plants mediated by specialized transmembrane proteins. Plants have two main systems for nitrogen uptake from soil and its transport within the system—a low-affinity transport system and a high-affinity transport system. Nitrate transporters are of special interest in cereal crops because large amount of money is spent on N fertilizers every year to enhance the crop productivity. Till date four gene families of nitrate transporter proteins; NPF (nitrate transporter 1/peptide transporter family), NRT2 (nitrate transporter 2 family), the CLC (chloride channel family), and the SLAC/SLAH (slow anion channel-associated homologues) have been reported in plants. In our study, in silico mining of nitrate transporter genes along with their detailed structure, phylogenetic and expression analysis was carried out. A total of 412 nitrate transporter genes were identified in hexaploid wheat genome using HMMER based homology searches in IWGSC Refseq v2.0. Out of those twenty genes were root specific, 11 leaf/shoot specific and 17 genes were grain/spike specific. The identification of nitrate transporter genes in the close proximity to the previously identified 67 marker-traits associations associated with the nitrogen use efficiency related traits in nested synthetic hexaploid wheat introgression library indicated the robustness of the reported transporter genes. The detailed crosstalk between the genome and proteome and the validation of identified putative candidate genes through expression and gene editing studies may lay down the foundation to improve nitrogen use efficiency of cereal crops.
Similar content being viewed by others
Introduction
Nitrogen is one of the essential elements required by plants. It is a constituent of nucleic acids, amino acids and proteins and therefore is of great importance in plant physiology and metabolic processes. Though N2 is abundant in atmosphere, only legumes are able to fix atmospheric N2 with the help of Rhizobium bacteria. All other plants mainly absorb N in the form of inorganic ions (ammonium (NH4+) and nitrate (NO3−)) from soil. Nitrate is mostly absorbed in aerobic soils, while ammonium is mostly absorbed in acidic soils and wet lands. After uptake, NO3− and NH4+ are assimilated, transformed and mobilized through various processes within plant system.
The agricultural systems focussed on the high-yield crop production remove nitrogen from the soil and depends mostly on the application of large quantities of nitrogenous fertilizers such as urea for the sustained productivity over time. Unfortunately, a large fraction of the applied nitrogen is not directly absorbed by the plants and is lost by the leaching1. Despite significant efforts made by the scientific community in the last 50 years, the nitrogen-use efficiency for the cereal crops has not been improved2. Beyond this, the economic losses and detrimental environmental consequences caused by the use of large quantities of fertilizers in agriculture are critical issues to be considered3,4. Unravelling the genomic regions or the putative candidate genes improving nitrogen-use efficiency will be the first step toward developing nutrient-efficient crop varieties.
To transport N from soil to roots and to other parts of plants, plasma membrane localized proteins known as transporters are essential. They are involved in regulation of N root uptake, root to shoot and leaf to sink transport5,6. Plants have evolved two systems for N uptake to cope with changes in N availability. These two systems are the low-affinity transport system (LATS) and high-affinity transport system (HATS). A low-affinity transport system (LATS) is involved where adequate amounts of nitrogen levels are present. A high-affinity transport system (HATS) is involved where limited amounts of N are present. Plants have two low-affinity and two high affinity N transport systems,for nitrate (NRT1- low-affinity NO3− transporters and NRT2-high-affinity NO3− transporters) and ammonium (AMT1-low-affinity NH4+ transporters and AMT2-high affinity NH4+ transporters). Majority of N in cereal crops such as wheat is taken up in form of nitrate (NO3−). Therefore, nitrate transporters are of great importance.
In plants four families of NO3- transporters have been identified named NPF (NRT1/PTR), NRT2, CLC (chloride channel) and SLAC1/SLAH (slow type anion channel associated homologs)7. NRT1.1 was first NO3- transporter to be identified in Arabidopsis8. The NRT1 transporter family which has been renamed as NPF family is the largest family of nitrate transporters and can further be classified into eight subfamilies9. In Arabidopsis NPF transporters have been well characterized and contain 53 members divided into eight subfamilies9. In rice (Oryza sativa) NPF transporters contain 93 members10. The majority of NPF transporters are involved in LATS with few exceptions of NRT1.1/NPF6.3 in Arabidopsis and MtNRT1.3 in Medicago truncatula, which are involved in both HATS and LATS11,12. Although majority of NPFs are involved in nitrate transport, several studies have suggested their role in transport of other substrates such as nitrite13, peptides14, amino acids15 and several plant hormones16,16,17,18,20. The second family known as NRT2 contains high affinity nitrate transporters. A total of seven NRT2 transporters in Arabidopsis21 and five NRT2 transporters in rice have been reported22,23. Most of NRT2 transporters require a partner protein—NAR2 (nitrate assimilation related protein) to function as high affinity nitrate transporters22,22,23,25. Third family of nitrate transporters, CLC (chloride channel) family is mainly associated with vacoular transport of NO3−26. In Arabidopsis, six CLC genes have been reported and are responsible for nitrate and chloride homoeostasis, thereby regulating stomatal movement and salt tolerance26,27,28. The fourth family—SLAC1/SLAH (slow type anion channel associated homologs) is anion channel family. In Arabidopsis this family contains four members-SLAC1, SLAH1, SLAH2 and SLAH3 which are involved in the nitrate transport in guard cells and roots and in chloride acquisition29. Together these four transporter families are involved in efficient nitrate uptake and utilization in plants.
To the best of our knowledge, the nitrate transporters in hexaploid wheat have not been characterized and explored completely. There are some studies conducted to access the effect of different nitrogen conditions on some of NPF and NRT2 genes30. Most of the studies in wheat have been conducted on members of TaNRT2 gene family. Overexpression of TaNRT2.5 has been associated with increased grain nitrate uptake and yield31. TaNRT2.1 has been associated with post flowering nitrate uptake in wheat32. Expression of TaNRT2.1 can be induced by nitrogen starvation and abscisic acid (ABA)33,32,33,34,37. Some phylogenetic studies and expression-based studies have been conducted on NPF and NRT2 genes recently34,33,36,38 but CLC and SLAC1/SLAH genes still remain uncharacterized. Structure of proteins play very important role in the functionality of transporter proteins but still no studies have been conducted on structure prediction of any of NPF, NRT2, CLC and SLAC1/SLAH genes in wheat. In our study we have identified and characterized genes belonging to all the four families of nitrate transporters. Our analysis includes gene composition, chromosomal location, phylogenetic relations with members of rice and Arabidopsis and expression analysis. We adopted a new nomenclature for identified genes as the earlier nomenclature systems do not include complete information about subgenome and homoeologs. We have classified the genes based on phylogeny and identified homoeologous pairs of the gene. Expression profiles of all the genes were studied for different developmental stages and different tissues. Further the structures of all the members of gene families were investigated.
Methodology
Sequence search and annotation of nitrate transporter genes
Two methods were used for the identification of NRT1, NRT2 genes in wheat. In the first method, the CDD IDs (conserved domain database IDs) specific to TaNPF, TaCLC, TaSLAC/TaSLAH and TaNRT2 genes (Table 1) were used as identifiers to retrieve genes from the wheat reference genome (IWGSC RefSeq V2.0) from the Ensembl Plants (https://plants.ensembl.org/index.html). In the second method, protein sequences were downloaded from the NCBI database using Nitrate/Nitrogen transporters, and NRT as queries. Incomplete, partial sequences, hypothetical, and predicted protein sequences were filtered out. The downloaded sequences were manually curated to remove duplicate sequences and incomplete sequences. The remaining protein sequences (1687 genes) were aligned using Clustal Omega, and the output Stockholm file was used to create the HMMER profile. The HMMER profile was used to search similar protein sequences in the wheat protein database downloaded from IWGSC. A total of 403 high confidence and 38 low confidence proteins were obtained. Separate searches were performed for TaCLC and TaSLAC1/TaSLAH genes using the same method. A total of 41 TaCLC and 43 TaSLAC1/TaSLAH high confidence genes and 10 TaCLC and 7 TaSLAC1/TaSLAH low confidence genes were obtained. The sequences from both the methods were combined, followed by the removal of low confidence proteins and duplicate sequences, and after manual curation, a final set of 412 genes belonging to all four nitrate transporter families were selected. The same methodology was used to identify sequences for Triticum dicoccoides (AABB), T. turgidum (AABB), T. urartu (AA), and Aegilops tauschii (DD) for comparative analysis.
Maximum likelihood phylogeny of nitrate transporter genes
The alignments of TaNRT1/TaNPF, TaCLC, TaSLAC1/TaSLAH and TaNRT2 sequences were created separately using wheat, rice, and Arabidopsis sequences by MAFT (E-INS-I algorithm). The evolutionary history was inferred by using the Maximum Likelihood method and JTT matrix-based model. Initial tree(s) for the heuristic search were obtained automatically by applying Neighbour-Join and BioNJ algorithms to a matrix of pairwise distances estimated using the JTT model and then selecting the topology having superior log-likelihood value. Evolutionary studies were conducted in MEGA X. The consistency of the phylogenetic estimate was evaluated by bootstraps (1000 replicates). The resulting tree was visualized using FIGTREE v.1.4.4 (http://tree.bio.ed.ac.uk/software/figtree/).
Gene structure prediction and identification of homoeologs
The genomic and CDS sequences of genes were downloaded from the Ensembl plants database. The sequence information was utilized to predict the intron/exon positions by using the GSDS server (Gene Structure Display Server, http://gsds.cbi.pku.edu.cn39). Separate phylogenies were generated for members of each subfamily to resolve the relationship between them. The analysis was performed in MEGA X by the method described previously. Homoeologous genes were identified based on the phylogenetic relationship between the members of subfamilies. The information regarding physical positions of genes were obtained from Ensembl Plants database. Genome wide distribution map of nitrate transporter genes was developed by web based online visualization tool PhenoGram (http://visualization.ritchielab.org/phenograms/plot).
Naming of TaNPF, TaNRT2, TaCLC and TaSLAC1/SLAH genes
We adopted the method proposed by Schilling et al.40 for the naming of NRT genes. The genes were named based on their phylogenetic relationships and subgenome location (A, B, or D). Each gene name started with the abbreviation for the species name Triticum aestivum (Ta), followed by the most closely related Arabidopsis gene name (i.e., NPF1-NPF8, NRT2), which was followed by the subgenome identifier (A, B, and D). Putative homoeologs were given identical gene names except for the subgenome identifier (TaNPF4-A1, TaNPF4-B1 and TaNPF4-D1). The genes belonging to the same subfamily in the same subgenome were consecutively numbered (Table 2).
Structure prediction of nitrate transporter proteins
Due to the unavailability of crystal structures, gene homology modelling was carried out to predict their three-dimensional (3D) structure. The sequences of TaNRT1, TaCLC, TaSLAC1/TaSLAH and TaNRT2 genes were submitted to web-based server Phyre241. Briefly, Phyre2 used PSI-BLAST to detect sequence homologues which was followed by Psi-pred and Diso-pred to predict secondary structure and disorder. Then Hidden Markov models (HMM) of sequences were generated based on homologues detected before. HMMs of query proteins were scanned against library of HMMs of proteins with experimentally solved structures to construct 3D models of query proteins. Transmembrane helix and topology prediction was carried by memsat-svm41.
Expression analysis of nitrate transporter genes
The RNAseq data of TaNPF, TaNRT2, TaCLC and TaSLAC1/TaSLAH genes of various tissues (root, shoot/leaf, spike, grain) at three developmental stages (seedling, vegetative and reproductive) for Chinese spring and Azhurnaya (cv) was downloaded from the wheat expression database (www.wheat-expression.com). Expression levels were downloaded as log2(transcripts per million) (log2tpm) for different tissues at different time points. Several tissue-specific (root, shoot, leaf, grain) genes were identified based on expression patterns. For triad expression analysis, a method described by Ramírez-González et al.42 was used. Briefly, the expression data from spring wheat (CS) and Azhurnaya was downloaded from the wheat expression database as TPM for root, leave, shoot spike and grain. For analysis, the triads with expression below one tpm were excluded. Expression values were normalized, triads were assigned balanced, A/B/D suppressed or A/B/D dominant profiles. To elucidate the role of Nitrate transporter genes towards N starvation and N recovery, the gene expression data set34,33,36 from wheat omics 1.0 database (http://wheatomics.sdau.edu.cn/) was analysed. The dataset contained expression data in roots of 10-day old wheat plants (Chinese Spring) treated for N-starvation for 5 days and then subjected for N-recovery34,33,36.
Development of validation panel to check the efficacy of the identified nitrate transporter genes
The nested synthetic hexaploid wheat (N-SHW) introgression library constituting a set of 352 breeding lines derived from four sub-populations (Pop1: 75 lines from PDW233/Ae. tauschii acc. pau 14,135 amphiploid //2*BWL4444; Pop2: 106 lines from PDW233/Ae. tauschii acc. pau 14,135 amphiploid //2*BWL3531; Pop3: 88 lines from PBW114/Ae. tauschii acc. pau 14,170 amphiploid //2*BWL4444; Pop4: 83 lines from PBW114/Ae. tauschii acc. pau 14,170 amphiploid //2*BWL3531) were developed43. These N-SHW library, six parents and two synthetic hexaploid wheats were assessed over 2 years in 2018 and 2019 at 3 nitrogen levels [i.e., zero N (0 kg ha−1), half N (60 kg ha-1) and full N (recommended, 120 kg ha−1]. The detailed phenotyping of the N-SHW introgression libraries for the nitrogen-use efficiency related traits was carried out across years and treatments43. High-density genotyping was performed using the 35 K Axiom® Wheat Breeder’s Array (Affymetrix UK Ltd., United Kingdom). The population structure of the 352 N-SHW lines was assessed on the basis of 9,474 SNPs distributed across all 21 wheat chromosomes. The most appropriate K explaining the population structure was K = 3 at MAF ≥ 5% (Supplementary Fig. 4A). The kinship heatmap suggested a weak relatedness in the panel (Supplementary Fig. 4B). The first three principal components (PCs) were most informative gradually decreasing (Supplementary Fig. 4C,D) until the tenth PC. The kinship and PCs were considered during the GWAS analysis to correct for population structure. The appropriate number of sub-populations was determined from the largest delta K value of 3 (Supplementary Fig. 4E). The kinship and PCs were considered during the GWAS analysis to identify population structure. Significant marker-trait associations were identified using CMLM (compressed mixed linear model)/P3D (population parameters previously defined) in GAPIT (Genome Association and Prediction Integrated Tool) executed in R. Over 322 marker trait associations for NUE were compared to nitrate transporter genes.
Results
The wheat genome consists of 412 nitrate transporter genes belonging to four different families
A total of 412 nitrate transporter sequences excluding splice variants were identified in IWGSC wheat genome assembly (RefSeq V2.0). The wheat genome consists of 292 TaNPF genes, 34 TaCLC genes, 40 TaSLAC1/TaSLAH genes and 46 TaNRT2 genes. The TaNPF genes could be divided into eight subgroups (TaNPF1 to TaNPF8) based on the presence of conserved domains (Table 1). TaNPF5 subgroup was the largest group consisting of 97 genes followed by TaNPF8 (70 genes), TaNPF2 (41 genes), TaNPF4 (33 genes), TaNPF6 (22 genes), TaNPF3 (12 genes) and TaNPF7 (11 genes). The NPF1 subgroup was the smallest one consisting of 6 genes present on homoeologous group chromosomes 3A, 3B and 3D. TaNRT1/TaNPF genes were present throughout the genome (Fig. 1). The location of genes across chromosomes varied according to the size of the subfamily. The genes belonging to larger subfamilies (e.g., TaNPF5, TaNPF8, TaNPF2) were predominantly located in tandem positions on the distal region of chromosomes. The genes belonging to smaller subfamilies (TaNPF1, TaNPHF7, TaNPF3) were located on proximal regions of chromosomes. The genes present near distal ends of chromosomes were found to be in the form of clusters in close vicinity to each other. The majority of TaNRT2 genes were present in the clusters on the distal end of homoeologous chromosomes 6A, 6B and 6D. TaCLC genes were distributed across the wheat genome. TaSLAC1/TaSLAH genes were only distributed on homoeologous chromosomes 1A,1B, 1D, 2A, 2B, 2D, 3A, 3B and 3D. The predicted gene structures contained several intron regions (Supplementary Fig. 1a–c) for many genes in TaNPF, TaCLC and TaSLAC1/TaSLAH families. All the TaNRT2 genes were intron less. The size of predicted genes ranged between 1 and 25 Kb. Several truncated and duplicated genes were also predicted.
Genome wide distribution of TaNPF, TaNRT2, TaCLC and TaSLAC1/TaSLAH genes in hexaploid wheat. Figure was generated by web-based software tool-Phenogram from Ritchie Lab44 (http://visualization.ritchielab.org/phenograms/plot).
Phylogenetic relationships among nitrate transporter genes
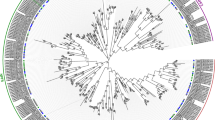
The maximum likelihood phylogenetic tree of all the nitrate transporter genes predicted that wheat contains all the major subfamilies present in Arabidopsis and rice (Oryza sativa) (Fig. 2a). The TaNRT1/TaNPF and TaNRT2 genes could be classified into five subclades. The subclades in the phylogenetic tree followed species phylogeny with Arabidopsis genes displaying sister group relationship with wheat genes. Based on the phylogenetic relationship, TaNRT1/TaNPF genes fitted well into eight subfamilies (TaNPF1 to TaNPF8) following the Arabidopsis model. The topology of larger subclades (TaNPF5, TaNPF8, TaNPF2) was more complex than smaller subclades as they were more expanded in wheat than Arabidopsis and rice (Fig. 2a, Supplementary Fig. 2). TaNRT2 genes were present as a separate subclade and were closely related to the TaNPF2 subfamily. The phylogenetic analysis of TaCLC and TaSLAC1/TaSLAH genes was carried out separately. The results showed TaCLC genes could be classified into 6 groups according to phylogenetic relation with Arabidopsis and rice genes (Fig. 2b). TaSLAC1/TaSLAH genes were divided into 4 subclades. The largest subclade in TaSLAC1/TaSLAH genes showed close relationship with rice SLAC1/SLAH genes but not with Arabidopsis genes (Fig. 2c).
Phylogenetic tree depicting relationship between (a) TaNPF and TaNRT2 genes in hexaploid wheat and Arabidopsis thaliana (b) TaCLC genes in wheat, rice and Arabidopsis thaliana (c) TaSLAC1/SLAH genes in hexaploid wheat, rice and Arabidopsis thaliana. Phylogenetic analysis was performed by MEGA X software45 and the results were edited and visualized by FIGTREE software v1.4.4. (http://tree.bio.ed.ac.uk/software/figtree/) to generate final images.
Homoeologs retention and gene duplication in nitrate transporter genes
The number of nitrate transporter genes in each family were significantly higher than those in Arabidopsis and rice (Table 1, Supplementary Table 1). The comparison with T. dicoccoides (AABB), T. turgidum (AABB), T. urartu (AA) and Ae. tauschii (DD) suggested that most of the homoeologs in hexaploid wheat were retained during evolution (Fig. 3, Supplementary Table 1). There was also evidence of gene duplications in tetraploids and hexaploid wheat, reflected in gene number and phylogenetic data (Fig. 2, Supplementary Fig. 1a–c). Most duplicated genes were present in subfamilies with a larger number of genes (TaNPF5, TaNPF8, TaNPF2 and TaNRT2). Nitrate transporters could be grouped into 13 triads, 26 diads, 2 tetrads and 48 singleton genes based on phylogeny (Table 3). Out of a total of 292 TaNPF genes, about 74% of TaNPF genes could be grouped into 72 triads of homoeologous genes (A, B, D) based on phylogenetic relationships. Similarly, 71% of TaNRT2 genes, 97% of TaCLC genes and 80% of TaSLAC1/TaSLAH genes could be grouped into homoeologous triads.
Synteny relationships of wheat nitrate transporter genes orthologous with (A) A. thaliana, (B) O. sativa, (C) T. urartu, (D) Ae. tauschii, (E) T. dicoccoides and, (F) T. turgidum. Circos plots were generated by web-based application- shinyCircos (https://venyao.xyz/shinycircos/)46.
Nitrate transporter proteins contain multiple transmembrane helices
To study the structural features of nitrate transporters, we predicted the 3D structures of all 412 protein sequences. All nitrogen transporters were predicted to be transmembrane proteins containing multiple transmembrane segments (Fig. 4i). The majority of proteins comprised of 12–14 transmembrane helices (TMs) with some variation. The basic structure of TaNRT/TaNPF proteins included N and C terminal segments followed by multiple transmembrane helices (TMs). The transmembrane helices were connected by alternating cytoplasmic and extracellular loop segments (Fig. 4ii). In TaNRT1/TaNPF family, approximately 67% of the proteins contained 14 TMs, 21% contained 13 TMs, 7% of proteins contained 12 TMs while 4% of proteins contained less than 12 TMs (Supplementary Table 2). Subfamily wise studies showed TaNPF1 proteins contained only 13 TMs and TaNPF7 contained only 14 TMs. In rest of subfamilies (TaNPF2-6, TaNPF8) majority of proteins contained 14 TMs but variation existed. Proteins with even number of TMs had both C and N terminals in cytoplasmic side of membrane. Proteins with odd number of TMs had one end in cytoplasmic side and other in extracellular side (Fig. 4ii). All TaNRT2 family members contained only 12 TMs (Supplementary Table 2) (Fig. 4ii). Both C and N terminals of TaNRT2 proteins were present in cytoplasmic side of the membrane. Both TaCLC and TaSLAC1/SLAH proteins contained 10 TMs with both N and C terminals in cytoplasmic side of membrane. TaCLC genes were characterized by presence of a 30–40 amino acids long re-entrant helix in cytoplasmic side (Fig. 4 ii) which was not observed in the proteins of other nitrate transporter gene families.
Protein structure prediction: (i) representative structures of TaNPF genes (A–H), TaNRT2 genes (I) TaCLC genes (J) and TaSLAC1/TaSLAH genes (K). (ii) Representative TMs structures of nitrate transporters containing (A) 14 TMs, (B) 13 TMs (C) 12 TMs and (D) CLC proteins containing 10 TMs and a re-entrant helix. Figures were developed by homology-based modelling by Phyre2 server41.
Expression patterns of nitrate transporter genes in development stages of wheat
To elucidate the expression patterns of nitrate transporter genes, we studied and compared the expression data of Chinese spring and Azhurnaya for different developmental stages. Approximately 77% of TaNPF genes, 30% of TaNRT2, 85% of TaCLC genes and 36% of TaSLAC1/TaSLAH genes were expressed at least at one developmental stage in wheat with a wide expression range of 1–103 tpm (Supplementary Table 3, Supplementary Fig. 3). The remaining genes showed very low or no expression (tpm < 1). Overall, we identified 20 triads in which 48 genes were showing tissue specific expression, out of which 8 triads were root specific, 5 triads were leaf/shoot specific and 7 triads were showing grain/ spike specific expression (Supplementary table 4). Tissue and developmental stage-specific expression were observed in TaNPF1 genes, which were only expressed in spike and grain at the reproductive stage (Fig. 5A). Similarly, TaNRT2 genes were predominantly expressed in roots in both vegetative and reproductive stages (Fig. 5A). TaSLAC1/TaSLAH genes were predominately expressed in roots and leaves with some genes showing expression in spikes also (Fig. 5B). TaCLC genes showed mostly ubiquitous expression (Fig. 5B). For the rest of the subfamilies, the genes within one subfamily differed considerably in their expression patterns. In TaNPF2 genes, spike/grain specific (3 genes), leaf, spike and grain specific (5 genes) and ubiquitous expression (6 genes) were observed (Fig. 5A). TaNPF3 genes showed spike/grain, leaf specific expression, TaNPF4 genes showed leaf/root-specific (4 genes) and ubiquitous expression (10 genes) (Fig. 5A). TaNPF5 and TaNPF8 genes mostly showed ubiquitous expression though the root-specific expression was observed in a few genes (Fig. 5A). TaNPF6 showed ubiquitous (6 genes), leaf and root-specific (6 genes), spike specific (3 genes) and root-specific expression (Fig. 5A). TaNPF7 showed ubiquitous expression in three genes, grain specific expression in two genes and root-specific expression in one gene (Fig. 5A).
Expression patterns of nitrate transporter gene triads in wheat (a) Tissue and development stage specific expression profiles of TaNPF and TaNRT2 genes (b) Tissue and development stage specific expression profiles of TaCLC and TaSLAC1/SLAH genes. The heat maps were generated by heatmap tool from wheat expression database42 (http://wheat-expression.com/).
To find out up to what extent homoeologs differ in the expression patterns, triad expression analysis was performed. Most of the triads showed balanced expression ranging from 55.6 to 65.2% in all the tissues (Fig. 6A). In roots, a total of 54 triads were showing expression out of total 83 triads. Out of which 55.6% showed balanced expression, 18.5% showed A suppressed, 11.1% showed D suppressed, 9.3% showed B suppressed expression. Three triads showed A, B and D dominant expression (1 each) (Fig. 6B). In leaf/shoot out of 51 triads, 64.7% showed balanced expression, 9.8% showed A suppressed and B suppressed each, 3.9% triads showed D suppressed expression. 5.8% triads showed A and D dominant expression each while no B dominant expression was observed (Fig. 6B). In spikes, 61.9% triads out of 42 triads showed balanced expression. Only D dominant expression was observed in 9.5% of triads while A suppressed, B suppressed, and D suppressed expressions were in about 16.7, 7.1% 4.7% triads (Fig. 6B). Only 23 triads were expressing in grains at the reproductive stage, out of which 65.2% showed balanced expression, 8.7% triads showed A, B, and D suppressed each and 4.3% triads showed B and D dominant expression (Fig. 6B).
Triad expression of nitrate transporters in wheat (A) Overall triad expression of all nitrate transporter genes (B) Tissue specific triad expression of nitrate transporter genes. Normalized expression values were used to generate ternary plots using online web-based tool (https://www.ternaryplot.com/).
Nitrate transporter genes are located in close proximity to the NUE associated SNPs
In a parallel study in our laboratory, the nested synthetic wheat introgression (N-SHW) libraries capturing novel genetic variation from wild wheat for the nitrogen use efficiency related traits were developed and genotyped using a high-density SNP array43. These libraries were phenotypically assessed for the root traits and agronomic performance under three nitrogen input conditions (N: 0 kg ha−1; N: 60 kg ha−1 and N:120 kg ha−1) in the field over two years in 2018 and 2019. Genome-wide association mapping was used to identify marker-trait associations for the root and agronomic traits to identify the marker-trait associations for traits improving nitrogen use efficiency in wheat (Supplementary Table 5). We compared 322 marker trait associations for NUE identified in this study43 to nitrate transporter genes identified during genome wide analysis. We identified 67 SNPs, which were in close proximity to nitrate transporter genes in the wheat genome. A total of 93 nitrate transporter genes could be located near NUE linked SNPs, out of which, 63 genes belonged to TaNPF family, 15 genes belonged to TaNRT2 family, 11 genes belonged to TaCLC and 4 genes belonged to TaSLAC1/TaSLAH family (Table 4, Supplementary Fig. 5).
Response of nitrate transporter genes during N-starvation and N-recovery
The response of all N transporter genes towards N starvation and N recovery was analysed from WheatOmics database34,33,36,47,48. The results suggested that the expression of N transporter genes towards N starvation and N recovery was variable. We specifically identified the genes whose expression patterns changed significantly in response to N starvation or N recovery. The expression values of TaNPF1 and TaNPF3 genes were not significant (Fig. 7A,C). Three genes in TaNPF2 showed increased expression in N starvation and their expression values returned to normal during N recovery (Fig. 7B). The expression values of most of TaNPF5 genes were slightly reduced during N starvation and increased significantly during N recovery (Fig. 7E,F). TaNPF6 genes expression reduced during both N starvation and N recovery (1 h) but their expression returned to normal 24 h after recovery (Fig. 7G). The expression of most of TaNPF7 genes was upregulated during N starvation and N recovery (1 h) and downregulated after 24 h of N recovery (Fig. 7H). The expression of TaNPF4 and TaNPF8 genes was variable (Fig. 7D,I,J). The expression of most of TaNRT2 and TaCLC genes was upregulated during N recovery (1 h) phase (Fig. 7K,L,M,N). The expression values of some TaSLAC1/TaSLAH genes were reduced in response to N starvation and increased during N recovery (24 h) (Fig. 7O,P). Specifically looking into the expression pattern of 93 genes in close proximity of NUE associated SNPs, we could identify 32 genes whose expression pattern changed in response to N starvation and N recovery (Supplementary Fig. 6, Supplementary Table 6). These genes can serve as candidate genes and may be further utilized in genomics-assisted breeding programs targeting improved nitrogen-use efficiency in wheat.
Discussion
The main aim of this study was to identify and analyse nitrate transporters belonging to all the four families and study their dynamics in wheat. The number of nitrate transporter genes detected in wheat was higher as compared to other plant species. This could be explained by a large genome (~ 18 Gb) and hexaploid nature of wheat. Presence of three homoeologous sub-genomes in wheat could allow multiple copies of nitrate transporters resulting in higher number of transporter genes. When comparing with diploid progenitors (Ae. tauschii and T. urartu) and tetraploid wheats (T. dicoccoides and T. turgidum) the number of genes in each subfamily were approximately proportional (Table 1). The genes were distributed randomly in the genome except for TaNRT2 genes which were predominantly present on group 6 homoeologous chromosome. Many genes were present in form of clusters and showed high percentage of similarity indicating gene-duplication events. There were genes with deleted segments present in the genome. The phylogenetic relationships with orthologues in other plants could be used to classify the genes in subfamilies. All the major subclades were conserved in wheat in comparison to other plant species indicating biological importance of the subfamilies. Based on phylogeny the genes could be grouped in homoeologous triads. Almost 73% of the genes could be assigned to 1:1:1 homoeologous groups which is very much above the average homoeologous retention rate (35.8%) in wheat (IWGSC 2018). Many genes were also grouped into tetrads and diads based on homology indicating gene duplication and deletion events in the genome. The overall results revealed that wheat nitrogen transporter families are much more complex than in other plant species. This complexity arises mostly due to presence of three sub-genomes (A B D) and gene duplication and deletion events.
The complexity of wheat genome also affects the expression patterns of genes. Due to presence of multiple sets of homoeologs on A, B and D genomes the buffering effects are observed in expression of genes. To study up to what extent these interactions affect the expression of nitrate transporters, triad expression analysis was performed. More than 55% of genes showed balanced expression in all the tissues which is comparable to genome-wide assessment of all transcripts in wheat42. The expression profiles of the genes identified in this study were in accordance to the previous studies in other plants. The expression patterns of nitrate transporter genes were similar to expression patterns of close orthologs in rice and Arabidopsis indicating the conservation of gene functions. CLC genes in previous studies in Arabidopsis showed ubiquitous expression which was observed in this study for wheat as well27,28. Several tissue specific nitrate transporter genes were identified which can be targeted for gene manipulation for wheat improvement. Several TaNRT2 and TaSLAC1/TaSLAH genes showed root specific expression suggesting their role in root nitrate uptake. Root specific expression of NRT2 and TaSLAC1/TaSLAH genes has already been reported in rice and Arabidopsis29,49. TaNPF1 genes and some TaSLAC1/SLAH genes showed grain and spike specific expression suggesting their role in nitrate transfer in developing seeds.
Structure plays a very important role in the function of transporter proteins. X-ray crystallographic structures of eukaryotic nitrate transporters have been elucidated50. All the nitrate transporter families belong to a much larger major facilitator superfamily (MFS) according to transporter classification database51. All the nitrate transporter proteins were predicted to have a typical MFS protein structure with multiple TMs. To the best of our knowledge our study is the first one to report homology-based models of nitrate transporter proteins belonging to all four families in wheat. The number of transmembrane segments play very important role in the optimal functioning MFS transporter proteins52. For an MFS transporter protein to have optimal transport properties pseudosymmetry is important which is provided by even number of TMs50. According to previous studies most of MFS proteins required 12 TMs to have optimal function53. In our study we predicted nitrate transporter families having variation in the number of TMs. TaNPF family being the largest of all showed most variation in the number of TMs with number ranging from 12 to 14. Several proteins with odd number of TMs were also observed. For example, all the members of TaNPF1 subfamily contain 13 TMs. All TaNRT2 proteins were highly conserved and contained 12 TMs. Most of the TaCLC and TaSLAC1/TaSLAH genes contained only 10 TMs. The variation in number of TMs between and within subfamilies and presence of odd number of TMs could not be corelated with expression data suggesting that a much more flexible criteria exists for the function of nitrate transporter proteins. The structural information presented in this study offer foundation for future work to identify molecular mechanisms responsible for functioning of nitrate transporters in wheat.
Previously in many studies overexpression of nitrate transporter genes has been linked to improved nitrogen use efficiency and yield in many plants54,51,52,57 and58. Overexpression of OsNRT2.1, OsNRT2.3b, OsNPF6.3 in rice and ZmNRT1.1A in maize has resulted in increased grain yield25,34,33,36,57,57,59. In wheat TaNRT2.1 is reported to be involved in post-flowering N uptake32 and is an important gene for improvement of nitrogen use efficiency. The CLC genes have been reported to be involved in nitrate accumulation in plants26 and many CLC genes have been reported to have role in stress responses. SLAC1 is a key player in regulation of stomatal closure. SLAH genes are involved in root nitrate and chloride acquisition and translocation to shoot. SLAC1/SLAH genes have also been reported to have important role in drought responses49. The genome wide analysis of TaCLC and TaSLAC1/TaSLAH genes in this study is the first reported study of these genes in wheat to the best of our knowledge. Nitrate transporters identified in this study can be promising candidates for gene manipulation to enhance productivity and nitrogen use efficiency in wheat. The identification of nitrate transporter genes in the close proximity to the marker-traits associations indicated the robustness of genome wide association mapping studies and the reliability of the reported transporter genes. The identified nitrate transporters could deepen the understanding of genetic and molecular mechanism behind improving nitrogen-use efficiency in wheat crop. The nutrient efficient improved breeding lines/accessions possessing identified potential nitrate transporters in the present study may have an effective and strong coordinated signal transduction network involving nitrate transceptor, nitrate response regulator and the master response regulator.
The in-silico mining of nitrate transporter genes along with their detailed structure, phylogenetic and expression studies reported a total of 412 nitrate transporter genes including 20 root specific, 11 leaf/shoot specific and 17 grain/spike specific putative candidate genes. The identification of nitrate transporter genes in the close proximity to the previously identified 67 marker-traits associations associated with the nitrogen use efficiency related traits in nested synthetic hexaploid wheat introgression library43 indicated the robustness of the reported transporter genes. The detailed crosstalk between the genome and proteome and the validation of identified putative candidate genes through expression and gene editing studies may lay down the foundation to improve nitrogen use efficiency of cereal crops. The existing genetic variability for 48 tissue specific genes and 93 genes in close proximity to NUE associated SNPs identified in the present study in different wild and cultivated wheat accessions/varieties may be further utilized in genomics-assisted breeding programs targeting improved nitrogen-use efficiency in wheat. A total of 32 genes out of these 93 genes show significant changes in expression patterns in response to N starvation and/ or N recovery suggesting their involvement in N uptake and assimilation. These genes can serve as initial candidates for targeting N use efficiency in wheat. The identification of improved breeding lines or the wild accessions possessing the potential nitrate transporters may serve as novel donors to be used in genomics-assisted introgression program developing nitrogen-efficient wheat varieties. The identified nitrate transporters may have potential for efficient nitrogen uptake and its transport from source to sink.
Once validated, the candidate genes may further be deployed in genomics-assisted breeding program to develop nutrient efficient wheat varieties. The present study provides important information on potential nitrate transporters that may lay foundation to develop a new breeding strategy for the sustainable agricultural development of cereal crops with less input—more output and the environmental protection. The identified nitrate transports may be of great significance both in the theory and in the genomics-assisted breeding application39,57,58,26.
Data availability
All data used in this research are included in this published article and its supplementary information files.
References
Hirel, B., Tétu, T., Lea, P. J. & Dubois, F. Improving nitrogen use efficiency in crops for sustainable agriculture. Sustainability 3(9), 1452–1485. https://doi.org/10.3390/su3091452 (2011).
Cassman, K. G., Dobermann, A. & Walters, D. T. Agroecosystems, nitrogen-use efficiency, and nitrogen management. AMBIO: A J. Human Environ. 31(2), 132–140. https://doi.org/10.1579/0044-7447-31.2.132 (2002).
Galloway, J. N. et al. Transformation of the nitrogen cycle: Recent trends, questions, and potential solutions. Science 320(5878), 889–892. https://doi.org/10.1126/science.1136674 (2008).
Hirel, B., Le Gouis, J., Ney, B. & Gallais, A. The challenge of improving nitrogen use efficiency in crop plants: Towards a more central role for genetic variability and quantitative genetics within integrated approaches. J. Exp. Bot. 58(9), 2369–2387. https://doi.org/10.1093/jxb/erm097 (2007).
Fan, X. et al. Plant nitrate transporters: From gene function to application. J. Exp. Bot. 68(10), 2463–2475. https://doi.org/10.1093/jxb/erx011 (2017).
Tegeder, M. Transporters involved in source to sink partitioning of amino acids and ureides: Opportunities for crop improvement. J. Exp. Bot. 65(7), 1865–1878. https://doi.org/10.1093/jxb/eru012 (2014).
Wang, Y., Hsu, P. & Tsay, Y. Uptake, allocation and signaling of nitrate. Trends Plant Sci. 17(8), 458–467. https://doi.org/10.1016/j.tplants.2012.04.006 (2012).
Tsay, Y., Schroeder, J. I., Feldmann, K. A. & Crawford, N. M. The herbicide sensitivity gene CHL1 of Arabidopsis encodes a nitrate-inducible nitrate transporter. Cell 72(5), 705–713. https://doi.org/10.1016/0092-8674(93)90399-b (1993).
Léran, S. et al. A unified nomenclature of nitrate transporter 1/Peptide transporter family members in plants. Trends Plant Sci. 19(1), 5–9. https://doi.org/10.1016/j.tplants.2013.08.008 (2014).
Von Wittgenstein, N. J., Le, C. H., Hawkins, B. J. & Ehlting, J. Evolutionary classification of ammonium, nitrate, and peptide transporters in land plants. BMC Evol. Biol. 14(1), 11. https://doi.org/10.1186/1471-2148-14-11 (2014).
Liu, K. & Tsay, Y. Switching between the two action modes of the dual-affinity nitrate transporter CHL1 by phosphorylation. EMBO J. 22(5), 1005–1013. https://doi.org/10.1093/emboj/cdg118 (2003).
Morere-Le Paven, M. et al. Characterization of a dual-affinity nitrate transporter MtNRT1.3 in the model legume Medicago truncatula. J. Exp. Bot. 62(15), 5595–5605. https://doi.org/10.1093/jxb/err243 (2011).
Sugiura, M., Georgescu, M. N. & Takahashi, M. A nitrite transporter associated with nitrite uptake by higher plant chloroplasts. Plant Cell Physiol. 48(7), 1022–1035. https://doi.org/10.1093/pcp/pcm073 (2007).
Komarova, N. Y. et al. AtPTR1 and AtPTR5 transport dipeptides in planta. Plant Physiol. 148(2), 856–869. https://doi.org/10.1104/pp.108.123844 (2008).
Zhou, J., Theodoulou, F. L., Muldin, I., Ingemarsson, B. & Miller, A. J. Cloning and functional characterization of a Brassica napus transporter that is able to transport nitrate and histidine. J. Biol. Chem. 273(20), 12017–12023. https://doi.org/10.1074/jbc.273.20.12017 (1998).
Chiba, Y. et al. Identification of Arabidopsis thaliana NRT1/PTR family (NPF) proteins capable of transporting plant hormones. J. Plant. Res. 128(4), 679–686. https://doi.org/10.1007/s10265-015-0710-2 (2015).
David, L. C. et al. N availability modulates the role of NPF3.1, a gibberellin transporter, in GA-mediated phenotypes in Arabidopsis. Planta 244(6), 1315–1328. https://doi.org/10.1007/s00425-016-2588-1 (2016).
Krouk, G. et al. Nitrate-regulated auxin transport by NRT1.1 defines a mechanism for nutrient sensing in plants. Dev. Cell 18(6), 927–937. https://doi.org/10.1016/j.devcel.2010.05.008 (2010).
Saito, H. et al. The jasmonate-responsive GTR1 transporter is required for gibberellin-mediated stamen development in Arabidopsis. Nat. Commun. https://doi.org/10.1038/ncomms7095 (2015).
Tal, I. et al. The Arabidopsis NPF3 protein is a GA transporter. Nat. Commun. https://doi.org/10.1038/ncomms11486 (2016).
Miller, A., Fan, X., Shen, Q. & Smith, S. Expression and functional analysis of rice NRT2 nitrate transporters. Comp. Biochem. Physiol. A: Mol. Integr. Physiol. 146(4), S241. https://doi.org/10.1016/j.cbpa.2007.01.558 (2007).
Feng, H. et al. Multiple roles of nitrate transport accessory protein NAR2 in plants. Plant Signal. Behav. 6(9), 1286–1289. https://doi.org/10.4161/psb.6.9.16377 (2011).
Feng, H. et al. Spatial expression and regulation of rice high-affinity nitrate transporters by nitrogen and carbon status. J. Exp. Bot. 62(7), 2319. https://doi.org/10.1093/jxb/erq403 (2011).
Kotur, Z. et al. Nitrate transport capacity of the Arabidopsis thaliana NRT2 family members and their interactions with AtNAR2.1. New Phytol. 194(3), 724–731. https://doi.org/10.1111/j.1469-8137.2012.04094.x (2012).
Yan, M. et al. Rice OsNAR2.1 interacts with OsNRT2.1, OsNRT2.2 and OsNRT2.3a nitrate transporters to provide uptake over high and low concentration ranges. Plant, Cell Environ. 34(8), 1360–1372. https://doi.org/10.1111/j.1365-3040.2011.02335.x (2011).
Zifarelli, G. & Pusch, M. CLC transport proteins in plants. FEBS Lett. 584(10), 2122–2127. https://doi.org/10.1016/j.febslet.2009.12.042 (2009).
Harada, H., Kuromori, T., Hirayama, T., Shinozaki, K. & Leigh, R. H. Quantitative trait loci analysis of nitrate storage in Arabidopsis leading to an investigation of the contribution of the anion channel gene, AtCLC-C, to variation in nitrate levels. J. Exp. Bot. 55(405), 2005–2014. https://doi.org/10.1093/jxb/erh224 (2004).
Jossier, M. et al. The Arabidopsis vacuolar anion transporter, AtCLCc, is involved in the regulation of stomatal movements and contributes to salt tolerance. Plant J. 64(4), 563–576. https://doi.org/10.1111/j.1365-313x.2010.04352.x (2010).
Hedrich, R. & Geiger, D. Biology of SLAC 1-type anion channels – from nutrient uptake to stomatal closure. New Phytol. 216(1), 46–61. https://doi.org/10.1111/nph.14685 (2017).
Buchner, P. & Hawkesford, M. J. Complex phylogeny and gene expression patterns of members of the nitrate transporter 1/Peptide transporter family (NPF) in wheat. J. Exp. Bot. 65(19), 5697–5710. https://doi.org/10.1093/jxb/eru231 (2014).
Li, W. et al. A wheat transcription factor positively sets seed vigour by regulating the grain nitrate signal. New Phytol. 225(4), 1667–1680. https://doi.org/10.1111/nph.16234 (2019).
Taulemesse, F., Le Gouis, J., Gouache, D., Gibon, Y. & Allard, V. Post-flowering nitrate uptake in wheat is controlled by N status at flowering, with a putative major role of root nitrate transporter NRT21. PLoS ONE 10(3), e0120291. https://doi.org/10.1371/journal.pone.0120291 (2015).
Guo, T. et al. Transcription analysis of genes encoding the wheat root transporter NRT1 and NRT2 families during nitrogen starvation. J. Plant Growth Regul. 33(4), 837–848. https://doi.org/10.1007/s00344-014-9435-z (2014).
Wang, H. et al. Phylogeny and gene expression of the complete nitrate transporter 1/Peptide transporter family in Triticum aestivum. J. Exp. Bot. 71(15), 4531–4546. https://doi.org/10.1093/jxb/eraa210 (2020).
Wang, M. et al. TaANR1-TaBG1 and TaWABI5-TaNRT2s/NARs link ABA metabolism and nitrate acquisition in wheat roots. Plant Physiol. 182(3), 1440–1453. https://doi.org/10.1104/pp.19.01482 (2020).
Wang, W., Hu, B., Li, A. & Chu, C. NRT1.1s in plants: Functions beyond nitrate transport. J. Exp. Bot. 71(15), 4373–4379 (2020).
Yin, L., Li, P., Wen, B., Taylor, D. & Berry, J. O. Characterization and expression of a high-affinity nitrate system transporter gene (TaNRT2.1) from wheat roots, and its evolutionary relationship to other NRT2 genes. Plant Sci. 172(3), 621–631. https://doi.org/10.1016/j.plantsci.2006.11.014 (2007).
Bajgain, P., Russell, B. & Mohammadi, M. Phylogenetic analyses and in-seedling expression of ammonium and nitrate transporters in wheat. Sci. Rep. https://doi.org/10.1038/s41598-018-25430-8 (2018).
Hu, B. et al. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 31(8), 1296–1297. https://doi.org/10.1093/bioinformatics/btu817 (2014).
Schilling, S., Kennedy, A., Pan, S., Jermiin, L. S. & Melzer, R. Genome-wide analysis of MIKC -type MADS -box genes in wheat: Pervasive duplications, functional conservation and putative neofunctionalization. New Phytol. 225(1), 511–529. https://doi.org/10.1111/nph.16122 (2020).
Kelley, L. A., Mezulis, S., Yates, C. M., Wass, M. N. & Sternberg, M. J. The Phyre2 web Portal for protein modeling, prediction and analysis. Nat. Protoc. 10(6), 845–858. https://doi.org/10.1038/nprot.2015.053 (2015).
Ramírez-González, R. H. et al. The transcriptional landscape of polyploid wheat. Science 361(6403), eaar6089. https://doi.org/10.1126/science.aar6089 (2018).
Sandhu, N., Kaur, A., Sethi, M., Kaur, S., Singh, V., Sharma, A., Bentley, A. R., Barsby, T., & Chhuneja, P. Genetic dissection uncovers genome wide marker trait associations for plant growth, yield and yield related traits under varying nitrogen levels in nested synthetic wheat introgression libraries. Front. Plant Sci. https://doi.org/10.21203/rs.3.rs-582649/v1(2021).
Wolfe, D., Dudek, S., Ritchie, M. D. & Pendergrass, S. A. Visualizing genomic information across chromosomes with PhenoGram. BioData Min. https://doi.org/10.1186/1756-0381-6-18 (2013).
Kumar, S., Stecher, G., Li, M., Knyaz, C. & Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35(6), 1547–1549. https://doi.org/10.1093/molbev/msy096 (2018).
Yu, Y., Ouyang, Y. & Yao, W. ShinyCircos: An R/Shiny application for interactive creation of Circos plot. Bioinformatics 34(7), 1229–1231. https://doi.org/10.1093/bioinformatics/btx763 (2017).
Ma, S. et al. WheatOmics: A platform combining multiple omics data to accelerate functional genomics studies in wheat. Mol. Plant 14(12), 1965–1968. https://doi.org/10.1016/j.molp.2021.10.006 (2021).
Ma, S. et al. WheatOmics: A platform combining multiple omics data to accelerate functional genomics studies in wheat. Mol. Plant 14, 1965–1968 (2021).
Zhang, A. et al. S-type anion channels SLAC1 and SLAH3 function as essential negative regulators of inward K+ channels and stomatal opening in Arabidopsis. Plant Cell 28(4), 949–965. https://doi.org/10.1105/tpc.15.01050 (2016).
Yan, H. et al. Structure and mechanism of a nitrate transporter. Cell Rep. 3(3), 716–723. https://doi.org/10.1016/j.celrep.2013.03.007 (2013).
Saier, M. H., Reddy, V. S., Tamang, D. G. & Västermark, Å. The transporter classification database. Nucl. Acids Res. 42(D1), D251–D258. https://doi.org/10.1093/nar/gkt1097 (2013).
Perland, E., Bagchi, S., Klaesson, A. & Fredriksson, R. Characteristics of 29 novel atypical solute carriers of major facilitator superfamily type: Evolutionary conservation, predicted structure and neuronal Co-expression. Open Biol. 7(9), 170142. https://doi.org/10.1098/rsob.170142 (2017).
Reddy, V. S., Shlykov, M. A., Castillo, R., Sun, E. I. & Saier, M. H. The major facilitator superfamily (MFS) revisited. FEBS J. 279(11), 2022–2035. https://doi.org/10.1111/j.1742-4658.2012.08588.x (2012).
Allen, S., Guo, m., Loussaert, D., Rupe, M., & Wang, H. (2016). Enhanced nitrate uptake and nitrate translocation by over-expressing maize functional low-affinity nitrate transporters in transgenic maize. US Patent 20160010101 A1.
Chen, J. et al. Agronomic nitrogen-use efficiency of rice can be increased by driving OsNRT2.1 expression with the OsNAR2.1 promoter. Plant Biotechnol. J. 14(8), 1705–1715. https://doi.org/10.1111/pbi.12531 (2016).
Fan, X. et al. Overexpression of a pH-sensitive nitrate transporter in rice increases crop yields. Proc. Natl. Acad. Sci. 113(26), 7118–7123. https://doi.org/10.1073/pnas.1525184113 (2016).
Wang, W. et al. Expression of the nitrate transporter gene OsNRT1.1A/OsNPF6.3 confers high yield and early maturation in rice. Plant Cell 30(3), 638–651. https://doi.org/10.1105/tpc.17.00809 (2018).
Fu, Y., Yi, H., Bao, J. & Gong, J. LeNRT2.3 functions in nitrate acquisition and long-distance transport in tomato. FEBS Lett. 589(10), 1072–1079. https://doi.org/10.1016/j.febslet.2015.03.016 (2015).
Luo, B. et al. Overexpression of a high-affinity nitrate transporter OsNRT2.1 increases yield and manganese accumulation in rice under alternating wet and dry condition. Front. Plant Sci. https://doi.org/10.3389/fpls.2018.01192 (2018).
Acknowledgements
We are thankful to IWGSC and Wheat-OMICS database for the data availability. We are thankful to the Department of Biotechnology, Govt. of India for providing DBT RA fellowship.
Funding
The work was compiled under projects funded by the Department of Biotechnology, Govt. of India (Grant No. BT/IN/UK-VNC/42/RG/2015–16 and BT/PR30871/BIC/101/1159/2018).
Author information
Authors and Affiliations
Contributions
N.S. and A.K. designed this study; A.K. conducted the in-silico studies and drafted the manuscript; N.S. conducted the field and genome wide association mapping studies and contributed to manuscript draft; P.K. helped in the in-silico analysis; G.P. and J.S. helped in visualisation; N.S., S.K. and P.C. provided resources and contributed to the critical revision of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Kumar, A., Sandhu, N., Kumar, P. et al. Genome-wide identification and in silico analysis of NPF, NRT2, CLC and SLAC1/SLAH nitrate transporters in hexaploid wheat (Triticum aestivum). Sci Rep 12, 11227 (2022). https://doi.org/10.1038/s41598-022-15202-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-022-15202-w
- Springer Nature Limited
This article is cited by
-
Overexpression of rice OsNRT1.1A/OsNPF6.3 enhanced the nitrogen use efficiency of wheat under low nitrogen conditions
Planta (2024)
-
Genome-wide systematic characterization of the NRT2 gene family and its expression profile in wheat (Triticum aestivum L.) during plant growth and in response to nitrate deficiency
BMC Plant Biology (2023)
-
Comprehensive in silico analysis of the underutilized crop tef (Eragrostis tef (Zucc.) Trotter) genome reveals drought tolerance signatures
BMC Plant Biology (2023)