Abstract
Bladder cancer has a high recurrence rate and low survival of advanced stage patients. Few genetic drivers of bladder cancer have thus far been identified. We performed in-depth structural variant analysis on whole-genome sequencing data of 206 metastasized urinary tract cancers. In ~ 10% of the patients, we identified recurrent in-frame deletions of exons 8 and 9 in the aryl hydrocarbon receptor gene (AHRΔe8-9), which codes for a ligand-activated transcription factor. Pan-cancer analyses show that AHRΔe8-9 is highly specific to urinary tract cancer and mutually exclusive with other bladder cancer drivers. The ligand-binding domain of the AHRΔe8-9 protein is disrupted and we show that this results in ligand-independent AHR-pathway activation. In bladder organoids, AHRΔe8-9 induces a transformed phenotype that is characterized by upregulation of AHR target genes, downregulation of differentiation markers and upregulation of genes associated with stemness and urothelial cancer. Furthermore, AHRΔe8-9 expression results in anchorage independent growth of bladder organoids, indicating tumorigenic potential. DNA-binding deficient AHRΔe8-9 fails to induce transformation, suggesting a role for AHR target genes in the acquisition of the oncogenic phenotype. In conclusion, we show that AHRΔe8-9 is a novel driver of urinary tract cancer and that the AHR pathway could be an interesting therapeutic target.
Similar content being viewed by others
Introduction
Bladder cancer is the fourth most common cancer in men with a 5-year survival rate of about 77%1. Despite risk stratification, the recurrence rate of bladder cancer is high and requires ongoing monitoring and treatment, which makes it also the costliest malignancy out of all cancers2. With recent advances, the therapeutic options have been expanded to include immune checkpoint inhibitors and antibody–drug conjugates, while clinical studies for targeted drug approaches are ongoing3,4. However, treatment and overall survival of cases with advanced stages of bladder cancer have not really improved in the last decades as compared to other cancers and there remains a need for the identification of drivers of bladder cancer tumorigenesis to effectively target early-stage disease and reduce tumor progression5,6.
Genomic analyses of bladder cancer of various stages and grades have shown that bladder cancer is a heterogeneous disease of which the high mutation burden is largely driven by activation of APOBEC mutagenesis7,8,9. Molecular characterization revealed activating FGFR3 alterations in ~ 16% of patients, with a higher prevalence in non-muscle invasive bladder cancer9,10. Furthermore, positive selection of mutated genes in the RTK-Ras-PI3K pathway (such as FGFR3, PIK3CA, and ERBB2), the p53-Rb pathway (such as TP53, RB1, and ATM), and genes involved in chromatin remodeling (KDM6A and KMT2D) has been identified in biopsies of patients with bladder cancer7,9.
A recent study identified a rare non-synonymous mutation (Q383H) in AHR as an APOBEC-associated hotspot mutation in bladder cancer, suggesting AHR as a potential driver of bladder cancer11. AHR is a ligand activated transcription factor involved in the cellular response to toxic aromatic hydrocarbons as well as cell type and context-specific physiological functions12. Increased AHR expression and activity have been associated with poor prognosis and survival for multiple cancers such as breast, lung, and upper urinary tract cancers13. However, the relevance and functional consequences of cancer-associated AHR mutations in bladder cancer are still poorly understood.
Here, we describe the identification of novel recurrent exon-deleting AHR alterations in the pan-cancer whole-genome sequencing database of metastatic cancers from Hartwig Medical Foundation. Using state-of-the-art data analysis tools with improved functionality for detecting structural variants we identified a previously unnoticed recurrent in-frame deletion of exons 8 and 9 in AHR. Together with the Q383H point mutation and AHR gene amplifications, AHR variants comprise ~ 19% of patients with urinary tract cancer. We demonstrate that the recurrent alterations lead to constitutively activated AHR signaling and induce an oncogenic phenotype in bladder cells. Our results suggest that aberrant AHR signaling is an important driver of urothelial tumorigenesis.
Results
AHR is frequently mutated in urinary tract cancers
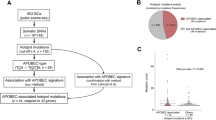
To identify genetic factors involved in urinary tract cancers, we analyzed the Hartwig database, which represents whole-genome sequencing data of solid metastatic tumors and normal tissue of 4500 patients14. Utilizing improved structural variant detection algorithms15,16, we detected a novel deletion spanning exons 8 and 9 of the AHR gene (AHRΔe8-9) in 22 (10.7%) out of the 206 urinary tract samples (Fig. 1a,b). The AHRΔe8-9 is identified in several urinary tract cancer subtypes, most of which are in bladder cancer (12/22) and pyelum (7/22) (Supplementary Table 1). The AHRΔe8-9 mutant is highly specific to urinary tract cancer, because in the complete pan-cancer data set of 4500 patients it was identified in only three non-urinary tract samples (2 × lung, 1 × skin; Supplementary Table 2). The identified exonic deletion was validated in RNA-sequencing, which was available for 129 out of 206 urinary tract samples. Read support for the AHR exon 7–10 splice junction was identified in all urinary tract samples harboring AHRΔe8-9 (n = 16; Supplementary Table 1) and confirmed the in-frame loss of exons 8 and 9 for six samples in which the genomic start position of the deletion is located within the coding sequence of exon 8.
AHR alterations in somatic metastatic urinary tract patients. (a) Schematic representation of AHR gene (ENST00000242057.4—GRCh37) with a magnification of the genomic region around exon 7–9 (Chr7:17372389–17378532). The point mutation Q383H (c.1149G>C) is marked with an asterisk. The black lines in the lower part of the figure represent the extent of the genomic deletions identified in the urinary tract cancer patients. Most of the right-hand breakpoints are localized to an AluYa5 element, which is part of an inverted repeat (indicated with orange boxes) that is also composed of an AluY element. See Supplementary Table 2 for genomic positions of the deletions. (b) Pie chart of the number of detected AHR alterations: AHR amplification (Amp), AHRΔe8-9, AHRQ383H or non-affected samples in the urinary tract patient cohort of the Hartwig database. (c) Schematic illustration of the AHR functional domains and the positions of AHRQ383H and AHRΔe8-9 (indicated in dark blue). The deletion (AHRΔe8-9) disrupts the PAS-B domain, ligand-binding domain (LBD), and XAP2 and HSP90 protein interaction domains. (d) Visualization of top 12 affected genes per mutation type in urinary tract cancer samples of the Hartwig database14. Gene names are colored for oncogenes (red) and tumor suppressor genes (blue). (e) Boxplot of the adjusted TPM values for AHR transcript. Asterisks indicate two samples that contain the deletion (AHRΔe8-9) but also have an amplification of the AHR gene. (f) Boxplot of the adjusted TPM values of AHR target genes.
The relatively small size of the AHR deletion (~ 3 kb) may explain why it has not been identified in previous studies9,17. Most of the 3’ breakpoint junctions are positioned in a narrow window of 30 bp between 2 Alu elements (AluYa5 and AluY) and overlap with the 3’ site of the AluYa5 element (Fig. 1a). Such inverted Alu repeats have been identified as strong initiators of genetic instability18 and we hypothesize that the inverted Alu repeats in the AHR gene facilitate the deletion of exons 8 and 9. The high prevalence in urinary tract cancer of such a rare event strongly suggests positive selection. Analysis of an independent RNA-sequencing dataset of urinary tract cancers from Weill Cornell Medicine confirmed the presence of the 7–10 splice junction in 8% (2/24) of samples. One of the bladder cancer samples with the detected splice junction was of primary origin (out of 8 primary tumors in this dataset). Together, these results demonstrate that the AHRΔe8-9 is present in primary and metastatic urinary tract cancer (Supplementary Table 3).
To identify additional events in the AHR gene we performed a targeted analysis of the Hartwig database. We identified recurrent gene amplifications and a recurrent point mutation, in 16 (7.8%) and 2 (1.0%) of the urinary tract cancer patients, respectively (Fig. 1a,b). The c.1149G>C (AHRQ383H) point mutation was previously reported as an APOBEC-associated hotspot mutation in bladder cancer based on the TCGA PanCancer Atlas data11. The AHRQ383H point mutation was found in 11 patients out of ~ 11,000 patients in the TCGA PanCancer Atlas, which are mainly of primary tumor origin19,20. The majority (n = 8) occurred in bladder urothelial carcinomas with the other three in hepatocellular carcinoma, papillary renal cell carcinoma, and lung adenocarcinoma. Thus, like the AHRΔe8-9 variant, the AHRQ383H mutation is highly specific for urinary tract cancer.
AHR is a ligand-activated transcription factor that contains a basic Helix-Loop-Helix/PER-ARNT-SIM (bHLH/PAS) motif. AHR contains two PAS domains of which the PAS-B domain contains the ligand binding domain (LBD) (Fig. 1c)12,21. AHR is part of a cytoplasmic protein complex containing HSP90, p23, and XAP2. Upon ligand binding, AHR dissociates from the complex and translocates to the nucleus where it forms a heterodimer with ARNT22. The AHR/ARNT complex subsequently binds to Xenobiotic Response Elements (XREs) in the genome to activate the transcription of target genes such as those coding for the phase I and II drug metabolizing enzymes CYP1A1 and CYP1B112. At the protein level, the AHRΔe8-9 deletion results in an in-frame protein coding sequence with the loss of 84 amino acids (p. 303–387), disrupting the PAS-B domain and the C-terminal part of the ligand-binding domain of the protein. The HSP90 and XAP2 protein binding domains are predicted to be affected, while the dimerization region of the protein likely remains intact (Fig. 1c).
Among the urinary tract cancer patients in the Hartwig database, AHR alterations are mutually exclusive with FGFR3 alterations and PIK3CA mutations, although the significance is impacted by low sample numbers (FGFR3: Odds ratio = 0.24 p = 0.21, PIK3CA: Odds ratio = 0.30 p = 0.32). The tendency of mutual exclusivity with genes from the RTK-Ras-PI3K pathway strengthens the idea that AHR is an independent driver of urinary tract cancer, with potential convergence on the RTK-Ras-PI3K pathway. AHR alterations do not show mutual exclusivity with genes involved in chromatin remodeling or the tumor suppressors TP53 and Rb1 (Fig. 1d, Supplementary Table 4).
For tumors with AHR amplifications, AHR expression is increased compared to the non-amplified urinary tract cancer samples (Fig. 1e). Urinary tract cancers with AHRΔe8-9 showed increased expression of AHR target genes, which was not observed in the AHR amplified or AHR non-affected samples (Fig. 1f). No RNA sequencing data was available for the 2 patients with the AHRQ383H mutation.
The hAHR Q383H mutation affects ligand binding affinity and specificity
The mouse ortholog of AHRQ383H (AhrQ377) can form hydrogen bonds with Ahr ligands in the ligand-binding domain and mutations that change this residue affect ligand binding affinities23,24,25. To examine if ligand binding affinities are also affected for the human AHR (hAHR), we measured the transcriptional activity of hAHRQ383H and hAHRWT in an XRE-luciferase reporter assay. Both the hAHRWT and hAHRQ383H expressing cells showed strong transcriptional activity with AHR ligands TCDD and B[a]P (Fig. 2a). Stimulation with B[a]P resulted in a higher induction of luciferase transcription for the hAHRQ383H mutant than for the hAHRWT, suggesting a difference in ligand affinity between both variants. The AHR antagonist CH-223191 reduced transcriptional activation in hAHRWT cells stimulated with TCDD or B[a]P (Fig. 2a)26,27. Surprisingly, incubation of hAHRQ383H with CH-223191 resulted in transcriptional activation and no antagonism was observed when CH-223191 was combined with TCDD or B[a]P (Fig. 2a). To examine this altered ligand-binding specificity, we analyzed protein localization in RPE1 cells, which is a genetically stable near diploid cell line well-suited for microscopy. In the absence of exogenous ligands, both hAHRQ383H and hAHRWT proteins are mainly localized to the cytoplasm and both proteins show nuclear translocation upon incubation with TCDD or B[a]P (Fig. 2b, Supplementary Fig. 1). Incubation with the antagonist CH-223191 also resulted in nuclear translocation of hAHRQ383H, while the hAHRWT remains localized to the cytoplasm (Fig. 2b). The difference in responses between the hAHRQ383H and hAHRWT to AHR agonists and antagonists demonstrate differential sensitivity to AHR pathway modulation.
Altered protein localization and transcriptional activation of hAHR mutants. (a) Luciferase reporter assay of HEK293T cells transiently transfected with empty control, hAHRWT, hAHRQ383H, or hAHRΔe8-9 vectors together with XRE-luciferase reporter construct. Assays were performed with different AHR ligands (TCDD and B[a]P) and AHR antagonist (CH-223191) conditions. Significance was calculated with Student’s T-test for stimulated versus untreated condition per construct. *p < 0.05, **p < 0.01. RLU = relative luciferase unit. (b) Immunofluorescence of RPE1 cells that constitutively express hAHRWT, hAHRQ383H, or hAHRΔe8-9 were incubated for 4 h with TCDD or CH-223191. AHR was detected with immunofluorescence (red) and DNA with Dapi (blue). Scale bar = 50 μm. The boxplots present the ratio of nuclear to cytoplasmic signal of AHR expression for all individual captured cells. Transgene free cells served as negative controls (left panels). Lowercase letters indicate significant differences between treatments (p < 0.05, Anova). (c) Luciferase reporter assay of HEK293T cells transiently transfected with increasing concentration of hAHRWT, hAHRΔe8-9, and empty control vectors (n = 4 per data point).
The hAHR Δe8-9 mutant leads to constitutive AHR pathway activation
To study the functional consequences of AHRΔe8-9, we also created transgenic hAHRΔe8-9 RPE1 cells. In contrast with hAHRWT, the hAHRΔe8-9 protein is localized in the nucleus regardless of the presence or absence of a ligand (Fig. 2b, Supplementary Fig. 1). In XRE-luciferase reporter assays, the degree of transcriptional activation by hAHRΔe8-9 was similar for all the conditions, irrespective of the presence of AHR ligands. Moreover, the AHR antagonist CH-223191 did not affect the transcriptional activation activity of hAHRΔe8-9 (Fig. 2a). Increased concentrations of hAHRWT and hAHRΔe8-9 constructs in the transfections resulting in increased activation of the luciferase reporter and revealed that in untreated conditions hAHRΔe8-9 induced higher transcriptional activation of the Luciferase reporter than hAHRWT (Fig. 2c).
To further characterize the consequences of hAHRΔe8-9 on a disease-relevant cell type, we created transgenic mouse bladder organoids expressing hAHRWT, hAHRΔe8-9 and a DNA binding deficient variant of hAHRΔe8-9 (hAHRΔe8-9 DBD). We expected similar protein expression levels between the cell lines, because these were produced in an identical manner. However, we observed weak protein expression of hAHRΔe8-9, but robust expression for the wildtype and the DNA binding deficient variant (Fig. 3a). These results suggest increased degradation of the DNA binding proficient hAHRe8-9 protein, possibly the result of negative feedback loops that depend on the DNA binding of AHR.
Transcriptome analysis of constitutively activated hAHRΔe8-9 mutant. (a) Western blot results for transgenic mouse cell lines, expressing Cas9 (negative control), hAHR WT, hAHRΔe8-9, or a DNA-binding deficient version of hAHRΔe8-9 (hAHRΔe8-9 DBD). (b) Rlog values of the 172 most differentially expressed genes between the hAHRWT and hAHRΔe8-9 transgenic mouse bladder organoids in untreated and TCDD treated conditions. (c) Boxplot of normalized counts of RNA reads of AHR target genes for the untreated mouse bladder organoids expressing hAHR mutants (n = 4). (d) Images of Dapi stained mouse bladder organoids constitutively expressing hAHR mutants. The lower panel shows the magnification of the boxed area. Scale bar 250 µm. (e) GO enrichment analysis of hAHRΔe8-9 mutant for the cellular component domain. (f) Rlog values of basal and luminal cell layer makers for hAHRΔe8-9 and hAHRWT transgenic mouse bladder organoids.
The hAHRWT and hAHRΔe8-9 bladder organoid lines were exposed to TCDD and together with untreated lines subjected to bulk RNA sequencing. Untreated hAHRΔe8-9 organoids differentially expressed several genes compared to the hAHRWT organoids, including the AHR target genes Cyp1a1, Cyp1b1, Ahrr, Gsta1, and Tiparp demonstrating constitutive activation of the AHR pathway (Fig. 3b,c). The expression pattern of up and down regulated genes is similar for hAHRΔe8-9 in untreated and TCDD treated conditions, confirming that loss of exons 8 and 9 leads to ligand-independent activation of the AHR pathway, which is in line with the upregulation of these genes in AHRΔe8-9 positive urinary tract cancers (Fig. 1f). Thus, AHRΔe8-9 leads to constitutive activation of the AHR pathway.
The hAHRWT organoids treated with TCDD also show upregulation of the canonical AHR target genes, but do not show the same downregulated genes (Fig. 3b). This observation indicates a different effect on the transcriptome between the constitutively active hAHRΔe8-9 and the 24 h TCDD stimulated hAHRWT condition. Moreover, overexpression of hAHRWT and hAHRQ383H without the addition of exogenous ligands already results in modest pathway activation when compared with control organoids that are not transgenic for hAHR (Fig. 3c). These observations are in line with the higher induction of luciferase transcription in the untreated condition for the hAHRWT and hAHRQ383H constructs compared to the control in the luciferase assay (Fig. 2a).
The hAHR Δe8-9 mutant induces cellular transformation
The bladder is a stratified epithelium, with stem cells that reside in the basal cell layer that support organ regeneration and renewal. Upon differentiation, the stem cells give rise to intermediate cells and luminal umbrella cells28. Mouse bladder organoids enriched for undifferentiated cells have a reduced diameter of the lumen and increased thickness of the epithelial layer when compared to organoids composed of differentiated cells29. We observed a mixture of cystic and compact organoids in the mouse bladder organoids expressing hAHRWT, indicating a heterogeneous population of differentiated and undifferentiated cells. No cystic organoids were observed in the organoids expressing hAHRΔe8-9 indicating a more basal stem-cell like phenotype (Fig. 3d).
We performed GO enrichment analysis on all differentially expressed genes in RNA-sequencing data of the mouse organoids to understand which processes are affected by hAHRΔe8-9. Most outstanding is the downregulation of genes related to the extracellular matrix and cell periphery organization (Fig. 3e, Supplementary Table 5). Moreover, hAHRΔe8-9 expressing mouse organoids show downregulation of the differentiation markers Upk3a, Upk1a, and Krt18 and upregulation of the stem cell markers Wnt5a, and Krt1 when compared to hAHRWT organoids (Fig. 3f)29,30,31. Together, these observations indicate that hAHRΔe8-9 promotes a basal stem-cell like phenotype in bladder cells.
Because RNA-seq was performed on bulk cultures, the transcriptional changes induced by hAHRΔe8-9 may reflect a shift in the composition of the cell types towards a more homogeneous population, or the transformation of cells towards a novel phenotype. To discriminate between these scenarios, we performed scRNA-seq on the mouse bladder organoid lines. Dimensional reduction and unsupervised clustering revealed the presence of 7 clusters (Fig. 4a). Based on the genes enriched in the different clusters, cluster 0 represents a basal phenotype (characterized by Krt14, Trp63, Bcam and Agrn), cluster 1 represents an intermediate/luminal phenotype (characterized by Krt19, Krt18, Upk1b, Cldn4, Cldn7, Ceacam1 and Alcam), and clusters 3 and 5 represent cells that are in the S-phase and M-phase of the cell cycle, respectively (Fig. 4a,b, Supplementary Table 5). Together, these clusters represent a classical stem cell system where stem cells divide to give rise to new stem cells or to cells that differentiate. The vast majority of all cells of the control lines fall into these clusters and in cluster 4, which we were not able to link to a particular cellular phenotype. Strikingly, two clusters (clusters 2 and 6) were almost exclusively occupied by mouse bladder cells expressing hAHRΔe8-9 (Fig. 4a,c). In addition to the canonical AHR target genes such as Cyp1b1 and Tiparp, cluster 2 expresses basal stem cell markers such as Krt17, Wnt5a, Itga6, Wnt429. In addition, cluster 2 is characterized by genes that are associated with urothelial cancer, such as: Htra132, Cyb5r133, Steap133,34, Ptgs235,36, and Trib337 (Fig. 4b). Cells in cluster 6 show upregulation of Dsp, Pkp1, Ppl, Jup and Krt13, which are involved in desmosome and intermediate filament cytoskeleton organization. This cluster has some overlap with markers expressed in the luminal-intermediate cluster (cluster 1) of the control cells, but with less apparent expression of the umbrella cell markers. This indicates that the cells in cluster 6 represent an intermediate cell type that however fails to differentiate towards luminal umbrella cells. Thus, the constitutive activation of the AHR pathway by hAHRΔe8-9 leads to a transformation of cells towards a less differentiated phenotype and the activation of genes linked to urothelial cancer.
Transformed phenotype of hAHRΔe8-9 mutant. (a) tSNE plot of single cell transcriptomics depicting clusters of the transgenic mouse bladder organoids cells. Plot is coloured for detected clusters and for the different transgenic mouse bladder organoid lines. (b) Heatmap depicting expression of selected markers, which are identified by expression analysis, representative for the different identified clusters. (c) Contribution of percentage of cells to the different clusters for each transgenic mouse bladder organoid line. (d) Soft agar assay of mouse bladder organoids expressing hAHR mutants. Results are presented as the mean ± SD of triplicate samples.
Cells of a DNA binding deficient variant of the oncogenic hAHRΔe8-9 (hAHRΔe8-9DBD) clustered among cells of the other control lines and apart from hAHRΔe8-9 cells (Fig. 4a). This shows that the cellular transformation of the hAHRΔe8-9 organoids depends on the transcriptional activation caused by AHR and is not the result of other transcription-independent effects caused by the deletion. To functionally explore if the transformed phenotype is accompanied by oncogenic properties, a soft agar growth assay was performed, a technique to measure cellular transformation in vitro. A higher number of colonies were counted for hAHRΔe8-9 organoids compared to hAHRWT organoids or control organoids demonstrating that hAHRΔe8-9 confers anchorage-independent growth to bladder cells (Fig. 4d).
Together, these observations demonstrate that transcriptional changes driven by the constitutively active hAHRΔe8-9 lead to a transformation of cells towards a cancerous phenotype that fails to differentiate and is able to grow independent of anchorage to the extracellular matrix.
Discussion
In this study, we show high prevalence of AHR alterations in urinary tract cancers and provide functional validations to support that these aberrations are oncogenic and drive urinary tract cancer. These findings are in line with mouse studies that have shown that AHR overexpression or pathway activation can lead to malignant transformation of epithelial cells38,39,40. A variety of molecular signaling pathways have been previously linked to AHR-mediated tumorigenesis whether or not in a cancer type-specific background and/or driven by AHR agonists13,41. This includes the biotransformation of hydrocarbons by CYP enzymes to mutagenic intermediates which can induce DNA adducts, cross-talk of AHR with other signaling pathways, and interaction of AHR with other binding partners than ARNT to promote transcription of non-canonical genes42,43,44,45,46,47. However, the underlying molecular signaling pathways driving the tumorigenesis of urinary tract cancers in patients with activating mutations in AHR remains unclear. The tendency of mutual exclusivity between AHR, FGFR3, and PIK3CA alterations may indicate convergence on the same pathway, although a fully independent parallel oncogenic pathway in bladder cancer driven by AHR cannot be excluded. The hAHRQ383H and hAHRΔe8-9 variants are almost uniquely detected in urothelial cancers, which suggests this tissue is particularly sensitive to deregulated AHR signaling. However, our results, nor information in literature, provides clues why AHR pathway activation is so specific for bladder cancer, so this will require further investigations.
We demonstrate that the hAHRQ383H mutation leads to increased ligand activation sensitivity which may lead to overactivity of the AHR pathway in the urinary tract, thereby driving tumorigenesis. This is mechanistically different from hAHRΔe8-9 which is constitutively active in a ligand-independent way. We presume that for hAHRΔe8-9 loss of the ligand binding domain leads to a conformational change that exposes a nuclear import signal that in the native protein is only exposed upon ligand binding dependent dissociation from the cytoplasmic complex. As for the hAHRQ383H mutation, increased sensitivity of the AHR pathway likely also holds true for AHR amplifications since overexpression of hAHRwt in different models shows a modest increase of AHR pathway activation compared to the controls. It is not known if the ligands that induce overactive AHR signaling in the tumors with AHR amplified and AHRQ383H backgrounds are of environmental origin (like components in tobacco smoke) or have an endogenous source (like metabolites)48. Surprisingly, we observed hAHRQ383H activation upon treatment with the AHR antagonist CH-223191. This contrasts with a recent study that demonstrated reduced viability upon treatment with CH-223191 of the bladder cancer cell line KMBC2 harboring the AHRQ383H mutation11. A possible explanation for the apparent discrepancy in results may lie in the different types of experiments that were conducted. The reduced viability of the KMBC2 cell line after CH-223191 incubation may also be independent of alterations in AHR pathway activity.
Patients that harbor AHR activating mutations could potentially benefit from AHR targeted therapies49. Here we show that the mode of action of AHR activation differs between the different mutations, which implies that tailored therapies depending on the underlying mutational event are required. Most classic AHR antagonists function by interference with the ligand-binding domain, but this domain is not targetable for the AHRΔe8-9 mutant as this domain is deleted. Therefore, functional screens to identify specific AHRΔe8-9 targeting compounds or the identification of essential downstream activated processes could be a next step towards the identification of novel treatment strategies for selected urinary tract cancer patients in the context of precision medicine.
Material and methods
Driver gene status urinary tract cohort
Variant detection and driver likelihood status are based on the Hartwig database14. Structural variation was detected and classified with the GRIDSS, PURPLE, LINX algorithms, using pairs of matched tumour/normal BAM files as input15,16. Driver likelihood is introduced to select for a sample specific likelihood of each gene based on the type of variant and taking mutation load per sample into account. To select for affected genes in the urinary tract cancer samples, the driver likelihood score is set to > 0.8 and detected gene fusions with a high impact are included. Visualized are the genes affected in more than 12.5% of the samples (top 12 genes). Mutually exclusivity is pairwise calculated with Fisher Exact Test based on Odd Ratio cut-offs as described in Gao et al.19.
Gene constructs
A plasmid containing the hAHRWT sequence was purchased from Origene (RC209832). The Q383H point mutation was introduced with site-directed mutagenesis with forward primer cattgtaactcacagaccactaacagatg and reverse primer gttagtggtctgtgagttacaatgatataatc. The hAHRWT, hAHRQ383H, and hAHRΔe8-9 sequences were cloned in a lentiviral plasmid (Addgene #52961) and subsequently to pcDNA3.1 vector with the In-fusion HD Eco-dry cloning (Takarabio). Primers for lentiviral plasmid are: forward N-flag caggaccggttctaggatatcgccaccatggattacaaagacgatgacgataagaacagcagcagcgccaac, forward caggaccggttctaggatatcgccaccatgaacagcagcagcgcc, reverse ttgttgcgccggatcgcaggaatccactggatgtcaaatcag, reverse C-flag ttgttgcgccggatcgcttatcgtcatcgtctttgtaatccaggaatccactggatgtcaaatcag, forward deletion tggttgtgatgccaaagatgaggaaggaacagagca and reverse deletion gttccttcctcatctttggcatcacaaccaatagg. Primers cloning of AHR sequences to pcDNA3.1 vector are forward taccgagctcggatcatatcgccaccatgaacag and reverse gatatctgcagaattttacaggaatccactggatgtcaaat. For the DNA binding deficient variant of AHRΔ8-9, mutations were introduced to substitute the amino acids Histidine 39 and Arginine 40 with Alanines50.
The luciferase reporter vector pGL3-XRE-Luc2 reporter vector was constructed using an in-house pGL3-minP-Luc2 vector containing DNA binding sequence “gggaggtacttggagcggc” with primers forward-1 gcgtgctagcccgggTTGCGTGACCctcgagatctgcgat, reverse-1 atcgcagatctcgagGGTCACGCAAcccgggctagcacgc, forward-2 aatcgataaggatccTTGCGTGACCgtcgaccgatgccct, reverse-2 agggcatcggtcgacGGTCACGCAAggatccttatcgatt.
Cell culture
All cells were cultured in a humidified atmosphere at 37 °C under 5%CO2 and 20%O2. Mouse bladder organoids were previously established and were a kind gift from dr Hans Clevers and dr Jasper Mullenders and were cultured as previously reported51. In short, mouse bladder organoids were plated in droplets of ice-cold Matrigel (Corning). After the matrigel had solidified, a complete mouse bladder medium was added consisting of Advanced DMEM/F-12 supplemented with HEPES, Glutamax, 1% penicillin-1% streptomycin (Pen-Strep), Nicotinamide (10 mM), N-Acetylcysteine (1.25 mM), FGF10 (100 ng/mL of Peprotech 100-26), FGF7 (25 ng/mL of Peprotech 100-19), A83-01 (500 nM), B27 (ThermoFisher 17504001) and primocin (Invivogen). Organoids were split weekly either through mechanical shearing with a pulled Pasteur’s pipet or by dissociation using TrypLE. To increase survival the medium was supplemented with ROCK inhibitor (Y-27632, 10 μM) after splitting.
RPE1 and HEK293T cells were cultured in DMEM, 10% fetal bovine serum, 1% penicillin, and 1% streptomycin. Cells were incubated with 10 nM TCDD (LGC), 10 µM B[a]P (sigma), or 10 µM CH-223191 (Sanbio) or vehicle alone (DMSO) for 24 h unless otherwise stated.
Lentivirus production and transduction
Lentivirus particles containing hAHR constructs were produced by transient calcium phosphate transfection of lentiviral transfer, packaging, and envelope plasmids into HEK293T cells. Virus particles were harvested 48 h posttransfection and concentrated with Lenti-X Concentrator (Takarabio) according to the manufacturer’s directions. Concentrated virus was resuspended in 250–500 µl Advanced DMEM/F-12 supplemented with HEPES, Glutamax, and 1% Pen-Strep. The day before transduction, RPE-1 cells were seeded in 6-well plates and mouse bladder organoids were seeded as single cells onto a layer of solidified matrigel in 96 well plates. Organoid culture medium was supplemented with 10 μM Y-27632 to enhance survival of single cells. The day after, RPE-1 cells and mouse bladder organoids were transduced with a dilution series ranging from 10–150 µl concentrated lentivirus. To enhance transduction efficiencies, the medium was supplemented with 8 µg/ml polybrene. After transduction, RPE1 cells and the mouse bladder organoids were placed on 10–15 µg/ml and 1 µg/ml puromycin selection, respectively. We continued with the transgenic lines that showed approximately 50% survival upon selection.
Immunofluorescence
RPE1 cells expressing hAHR mutants were plated on coverslips and incubated with TCDD, B[a]P, CH-223191 or untreated (DMSO) for 4 or 24 h. Cells were fixed in 4% formaldehyde, permeabilized with 0.3% Triton X-100 in PBS, and blocked with 4% BSA in PBS. AHR was stained with Anti-AHR antibody (Abcam ab190797) in blocking buffer. After incubation with the primary antibody, cells were washed followed by incubation with secondary antibodies conjugated with Alexa fluor dyes. Subsequently, the cells were washed, counterstained with DAPI, and mounted on microscope slides. Images were taken with predefined settings on Zeiss LSM510 microscope and analyzed with Fuij/imageJ. Ratios of AHR signal intensities were calculated by determining average grey value intensities for each nucleus, based on dapi staining, and for each corresponding cytoplasm, as 1 µm band around each nucleus.
Luciferase reporter gene assay
HEK293T cells were cultured in 24-wells plates and transiently transfected with pcDNA3.1 empty vector or pcDNA3.1 vector containing the expression cassette of hAHRWT, hAHRQ383H, or hAHRΔe8-9 (10 ng), together with the pGL3-XRE-Luc2 reporter vector (1000 ng) and pRL-CMV Renilla vector (2 ng). 24 h after transfection, cells were treated with ligand TCDD (50 nM) or B[a]P and/or antagonist CH223191 for approximately 24 h. Luciferase activity was measured with the Dual-Luciferase reporter assay system and normalized for Renilla activity (Promega, Madison, WI). The results are averages of at least three independent experiments assayed in duplicate ± SEM. Results are presented as change relative to the empty vector in the untreated (DMSO) condition.
RNA-sequencing
The RNA from the transgenic mouse bladder organoids was isolated with Trizol (ThermoFisher) according to the manufacturer’s instructions. RNA-seq libraries were prepared using TruSeq Stranded Total RNA Library Prep Kit (Illumina) according to the manufacturer’s protocol. RNA-seq libraries were pooled and sequenced on a NextSeq2000 (Illumina) in 1 × 50 bp single end mode. RNA sequencing reads were aligned against mouse reference genome GRCm38 using STAR and the number of reads mapping to genes was counted using featureCounts all by using a custom in-house pipeline (https://github.com/UMCUGenetics/RNASeq-NF). The Bioconductor package DESeq2 was used to normalize raw read counts and to perform differential gene expression analysis with apeglm shrinkage52,53. The analyses were performed with significant (P.adjust < 0.05) and differentially expressed (Log2FoldChange > 2.5) genes with exception of luminal and basal marker analyses where smaller differences were included (Log2FoldChange > 1.5). The Bioconductor package clusterProfiler and Revigo were used for GO enrichment analysis54,55.
scRNA-seq
The transgenic mouse bladder organoids were cultured in 2D and dissociated to single cells using a 10 min incubation with TryLE. Cells were sorted into 384-well capture plates (1 plate per condition) and the scRNA library preparation and sequencing were performed according to the SORT-seq protocol by Single Cell Discoveries B.V.56. For all single cells, reads were aligned to the mouse reference genome GRCm38 and Sort-seq read counts were filtered to exclude reads with identical library-, cell- and molecules56. With the Seurat R package, low quality cells were removed by a cut-off of 10,000 transcripts per cell and the data was normalized and scaled57. The top 2000 most variable genes in the dataset were identified and used for principal component analysis to determine dimensionality and clustering of the dataset. Cluster gene markers were detected using a Wilcoxon rank sum test between each cluster and the rest of the cells in the dataset with a bonferroni correction for multiple testing.
Soft agar assay
3% agarose (REF11388991001) was dissolved and autoclaved in 100 ml EBSS. One volume of melted 3% agarose was mixed with four volumes of Advanced DMEM/F12 to obtain a 0.6% solution. This mixture was added to 6-well plates (1.5 ml/well) in which the gels were allowed to solidify. Subsequently, Tryple was used to prepare single-cell suspensions of the mouse bladder organoids. The cells were counted and for each condition, 2.5 ml of cell suspension was prepared at a concentration of 1 × 10e4 cells/ml in complete medium. The cell suspension was mixed 1:1 with a warm 0.6% agarose solution to get 0.5 × 10e4 cells/ml in 0.3% agarose solution. Per well 1.5 ml agarose/cell mixture was plated. The next day 300 µl of the medium was added and the cells were refed every 2–4 days. After 3 weeks the cells were stained with nitroblue tetrazolium chloride solution and pictures were made. Colonies were counted using ImageJ.
Data availability
The bulk RNA-sequencing and the scRNA-seq data of the mouse bladder organoids have been deposited in ENA with the accession code PRJEB49233.
References
American Cancer Society. Cancer Facts and Figures 2020 (American Cancer Society, 2020).
James, A. C. & Gore, J. L. The costs of non-muscle invasive bladder cancer. Urol. Clin. N. Am. 40, 261–269 (2013).
Lenis, A. T., Lec, P. M., Chamie, K. & Mshs, M. D. Bladder cancer: A review. JAMA 324, 1980–1991 (2020).
Loriot, Y. et al. Erdafitinib in locally advanced or metastatic urothelial carcinoma. N. Engl. J. Med. 381, 338–348 (2019).
Berdik, C. Unlocking bladder cancer. Nature 551, S34–S35 (2017).
Abdollah, F. et al. Incidence, survival and mortality rates of stage-specific bladder cancer in United States: A trend analysis. Cancer Epidemiol. 37, 219–225 (2013).
Lawson, A. R. J. et al. Extensive heterogeneity in somatic mutation and selection in the human bladder. Science 370, 75–82 (2020).
Nakauma-González, J. A. et al. Molecular characterization reveals genomic and transcriptomic subtypes of metastatic urothelial carcinoma. BioRxiv 2021.03.17.435757 (2021).
Robertson, A. G. et al. Comprehensive molecular characterization of muscle-invasive bladder cancer. Cell 174, 1033 (2018).
Cancer Genome Atlas Research Network. Comprehensive molecular characterization of urothelial bladder carcinoma. Nature 507, 315–322 (2014).
Shi, M.-J. et al. Identification of new driver and passenger mutations within APOBEC-induced hotspot mutations in bladder cancer. Genome Med. 12, 85 (2020).
Denison, M. S., Soshilov, A. A., He, G., DeGroot, D. E. & Zhao, B. Exactly the same but different: Promiscuity and diversity in the molecular mechanisms of action of the aryl hydrocarbon (dioxin) receptor. Toxicol. Sci. 124, 1–22 (2011).
Wang, Z. et al. How the AHR became important in cancer: The role of chronically active AHR in cancer aggression. Int. J. Mol. Sci. 22, 387. https://doi.org/10.3390/ijms22010387 (2020).
Priestley, P. et al. Pan-cancer whole-genome analyses of metastatic solid tumours. Nature 575, 210–216 (2019).
Shale, C. et al. Unscrambling cancer genomes via integrated analysis of structural variation and copy number. BioRxiv 2020.12.03.410860 (2020).
Cameron, D. L. et al. GRIDSS2: Comprehensive characterisation of somatic structural variation using single breakend variants and structural variant phasing. Genome Biol. 22, 202 (2021).
ICGC/TCGA Pan-Cancer Analysis of Whole Genomes Consortium. Pan-cancer analysis of whole genomes. Nature 578, 82–93 (2020).
Lobachev, K. S. et al. Inverted Alu repeats unstable in yeast are excluded from the human genome. EMBO J. 19, 3822–3830 (2000).
Gao, J. et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 6, l1 (2013).
Cerami, E. et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2, 401–404 (2012).
Larigot, L., Juricek, L., Dairou, J. & Coumoul, X. AhR signaling pathways and regulatory functions. Biochim. Open 7, 1–9 (2018).
Seok, S.-H. et al. Structural hierarchy controlling dimerization and target DNA recognition in the AHR transcriptional complex. Proc. Natl. Acad. Sci. U. S. A. 114, 5431–5436 (2017).
Xing, Y. et al. Identification of the Ah-receptor structural determinants for ligand preferences. Toxicol. Sci. 129, 86–97 (2012).
Pandini, A., Denison, M. S., Song, Y., Soshilov, A. A. & Bonati, L. Structural and functional characterization of the aryl hydrocarbon receptor ligand binding domain by homology modeling and mutational analysis. Biochemistry 46, 696–708 (2007).
Bisson, W. H. et al. Modeling of the aryl hydrocarbon receptor (AhR) ligand binding domain and its utility in virtual ligand screening to predict new AhR ligands. J. Med. Chem. 52, 5635–5641 (2009).
Kim, S.-H. et al. Novel compound 2-methyl-2H-pyrazole-3-carboxylic acid (2-methyl-4-o-tolylazo-phenyl)-amide (CH-223191) prevents 2,3,7,8-TCDD-induced toxicity by antagonizing the aryl hydrocarbon receptor. Mol. Pharmacol. 69, 1871–1878 (2006).
Zhao, B., DeGroot, D. E., Hayashi, A., He, G. & Denison, M. S. CH223191 Is a ligand-selective antagonist of the Ah (dioxin) receptor. Toxicol. Sci. 117, 393–403 (2010).
Yamany, T., Van Batavia, J. & Mendelsohn, C. Formation and regeneration of the urothelium. Curr. Opin. Organ. Transplant. 19, 323–330 (2014).
Santos, C. P. et al. Urothelial organoids originating from Cd49f mouse stem cells display Notch-dependent differentiation capacity. Nat. Commun. 10, 4407 (2019).
Yu, Z. et al. Single-cell transcriptomic map of the human and mouse bladders. J. Am. Soc. Nephrol. 30, 2159–2176 (2019).
Guo, C. C. et al. Assessment of luminal and basal phenotypes in bladder cancer. Sci. Rep. 10, 9743 (2020).
Lorenzi, T. et al. HtrA1 in human urothelial bladder cancer: A secreted protein and a potential novel biomarker. Int. J. Cancer https://doi.org/10.1002/ijc.28280 (2013).
Stroggilos, R. et al. Proteome-based classification of nonmuscle invasive bladder cancer. Int. J. Cancer 146, 281–294 (2020).
Azumi, M. et al. Six-transmembrane epithelial antigen of the prostate as an immunotherapeutic target for renal cell and bladder cancer. J. Urol. 183, 2036–2044 (2010).
Wild, P. J. et al. High-throughput tissue microarray analysis of COX2 expression in urinary bladder cancer. Int. J. Oncol. 27, 385–391 (2005).
Agrawal, U., Kumari, N., Vasudeva, P., Mohanty, N. K. & Saxena, S. Overexpression of COX2 indicates poor survival in urothelial bladder cancer. Ann. Diagn. Pathol. 34, 50–55 (2018).
Yang, J. et al. TRIB3 promotes the malignant progression of bladder cancer: An integrated analysis of bioinformatics and in vitro experiments. Front. Genet. 12, 406. https://doi.org/10.3389/fgene.2021.649208 (2021).
Brooks, J. & Eltom, S. E. Malignant transformation of mammary epithelial cells by ectopic overexpression of the aryl hydrocarbon receptor. Curr. Cancer Drug Targets 11, 654–669 (2011).
Andersson, P. et al. A constitutively active dioxin/aryl hydrocarbon receptor induces stomach tumors. Proc. Natl. Acad. Sci. U. S. A. 99, 9990–9995 (2002).
Dantsuka, A. et al. Histopathological features of the proper gastric glands in FVB/N-background mice carrying constitutively-active aryl-hydrocarbon receptor. BMC Gastroenterol. 19, 102 (2019).
Dietrich, C. & Kaina, B. The aryl hydrocarbon receptor (AhR) in the regulation of cell–cell contact and tumor growth. Carcinogenesis 31, 1319–1328 (2010).
Shimizu, Y. et al. Benzo[a]pyrene carcinogenicity is lost in mice lacking the aryl hydrocarbon receptor. Proc. Natl. Acad. Sci. U. S. A. 97, 779–782 (2000).
Wilson, S. R., Joshi, A. D. & Elferink, C. J. The tumor suppressor Kruppel-like factor 6 is a novel aryl hydrocarbon receptor DNA binding partner. J. Pharmacol. Exp. Ther. 345, 419–429 (2013).
Vogel, C. F. A. et al. Interaction of aryl hydrocarbon receptor and NF-κB subunit RelB in breast cancer is associated with interleukin-8 overexpression. Arch. Biochem. Biophys. 512, 78–86 (2011).
Xie, G., Peng, Z. & Raufman, J.-P. Src-mediated aryl hydrocarbon and epidermal growth factor receptor cross talk stimulates colon cancer cell proliferation. Am. J. Physiol. Gastrointest. Liver Physiol. 302, G1006–G1015 (2012).
Fritsche, E. et al. Lightening up the UV response by identification of the arylhydrocarbon receptor as a cytoplasmatic target for ultraviolet B radiation. Proc. Natl. Acad. Sci. U. S. A. 104, 8851–8856 (2007).
Baker, J. R., Sakoff, J. A. & McCluskey, A. The aryl hydrocarbon receptor (AhR) as a breast cancer drug target. Med. Res. Rev. 40, 972–1001 (2020).
Denison, M. S. & Nagy, S. R. Activation of the aryl hydrocarbon receptor by structurally diverse exogenous and endogenous chemicals. Annu. Rev. Pharmacol. Toxicol. 43, 309–334 (2003).
Kolluri, S. K., Jin, U.-H. & Safe, S. Role of the aryl hydrocarbon receptor in carcinogenesis and potential as an anti-cancer drug target. Arch. Toxicol. 91, 2497–2513 (2017).
Dong, L., Ma, Q. & Whitlock, J. P. DNA binding by the heterodimeric Ah receptor. J. Biol. Chem. 271, 7942–7948 (1996).
Mullenders, J. et al. Mouse and human urothelial cancer organoids: A tool for bladder cancer research. Proc. Natl. Acad. Sci. U. S. A. 116, 4567–4574 (2019).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Zhu, A., Ibrahim, J. G. & Love, M. I. Heavy-tailed prior distributions for sequence count data: Removing the noise and preserving large differences. Bioinformatics 35, 2084–2092 (2019).
Yu, G., Wang, L.-G., Han, Y. & He, Q.-Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS J. Integr. Biol. 16, 284–287 (2012).
Supek, F., Bošnjak, M., Škunca, N. & Šmuc, T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS ONE 6, e21800 (2011).
Muraro, M. J. et al. A single-cell transcriptome atlas of the human pancreas. Cell Syst. 3, 385–394.e3 (2016).
Hao, Y. et al. Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587.e29 (2021).
Acknowledgements
We are grateful to Jasper Mullenders and Hans Clevers for providing the mouse bladder organoids. We thank Livio Kleij for his support with microscopy, Single Cell Discoveries for support with single cell RNA sequencing, the Utrecht Sequencing Facility for providing sequencing service, and the UMC Utrecht Bioinformatics Expertise Core for data analysis and data handling. The Utrecht Sequencing Facility is subsidized by the University Medical Center Utrecht, Hubrecht Institute, Utrecht University, and The Netherlands X-omics Initiative (NWO project 184.034.019). The UMC Utrecht Bioinformatics Expertise Core is subsidized by the University Medical Center Utrecht, Center for Molecular Medicine. This publication and the underlying study have been made possible partly based on the data that Hartwig Medical Foundation and the Center of Personalised Cancer Treatment (CPCT) have made available to the study.
Author information
Authors and Affiliations
Contributions
J.M.V., E.Kuijk, N.B., D.W. and E.C. designed and performed experiments including data analysis and interpretation. A.B., and E.Kalkhoven designed and performed the luciferase assay experiments. C.S. and P.P. conducted Hartwig database analysis. B.M.F. provided Weill Cornell Medicine data analysis. P.P. and E.C. conceived the original idea. E.C. and E.Kuijk supervised the project. J.M.V., E.Kuijk, and E.C. wrote the manuscript with input from all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Vlaar, J.M., Borgman, A., Kalkhoven, E. et al. Recurrent exon-deleting activating mutations in AHR act as drivers of urinary tract cancer. Sci Rep 12, 10081 (2022). https://doi.org/10.1038/s41598-022-14256-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-022-14256-0
- Springer Nature Limited