Abstract
microRNAs (miRNAs) are crucial for normal development and physiology. To identify factors that might coordinate with miRNAs to regulate gene expression, we used 2′O-methylated oligonucleotides to precipitate Caenorhabditis elegans let-7, miR-58, and miR-2 miRNAs and the associated proteins. A total of 211 proteins were identified through mass-spectrometry analysis of miRNA co-precipitates, which included previously identified interactors of key miRNA pathway components. Gene ontology analysis of the identified interactors revealed an enrichment for RNA binding proteins, suggesting that we captured proteins that may be involved in mRNA lifecycle. To determine which miRNA interactors are important for miRNA activity, we used RNAi to deplete putative miRNA co-factors in animals with compromised miRNA activity and looked for alterations of the miRNA mutant phenotypes. Depletion of 25 of 39 tested genes modified the miRNA mutant phenotypes in three sensitized backgrounds. Modulators of miRNA phenotypes ranged from RNA binding proteins RBD-1 and CEY-1 to metabolic factors such as DLST-1 and ECH-5, among others. The observed functional interactions suggest widespread coordination of these proteins with miRNAs to ultimately regulate gene expression. This study provides a foundation for future investigations aimed at deciphering the molecular mechanisms of miRNA-mediated gene regulation.
Similar content being viewed by others
Introduction
Developmental and physiological processes require precise spatio-temporal regulation of gene expression. One post-transcriptional gene regulatory mechanism is directed by a class of small non-coding RNAs called microRNAs (miRNAs). miRNAs regulate a wide range of developmental and cellular processes, with dysregulated miRNA activity prevalent in diseases1,2. To exert their regulatory roles, miRNAs are loaded into Argonaute (AGO) proteins to form a miRNA Induced Silencing Complex (miRISC), which ultimately associates with an effector protein GW182. miRISC binding to the target mRNA via partial sequence complementarity between a miRNA and 3′ UTR of mRNA triggers a series of gene silencing mechanisms including translation inhibition, decapping, and mRNA decay3,4.
miRNAs are produced by a complex biogenesis process which involves enzymatic processing of miRNA intermediates in the nucleus and cytoplasm. Primary miRNAs are first cleaved by the Microprocessor complex (Drosha and DGCR8) to form pre-miRNAs5,6. After export from nucleus into cytoplasm, pre-miRNAs are further processed by Dicer to generate a miRNA duplex7,8. The miRNA duplex bound by Argonaute is then unwound, with the guide miRNA strand retained to form the mature miRISC, and the passenger strand released and degraded9,10. Each of the steps in miRNA biogenesis process can be regulated by RNA binding and other auxiliary factors thereby modulating the final gene-regulatory impact of miRNAs. These factors could bind miRNA intermediates or miRISC protein components to affect miRNA activity. For example, several RNA binding proteins including RBFOX3 and HnRNP A1 have been identified to bind to the hairpin structures of primary miRNAs and modulate their processing11,12. Other proteins, such as NHL-2 and CGH-1, associate with ALG-1 and AIN-1 to promote mRNA targeting13. RNA binding proteins Staufen14 and HuR15,16 indirectly affect miRNA-mediated gene silencing by competing for binding of the 3′UTRs of target mRNAs. Characterizing miRISC-associated protein complexes followed by functional analyses and mechanistic studies has high potential to identify additional mechanisms by which miRNA activity may be regulated.
Identifying proteins that associate with Argonaute proteins or miRNAs has been a productive approach to begin to unravel the mechanisms by which aspects of miRNA biogenesis and activity are regulated17,18,19,20. In human cells, investigation of proteomic profiles of AGO complexes led to identification of common protein interactors of all four AGO proteins, which included heat shock proteins, helicases, and components of translational machinery17. Some proteins identified in this study, including Hsc70/Hsp90 chaperone machinery, were later characterized for their roles in RISC loading of small RNA duplexes21. In mice, exploration of Dicer dependent and independent interactions of Ago218 identified proteins that participate in miRISC-mediated decapping22, among other mechanisms. However, while proteomic approaches have characterized miRNA-associated complexes, it has been challenging to identify which of these co-factors are functionally important for miRNA activity, especially in tissue culture. In contrast, functional assays that quantitatively assess miRNA activity are available in model organisms such as C. elegans.
To identify miRNA/miRISC auxiliary cofactors important for miRNA gene regulatory activity, we took a functional proteomics approach. Specifically, we used 2′O-methylated biotinylated oligonucleotides to pull down three miRNAs of interest (let-7, miR-58, and miR-2) and subjected the associated protein complexes to proteomic analysis. Comparative analysis of miRNA pulldown and ALG-1 immunoprecipitation precipitates19 identified high confidence interactors common to all four datasets. In addition, we identified a unique set of interactors in each miRNA pulldown dataset. To assess whether the co-precipitated proteins are functionally important for miRNA activity, we performed RNAi knockdown of genes encoding for the putative physical interactors in multiple miRNA sensitized genetic backgrounds. Of the 39 interactors tested, depletion of 25 factors modified miRNA reduction of function phenotypes in one or more assays. Overall, we demonstrate that capturing physical interactors of miRNA machinery followed by in vivo functional assays is an efficient approach to identify novel players in miRNA-mediated gene regulation. While further mechanistic characterizations are necessary to determine the extent of the physical and functional interactions, this study identifies a functional requirement for a subset of potential ALG-1 and miRNA co-factors.
Methods
C. elegans maintenance, strains, and RNAi
All C. elegans strains were maintained on NGM and fed with E. coli OP50. Strains were maintained at 20 °C unless otherwise noted. RNAi knockdown was performed by feeding as previously described23.
The following strains were used in this study: N2 (wild type), MT7626 (let-7(n2853)), HW1113 [Pdpy-30::GFP(PEST)-H2B::lin-41 3’ UTR (xeSi78); Pdpy-30::mCherry::H2B::artificial 3' UTR (xeSi36)], HW1114 [Pdpy-30::GFP (PEST)-H2B::lin-41 3’ UTR (xeSi78); Pdpy-30::mCherry::H2B::artificial 3’ UTR (xeSi36), let-7(n2853)], VT1367 (col-19::gfp (maIs105)), VT1296 (mir-48 mir-241(nDf51) col-19::gfp (maIs105)), BW1932 [hbl-1p::gfp::NLS::hbl-1 3’ UTR (ctIS39)] and UY458 (mir-48 mir-241(nDf51); hbl-1p::gfp::NLS::hbl-1 3′ UTR (ctIS39)]), OH812 (otIs114 [Plim-6-gfp + rol-6(su1006)]), OH3646 (lsy-6(ot150); otIs114 [Plim-6-gfp + rol-6(su1006)]), PS3662 (syIs63[cog-1::gfp + unc-119(+)]), OH7310 (otIs193 [cog-1p::lsy-6 + rol-6(su1006)] syIS63[cog-1::gfp + unc-119(+)]).
2′O-Methyl oligo pulldowns and mass spec analysis
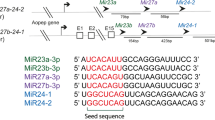
All experiments were performed on mixed-stage animals. Whole worm extracts24 and 2′O-methyl oligo pulldowns25 were performed as previously described. For mass spectrometry, each sample contained 20 mg of total protein. miRNA pulldowns were performed in two biological replicates using 2′O-methylated oligos with perfect complementation to miR-58, let-7, and miR-2, and scrambled oligo control (IDT). Sequences of the 2′O-methylated, biotinylated oligonucleotides are as follows: miR-58 oligo (5′-CAUCAUUGCCGUACUGAACGAUCUCAAGUC-3′), miR-2 oligo (5′-AUUCAGCACAUCAAAGCUGGCUGUGAUAUUCCA-3′), let-7 oligo (5′-UCUUCACUAUACAACCUACUACCUCAACCUU-3′), and scrambled oligo (5′-CAUCACGUACGCGGAAUACUUCGAAAUGUC-3′).
Mass spectrometric analysis of pulldown factors was performed as previously described19. Briefly, DTASelect26 was used to filter the proteins identified by applying a criterion that required proteins to have at least two unique peptides with total spectral intensities greater or equal to four in both replicates. To determine enrichment of protein association in a miRNA pulldown, the Normalized Spectral Abundance Factor (NSAF) values in miRNA pulldown were divided by that in control pulldown. NSAF value of zero in control was replaced by 1. Proteins with the pulldown/control ratio of ≥ 4 in all replicates were considered putative physical interactors.
GO term and network analysis
Gene ontology analysis was performed using Database for Annotation, Visualization and Integrated Discovery (DAVID)27. Factors that had a fold change ≥ 4 in both replicates of the miRNA pulldowns were used for this analysis. For comparison, we included factors identified in atleast two replicates of ALG-1 IP for GO term analysis (as previously described19). Protein domain information, domain enrichment analysis, and the associated statistics were retrieved using STRING28. Enrichment for proteins harboring an RNA binding domain (RBD) among the proteins that passed our criteria was determined against a background set of C. elegans proteins that harbor the same RNA binding domain. Statistically significant enrichment was determined by applying Benjamini–Hochberg procedure on p-values to correct for multiple-testing. Network analysis was performed on the top 40 most enriched factors using STRING28 after excluding ribosomal proteins.
Functional assays
let-7(n2853) vulval bursting assay
Vulval bursting assay was performed as previously described29. Briefly, let-7(n2853) and N2 worms were grown and maintained at 15℃. Embryos obtained through bleaching30 were plated on RNAi plates23 and grown until L4 larval stage. L4 animals were shifted to new RNAi plates and scored as day 1 adults for vulval bursting using a Leica dissecting microscope. Total number of worms (n) scored for this assay across two to four independent RNAi experiments ranged from 45 to 330.
col-19::gfp expression and seam cell number assay
mir-48 mir-241(nDf51) col-19::gfp (maIs105) animals were transferred to RNAi plates as L3 stage larvae and their progeny were scored for heterochronic phenotypes for hypodermal col-19::gfp expression. Worms with seam-only reporter expression were classified as having “delayed hypodermal col-19::gfp expression”. Seam cell numbers were scored by counting the number of seam cells expressing col-19::gfp between pharynx and anus. For most candidates, the total number of worms scored (n) across two to four replicates ranged from 22 to 80. For genes whose knockdowns resulted in severe developmental defects such as snr-4, snr-6, let-363, and rnp-7, the (n) was either 18 or 19.
lsy-6(ot150) ASEL cell fate assay
lsy-6(ot150); plim-6::gfp and plim-6::gfp worms were transferred onto RNAi plates as embryos. Their progeny were scored as L4s for specification of ASEL cell fate based on plim-6::gfp reporter expression. Worms lacking the reporter expression in ASEL neurons were scored as cell fate defective. Across two to four replicates, a total of 90 to 286 worms were scored.
pdpy-30::gfp::lin-41 reporter assay
pdpy-30::GFP(PEST)-H2B::lin-41 3' UTR (xeSi78); pdpy-30::m Cherry::H2B::artificial 3' UTR (xeSi36) and pdpy-30::GFP (PEST)-H2B::lin-41 3' UTR (xeSi78); pdpy-30::mCherry::H2B::artificial 3' UTR (xeSi36), let-7(n2853) embryos obtained by bleaching were plated on RNAi plates and grown at 15 °C. Reporter expression was measured in L4 stage animals by imaging the vulva at 63× magnification. To quantify expression levels in six vulval cells, ROIs were manually drawn and signal intensities within the ROI were measured using the Leica image analysis software. For each vulval cell, GFP signal intensity was divided by mCherry signal intensity and relative signal intensities were averaged across the six cells imaged in an individual animal. Representative images were equally adjusted after quantification to make the fluorescence more observable.
hbl-1p::gfp reporter assay
hbl-1p::gfp::NLS::hbl-1 3′ UTR (ctIS39) and mir-48 mir-241(nDf51); hbl-1p::gfp::NLS::hbl-1 3′ UTR (ctIS39) embryos were obtained by bleaching. Embryos were transferred to RNAi plates and animals were scored for hbl-1p::gfp expression in hypodermal cells at early to mid L3 stage. Animals were staged by time of development, and gonad size and shape.
Uterine cog-1 reporter expression
cog-1::gfp and pcog-1::lsy-6; cog-1::gfp animals were transferred onto RNAi plates as embryos and their F1 progeny were scored at L4 stage for cog-1::gfp expression in the uterine cells. Worms expressing cog-1::gfp in both uterine cells and vulval cells were scored as wild type. Worms that lacked cog-1::gfp reporter expression in either of the two uterine cells were scored as abnormal.
Fluorescence microscopy, image capture and illustrations
Fluorescence equipped Zeiss Axioplan 2 or Leica DM6 upright microscopes were used for scoring phenotypes. Images were captured using the Leica DM6B camera and processed using the Leica Application Suite X (3.4.1.17822) software (https://www.leica-microsystems.com/products/microscope-software/p/leica-las-x-ls/). Illustrations in Figs. 1a and 6e–g were drawn using BioRender (biorender.com).
miRNA pulldowns (PDs) identify putative physical interactors of miRNA complexes. (a) Protein complexes associated with miRNAs were isolated using 2′-OMe modified oligonucleotides complementary to the miRNA of interest and subjected to MudPIT mass spectrometry analysis (drawn using BioRender (biorender.com)). (b–d) Average Normalized Spectral Abundance Factor (NSAF) values of factors identified in pulldown experiments (Y-axis) are plotted against that of corresponding scrambled oligo controls (X-axis) for (b) let-7 PD, (c) miR-58 PD and (d) miR-2 PDs. Highlighted in black are key miRISC components and components of miRNA biogenesis machinery. (e) Venn diagram showing the number of factors that passed a set of criteria to qualify as a putative interactor (for description of criteria, see “Methods”). Percentages shown here are percentage of total number of proteins (308) that passed the criteria in interaction datasets including ALG-119. Proteins commonly co-precipitated with †ALG-1, let-7, miR-58, and miR-2, ǂALG-1 IP, let-7 PD, and miR-58 PD, §let-7 PD, miR-58 PD, miR-2 PD, and ¶ALG-1 IP, miR-58 PD, and miR-2 PD. (f) Factors commonly identified in two or more interaction datasets include core miRNA machinery components. Symbols in the left column correspond to overlapping protein populations in (e). Colored boxes show which IP or PD precipitated the proteins listed in the left column. (g) NSAF values of proteins identified in pulldowns with let-7 complementary oligonucleotide from mir-48 mir-241(nDf51); mir-84(n4037) mutant animals, plotted against NSAF of proteins identified in scrambled oligo control. (h) let-7 complementary oligonucleotide precipitates a partially overlapping set of factors from let-7-family miRNA deletion backgrounds.
Statistical analysis
All statistics were done using GraphPad Prism (9.2.0 (332)) software. Statistical significance was determined using a one-way ANOVA test with predetermined comparisons. Bonferroni correction was applied as a post hoc analysis. T-test was used to determine statistical significance of pdpy-30::gfp::lin-41, hbl-1p::gfp, and cog-1::gfp reporter assays.
Results
miRNA pulldowns (PDs) identify overlapping sets of putative physical interactors of miRNA-centered complexes
To identify factors that may regulate miRNA activity, we sought to determine the molecular composition of protein complexes associated with let-7, miR-58, and miR-2 miRNAs. let-7 is highly conserved across all bilateral animals31 and is required for the larval to adult transition in C. elegans32. miR-58 is a highly abundant miRNA that regulates lifespan and dauer formation33, primarily by coordinating with the TGF-β pathway34,35. miR-2, a neuronal miRNA conserved among invertebrates36, is necessary for proper neuromuscular junction function in C. elegans37. We used biotinylated, 2′O-methylated oligonucleotides with perfect sequence complementarity to mature miRNA sequences to pulldown miRNAs of interest and characterized the precipitates using a shotgun proteomics approach (Fig. 1a) to identify proteins associated with miRNAs of interest compared to scrambled control (Fig. 1b–d, Supplementary Table S1). To identify high confidence interactors, we retained only the proteins that were ≥ fourfold enriched in miRNA pulldowns over the scrambled control, had a minimum NSAF value of 4, and were identified in all replicates. Overall, a total of 211 proteins passed the criteria we set (Fig. 1e), with 136 factors co-precipitating with let-7, 54 factors co-precipitating with miR-58, and 25 factors co-precipitating with miR-2 (Fig. 1e, Supplementary Table S1). Among the proteins enriched in miRNA co-precipitates were known miRISC components ALG-1 and ALG-2, the two major miRNA-associated Argonautes in C. elegans38 and AIN-1 and AIN-2, GW182 homologs and miRISC effectors39,40 (Fig. 1b–f). In addition, DCR-1 nuclease, responsible for pre-miRNA processing, was detected in all pulldown experiments performed, but did not meet our stringent interaction criteria in the let-7 pulldown (Fig. 1e,f, Supplementary Table S1).
To determine the overlap between complexes precipitated by miRNA pulldowns and those previously found to associate with ALG-119, we compared miRNA and ALG-1 co-precipitated factors (Fig. 1e,f, Supplementary Table S1). Eleven (8%) let-7 interactors, ten (18.5%) miR-58 interactors, and five (24%) miR-2 interactors were found to overlap with the ALG-1 co-immunoprecipitated dataset (Fig. 1e, Table 1). Overall, 41 proteins were present in at least two interaction datasets (Fig. 1e,f, Table 1), potentially representing general miRNA-associated co-factors. In addition, four proteins, HRPK-1, SMG-2, IMPH-1, and SUP-26, were present in 2 out of 3 pulldowns and the ALG-1 IP (Fig. 1e,f, Table 1). Their homologs were also found to co-immunoprecipitate with human and/or mouse Argonautes17,18,19, suggesting that they may have a conserved function in miRNA-mediated gene regulation. In fact, we previously confirmed a physical HRPK-1 interaction with ALG-1 and reported hrpk-1 to genetically interact with multiple miRNAs41. Five proteins were commonly captured in all the miRNA pulldowns (Fig. 1e,f, Table 1). Interestingly, one such protein was LET-363, an mTOR homolog42 (Fig. 1e,f, Table 1). Similarly, we observed overlaps between our miRNA co-precipitates and previously reported miRNA physical interactors43,44 (Supplementary Table S2). Overlaps among our miRNA interaction datasets and AIN-1 and AIN-2 co-precipitates40,44 were also observed, further emphasizing that our approach captured potential miRISC interactors (Supplementary Table S2). Finally, candidates identified in genetic screens for miRNA and siRNA pathway genes also intersected with many of our miRNA co-precipitates29,45,46 (Supplementary Table S2). The observed overlaps among various groups of physical and genetic interactors support the idea that we are detecting real physical interactors of miRNA-centered complexes.
Due to a high level of sequence similarity amongst the let-7 miRNA family members, the let-7 complementary oligonucleotide precipitates other members of the miRNA family, albeit with reduced efficiency47. To determine whether distinct populations of proteins might associate with let-7 miRNA family members, we performed additional let-7 pulldown experiments in mir-48 mir-241(nDf51); mir-84(n4037) mutant animals (Fig. 1g, Supplementary Table S1). 55 (29.4%) proteins were in common among both let-7 pulldowns, suggesting that these factors may interact with let-7 itself (Fig. 1h). The 51 (27.3%) proteins that precipitated with the let-7-complementary oligonucleotide from the mir-48 mir-241(nDf51); mir-84 (n4037) animals may similarly represent let-7-interacting factors, having been enriched in the let-7 pulldown in the absence of miR-48, miR-241, and miR-84 (Fig. 1h). In contrast, 81 (43.3%) proteins were present only in wildtype background pulldowns (Fig. 1h), suggesting that these factors may normally associate with miR-48, miR-241 and miR-84 (Fig. 1h). While we cannot rule out an association with the remaining let-7 family miRNAs, miR-793–795, the low relative abundance of these miRNAs19 suggests that miR-793–795 interactors are unlikely to represent significant fractions of the observed co-precipitates. Finally, some co-precipitates could represent non-specific interactions.
Ribonucleoprotein complex components are enriched among miRNA interactors
To understand what biological processes and functions are represented in the miRNA-precipitated complexes, we performed Gene Ontology (GO) analysis on putative miRNA interactors (Fig. 2a–d, Supplementary Table S3). Factors implicated in embryonic and larval developmental processes were commonly enriched in all interaction datasets (Fig. 2a–c; ALG-1 interactome analysis is shown in Fig. 2d for comparison19, Supplementary Table S3). Selective enrichment for splicing associated factors was observed in let-7 and miR-58 PD datasets (Fig. 2a,c, Supplementary Table S3). Components of intracellular ribonucleoprotein complexes were consistently captured in all datasets (Fig. 2a–d and Ref.19). Enrichment for ribosomal components was observed only in let-7 and ALG-1 datasets (Fig. 2a,b,d and Ref.19). Unsurprisingly, the RNA/nucleic acid binding term was commonly overrepresented in all the datasets in the molecular function category (Fig. 2a–d and Ref.19). miR-2 interactors did not show an enrichment for any of the GO terms (Supplementary Table S3), potentially due to the low number of interactors captured in our pulldown and were therefore excluded from further analysis.
miRNA pulldowns identify components of translation machinery and mRNA processing factors, which may form a functional network. (a–d) Terms identified through GO analysis for biological processes (top four), cellular components (top four) and molecular function categories (top three) among let-7 PDs in (a) wild type, (b) mir-48 mir-241(nDf51); mir-84(n4037) backgrounds, (c) miR-58 PD and (d) ALG-1 IP19. Total number of proteins classified under each term shown adjacent to respective bars. (e–h) RNA binding domains identified in factors captured in let-7 PDs from (e) wild type, and (f) mir-48 mir-241(nDf51); mir-84(n4037)) backgrounds, (g) miR-58 PD and (h) ALG-1 IP. The FDR adjusted p-values for enrichment of proteins harboring individual domains are shown within the respective bars. (i–l) The reproducibly enriched proteins form functional network. Network analysis was performed using STRING28 on top 40 enriched interactors in (i) let-7 PD (wild type), (j) let-7 PD (mir-48 mir-241(nDf51); mir-84(n4037)) (k) miR-58 PD and (l) ALG-1 IP. Ribosomal proteins were excluded from this analysis. Thickness of edges represents degree of confidence of functional linkages. The number of edges is significantly higher than expected, with a p-value < 1.0e−16.
As RNA binding proteins (RBPs) were enriched in miRNA co-precipitates (Fig. 2a–d) and RBPs carry distinct domains critical for their RNA binding activity, we examined what RNA binding domains (RBDs) were present in proteins identified in our pulldowns and ALG-1 IP (Fig. 2e–h, Supplementary Table S3). At least 5 different RBDs were observed among RBPs in all the datasets. Notably, RBDs such as RNA Recognition Motif, KH, PAZ, and Nucleotide-binding alpha–beta plait domain superfamily were present in RBPs in one or more datasets (Fig. 2e–h, Supplementary Table S3). Overall, miRNA pulldowns captured factors that may play critical roles during the lifecycle of RNA.
To determine whether factors identified through our proteomics approach form a functional network, we performed network analysis using STRING28 (Fig. 2i–l). STRING predicts candidate protein interactions by utilizing both known and predicted protein–protein interactions sourced from databases, text mining, experimental, and co-expression data28. Top 40 enriched putative interactors in each dataset, minus the ribosomal proteins, were chosen for this analysis. Interestingly, we observed that in all the datasets proteins formed functional networks with a significant number of edges (p-value < 1.0e−16, as determined by STRING) (Fig. 2i–l), further supporting the idea that miRNA pulldown-captured proteins form functional complexes that may coordinate with miRNAs to regulate gene expression.
Functional analysis of putative miRNA interactors
To identify which putative interactors might functionally coordinate with miRNAs to regulate gene expression, we took advantage of sensitized genetic backgrounds with reduced miRNA or miRNA family activity. These functional assays with quantifiable phenotypic outputs allow for assessment of a gene’s role in miRNA-mediated gene repression. Our pulldown experiments targeted miRNAs with varied functions and spatio-temporal expression patterns. Some of the identified interactors of miRNA- or ALG-1-centered complexes could have broad functional requirements, while others could be specific to a particular tissue or a developmental time. We hypothesized that knockdown of generally-required factors in multiple sensitized miRNA backgrounds would modulate phenotypes in multiple functional assays. In contrast, spatio-temporal specificity of the putative interactors may limit their functional relevance to specific miRNAs and may not result in a phenotype in some, or all, of our assays. In addition, the miRNA-centered protein complex analyses potentially identified interactors that may positively or negatively modulate microRNA activity. Knockdown of these factors in sensitized genetic backgrounds may therefore result in an enhancement or a suppression of the phenotype associated with reduction of miRNA function.
For our functional assessment, we prioritized factors that were highly enriched in our pulldown and/or ALG-1 IP experiments19 and were captured in multiple datasets. We excluded ribosomal proteins and factors lacking RNAi clones. The 39 candidates assayed ranged from common interactors of miRNA(s) and ALG-1 (6), common miRNA interactors (6), ALG-1 interactors (16), and specific miRNA interactors [let-7 (10), and miR-58 (1)] (Supplemental Table S4). Among the ALG-1 interactors, we assayed genes encoding for six proteins consistently identified in human and mouse AGO IP (referred to as conserved AGO interactors from hereon).
RNAi knockdown of genes of let-7 and ALG-1 interactors alters let-7(n2853) mutant phenotype
let-7 is essential for C. elegans development and promotes transition from the fourth larval stage (L4) to adulthood32. Loss of let-7 function results in vulval bursting and failure of seam cells to differentiate during the L4 to adult transition32. let-7(n2853) is a temperature-sensitive reduction of function mutation that impairs regulation of let-7 targets, including lin-4148. let-7(n2853) mutants have a partially penetrant vulval bursting phenotype at permissive temperature32 (15 °C) (Fig. 3a). To determine whether the identified let-7 and ALG-1 interactors are functionally important for let-7 miRNA activity, we used this well-established genetic background to assay the effects of gene knockdown on let-7(n2853) bursting phenotype. RNAi of six genes enhanced vulval bursting of let-7(n2853) mutant (Fig. 3b,c, Supplementary Table S4). One such gene (pab-1) encoded a conserved AGO interactor19 (Fig. 3b, Supplementary Table S4) and five genes including C04E6.11, ech-5, and rbd-1, which code for let-7 interactors (Fig. 3c, Supplementary Table S4). RNAi of cey-1 suppressed the bursting (Fig. 3c, Supplementary Table S4). Knockdown of ifg-1 also mildly suppressed let-7(n2853) vulval bursting from 30 to 6% (Fig. 3b, Supplementary Table S4), although the suppression did not reach a statistically significant level (Anova p-value = 0.078). RNAi knockdown of these genes in the wild type background did not result in vulval bursting, suggesting that these genes do not play a central role in gene regulation, only revealing the function in the sensitized let-7(n2853) background (Fig. 3b,c). We cannot, however, rule out the possibility that RNAi knockdown in wild type background may have been ineffective. Overall, these findings support our hypothesis that the identified let-7 physical interactors play a role in let-7-mediated gene repression.
RNAi knockdown of genes encoding ALG-1 and let-7-associated factors genetically modifies let-7(n2853) vulval bursting phenotype. (a) let-7(n2853) mutants show partially penetrant vulval bursting phenotype at 15 °C. RNAi of (b) genes of previously reported conserved interactors of AGO19 and (c) let-7-precipitated factors in wild type and let-7(n2853) mutants. Each dot represents an independent RNAi experiment. Statistical significance was determined by one-way ANOVA with post hoc Bonferroni correction. (d) Effects of vector and rbd-1 RNAi on pdpy-30::gfp::lin-41 3′UTR reporter expression in the vulval cells of wild type and let-7(n2853) L4 larvae. Compromised miRNA activity in let-7(n2853) background leads to de-repression of pdpy-30::gfp::lin-41 3′ UTR reporter expression levels (quantified in e). (e) RNAi of genes of modifiers of let-7(n2853) vulval bursting phenotype affects pdpy-30::gfp::lin-41 3ʹUTR reporter expression levels in let-7(n2853) background. Each dot represents the relative intensity in an individual worm. Vulval precursor cells used for fluorescence quantification are highlighted with dashed circles. T-test was used to determine statistical significance. Vector = empty vector RNAi control.
Since dysregulation of let-7 target gene lin-41 in vulval-uterine system is sufficient to cause vulval rupturing49, we wanted to determine how depletion of putative physical and genetic let-7 interactors affects lin-41 expression in the relevant cells. To do this, we used an established let-7-lin-41 reporter system49. We performed RNAi knockdown of genes that enhance (ech-5, rbd-1, C04E6.11, and let-363) and suppress (cey-1) let-7(n2853) vulval bursting in the background of two reporter strains: pdpy-30::gfp::lin-41 3ʹUTR and pdpy-30::gfp::lin-41 3ʹUTR; let-7(n2853)49. RNAi depletion of these genes did not alter pdpy-30::gfp::lin-41 3ʹUTR reporter levels in the wildtype background (Fig. 3d,e, Supplementary Table S5), suggesting that these genes do not have a major effect on lin-41 levels on their own. However, in let-7(n2853) background at 15 °C, knockdown of ech-5 or rbd-1 substantially increased lin-41 levels, while cey-1 depletion reduced pdpy-30::gfp::lin-41 3ʹUTR reporter levels in the vulval cells (Fig. 3d,e, Supplementary Table S5). RNAi of let-363 led to a mild increase in the reporter levels in let-7(n2853) at 15 °C, although the increase was not statistically significant (Fig. 3e, Supplementary Table S5). Interestingly, let-363 depletion was previously reported to increase levels of let-7 target reporters hbl-1p::gfp::hbl-1 in VNC and col-10::gfp::lin-41 3′UTR in hypodermal cells50. These observations suggest that ech-5, rbd-1, cey-1 and perhaps even let-363 may be contributing to regulation of vulval bursting by modulating let-7 miRNA activity.
RNAi knockdown of genes of let-7 and ALG-1 interactors alters mir-48 mir-241(nDf51) mutant phenotype
We next asked whether miRNA and ALG-1 interactors might functionally coordinate with other members of the let-7 family of miRNAs. let-7 family members mir-48, mir-84 and mir-241 specify developmental timing in C. elegans, regulating seam cell divisions and contributing to L2–L3 larval developmental transition51 (Fig. 4a). Deletion of all three miRNAs (mir-48 mir-241(nDf51); mir-84(n4037)) results in reiteration of L2 stage seam cell divisions, resulting in an increased number of seam cells and delayed terminal cell differentiation in young adults51 (Fig. 4a). Partial deletion (mir-48 mir-241(nDf51)) mutants display incompletely penetrant heterochronic phenotype which can be monitored using an adult stage marker, col-19::gfp, expressed in seam and hypodermal cells (Fig. 4b). RNAi of 11 genes enhanced the abnormal col-19::gfp expression in hypodermal cells (Fig. 4c,d, Supplementary Table S4), with five genes, pab-1, dlst-1, C43E11.9, snr-4, and rbd-1, enhancing the phenotype to > 50% (Fig. 4c,d Supplementary Table S4). This suggests that these potential miRNA interactors are required for regulation of developmental timing programs. We also examined the effects of gene knockdown on seam cell number. RNAi of three genes, pqn-70, dlst-1, and cey-1, increased the seam cell number of mir-48 mir-241(nDf51) mutants (Fig. 4f, Supplementary Table S4). RNAi of nine genes suppressed seam cell lineage defect, with knockdown of let-363, rbd-1, snr-6, and snr-4, restoring the seam cell number to an average of 13 or lower (Fig. 4e,f, Supplementary Table S4). We should note that while most genes were assayed across a minimum of two independent RNAi experiments, four genes (snr-6, snr-4, let-363, and rnp-7) were tested only once, as knockdown of these genes caused lethality, reduced brood size, and slowed growth. Depletion of some genes had varied effects on hypodermal col-19::gfp expression and seam cell lineage. Knockdown of C43E11.9, pdi-2, and Y71F9AL.9 modified col-19::gfp expression but not seam cell number, while knockdown of pqn-70 and C28H8.3 modified the seam cell number of mir-48 mir-241(nDf51) animals without affecting hypodermal col-19::gfp expression (Fig. 4d,f). This could perhaps be explained by distinct roles these genes may play during proliferative seam cell divisions and terminal hypodermal cell fate specification.
RNAi knockdown of genes encoding for putative ALG-1 and miRNA interactors modulates mir-48 mir-241(nDf51) seam cell lineage and hypodermal cell fate defects. (a) A schematic representation of seam cell lineages of wild type and mir-48 mir-241(nDf51) mutant animals throughout C. elegans larval development. (b) Wild type young adult animals express adult cell fate marker, col-19::gfp, in seam and hypodermal cells, while mir-48 mir-241(nDf51) mutants show lack the col-19::gfp expression in the hypodermis as young adults some of the time (quantified in c). (c,d) Effects of RNAi knockdown of genes of (c) conserved AGO19 and (d) other miRISC interactors on hypodermal col-19::gfp expression in wild type and mir-48 mir-241(nDf51) mutants. Each dot represents an independent RNAi experiment. (e,f) Effects of RNAi knockdown of genes of (e) conserved AGO19 and (f) other miRISC interactors on the seam cell number in mir-48 mir-241(nDf51) mutant young adults. Each dot represents seam cell number in an individual worm. Statistical significance was determined by One-way ANOVA with post hoc Bonferroni correction. (g) Expression of hbl-1::gfp::hbl-1 3′ UTR reporter in control and pdi-2 RNAi in wild type and mir-48 mir-241(nDf51) L3 larvae. Arrows indicate hypodermal cells and arrowheads indicate seam cells. (h) RNAi of most genes that genetically modified heterochronic defects in mir-48 mir-241(nDf51) mutants significantly affected hbl-1::gfp::hbl-1 3′ UTR reporter expression in both wildtype and mir-48 mir-241(nDf51) backgrounds. Each dot represents an independent RNAi experiment. T-test was used to determine statistical significance. Vector = empty vector RNAi control.
Since let-7 family miRNAs promote L3 cell fates by repressing hbl-151, we sought to determine whether the genes that affect heterochronic phenotypes in mir-48 mir-241(nDf51) background do so by regulating hbl-1. We used the hbl-1p::gfp::hbl-1 3′UTR fusion construct as a reporter to assess the effects of gene knockdown on levels of HBL-152. Strong hbl-1 expression can be seen during embryogenesis with hypodermal hbl-1::gfp::hbl-1 3′UTR expression decreasing beyond detection at the L3 stage52 (Fig. 4g). RNAi knockdown of pdi-2, rbd-1,Y71F9AL.9, and cey-1 resulted in higher percentages of L3 animals expressing the reporter in the wild type background, potentially indicating a miRNA-independent effect (Fig. 4h, Supplementary Table S5). Since depletion of these genes did not affect col-19:gfp expression or other heterochronic defects in the wild type (Fig. 4d), perhaps the extent of hbl-1 derepression was not strong enough to impact developmental timing. This notion is supported by the observation that hbl-1 reporter expression in mir-48 mir-241(nDf51) background is observed at a higher rate (75%, Fig. 4h). Knockdown of enhancers of mir-48 mir-241(nDf51) mutant phenotype derepressed hbl-1 reporter expression in the mir-48 mir-241(nDf51) background (Fig. 4h, Supplementary Table S5). Overall, these findings suggest that our proteomics approach captured proteins that may co-ordinate with let-7 family miRNAs to repress their target, hbl-1, and ultimately coordinate developmental timing.
Depletion of ALG-1 physical interactors altered lsy-6(ot150) phenotype
We hypothesized that some of the ALG-1 and/or miRNA putative physical interactors could be factors that are generally required for miRISC activity. To test this, we RNAi depleted them in lsy-6(ot150) background. lsy-6 is essential for cell fate determination of chemosensory ASE neurons53. lsy-6 represses an ASER cell fate promoting transcription factor cog-1, leading to an ASEL neuronal specific gene expression pattern. Loss of lsy-6 leads to dysregulated gene expression of cog-1 and downstream effectors resulting in defective ASEL cell fate which leads to lack of plim-6::gfp reporter. However, the reduction of function mutant lsy-6(ot150) shows partially penetrant cell fate defective phenotype in approximately 20% of animals53 (Fig. 5a). Knockdown of 8 genes modified lsy-6(ot150) defective phenotype (Fig. 5b,c, Supplementary Table S4) including previously reported conserved AGO interactors pab-1, and larp-119 (Fig. 5b). Interestingly, we identified two suppressors (ifg-1, and F28B4.3) of ASEL cell fate defect (Fig. 5b,c, Supplementary Table S4). To determine whether these candidate factors can influence ASEL cell fate independent of lsy-6 miRNA, we knocked them down in wild-type worms and observed no change in plim-6::gfp reporter expression in ASEL cells (Fig. 5b,c).
ALG-1 and miRNA interactors genetically interact with lsy-6(ot150). (a) lsy-6 determines the asymmetric expression of chemoreceptors in ASER and ASEL neurons, with plim-6::gfp expression marking ASEL neuronal cell fate. ASEL cell defective phenotype in the lsy-6(ot150) animals can be observed by lack of plim-6::gfp expression some of the time (quantified in b). RNAi knockdown of (b) conserved AGO19 and (c) miRNA/ALG-1 interactors in plim-6::gfp or lsy-6(ot150); plim-6::gfp animals. Each dot represents an independent RNAi experiment. Statistical significance was determined by One-way ANOVA with post hoc Bonferroni correction. (d) cog-1 expression in uterine cells is repressed by pcog-1::lsy-6. RNAi of F33D11.10 depresses cog-1::gfp in that background (quantified in e). (e) RNAi of genes that modify lsy-6(ot150) phenotype alleviates lsy-6-mediated repression of cog-1::gfp in uterine cells. Each dot represents an independent RNAi experiment. T-test was used to determine statistical significance. Vector = empty vector RNAi control.
We next determined whether genes that modified lsy-6 phenotype upon knockdown were important for lsy-6 mediated target activity using cog-1 reporter system54 (Fig. 5d). cog-1 is expressed in vulval and uterine cells where lsy-6 is absent (Fig. 5d, top left panel). When lsy-6 is ectopically expressed in these tissues under cog-1 promoter, there is reduced expression of cog-1 as a result of lsy-6 mediated repression54 (Fig. 5d, top right panel). We performed RNAi knockdown of top hits from lsy-6 assay in the cog-1 reporter strain and observed no difference in cog-1 expression (Fig. 5e, Supplementary Table S5), suggesting that these factors do not regulate cog-1 directly in uterine cells. RNAi knockdown of F33D11.10 and psf-1 (enhancers of lsy-6(ot150) phenotype) restored cog-1 expression in the presence of lsy-6, suggesting their requirement for lsy-6 mediated cog-1 repression (Fig. 5e). Knockdown of larp-1 did not restore cog-1 expression to a statistically significant level (Fig. 5e, Supplementary Table S5). This could be due to RNAi variability among replicates, or possibly because lsy-6 activity was initially assessed in ASE neurons, while cog-1 reporter expression was assessed in uterine tissue. Previously reported tissue-specific composition of miRNA-centered complexes and distinct mechanisms of target suppression44 support this potential explanation for the observed discrepancy in larp-1 effects. lsy-6(ot150) and cog-1 reporter assays collectively demonstrate that we identified factors that may directly or indirectly coordinate with lsy-6, affecting its target cog-1 expression.
Overall, depletion of miRNA complex interactors did not produce a phenotype in the absence of the sensitized miRNA mutations (Figs. 3b,c, 4c,d, 5b,c). While we cannot rule out inefficient RNAi knockdown as a possible explanation, we hypothesize that the tested factors are not critical for regulation of miRNA target gene expression, but rather play a modulatory role in miRNA production and/or activity, or influence gene expression downstream of miRNA activity.
Discussion
To better understand miRNA mediated gene regulation, we performed miRNA pulldowns to identify components of miRNA-centered complexes. Our proteomics approach captured 211 miRNA-interacting proteins, some of which were previously reported to precipitate with other miRISC components (Supplementary Table S2). Knockdown of 25 out of 39 genes significantly modulated miRNA mutant phenotypes in one or more assays, suggesting that our pulldowns captured proteins that coordinate with miRNAs to affect gene regulation (Fig. 6a, Supplementary Table S4). Of the 25 hits, knockdown of five genes (pab-1, let-363, rbd-1, cey-1, and lys-8) and dcr-1, a positive control, consistently modified miRNA phenotypes in two or more assays (Fig. 6a, Supplementary Table S4). Of the 22 candidate genes tested, RNAi of six genes modulated let-7(n2853) vulval bursting phenotype (Fig. 6a,b, Supplementary Table S4). Five of these functional interactors were identified in let-7 PD experiments, either in let-7 PD alone or in let-7 PD plus additional precipitation experiments (Fig. 6b, Supplementary Table S4), suggesting that let-7 interacting factors indeed functionally coordinate with let-7 activity. Knockdown of 16 putative interactors did not modify vulval bursting phenotype of let-7(n2853) (Fig. 6b), perhaps due to tissue or time specific physical interactions of these proteins with let-7 miRNA or ALG-1 complexes. Such spatio-temporal complex compositions could explain the corresponding lack of activity in vulval tissue. We cannot, however, rule out insufficient RNAi knockdown or non-specific interactions of these proteins with anti-let-7 oligonucleotide. Knockdown of 18/38 candidate genes genetically modified hypodermal and/or seam cell lineage defects of mir-48 mir-241(nDf51) mutants (Fig. 6c, Supplementary Table S4). 12 of these factors were identified in let-7 pulldowns from wild type and/or mir-48 mir-241; mir-84 mutant backgrounds, suggesting that let-7 PD proteomics captured factors that support let-7 family miRNA activity in developmental timing.
Summary of functionally relevant interactors and models for possible mechanisms through which miRNA and ALG-1 interactors affect gene regulation. (a) Venn diagram showing the overlap of functional hits among the three functional assays. (b–d) Percentages of assayed factors that modulated phenotypes upon their respective gene knockdowns in (b) let-7(n2853), (c) mir-48 mir-241(nDf51), and (d) lsy-6(ot150) assays, with protein physical interaction status shown on the x-axis. *One ALG-1 interactor was also identified in miR-2 pulldown in addition to ALG-1 IP. The number of genes that modulated miRNA phenotypes over total number of genes tested is shown above respective bars. (e–f) RNA binding proteins found interacting with ALG-1 as well as miRNAs could be involved in (e) miRNA duplex processing and/or (f) facilitating miRISC activity on the targets by associating with downstream effectors. (g) Metabolic enzymes with putative RNA binding ability, such as ECH-5, could bind target mRNAs and promote miRISC activity by recruiting deadenylases and decapping proteins. Panels (e,f), and (g) were drawn using BioRender (https://biorender.com).
As lsy-6 miRNA activity is highly localized and unrelated to miRNAs precipitated in our PD experiments, lsy-6(ot150) mutation provided a convenient genetic background to identify which factors may be broadly involved miRNA-mediated gene regulation. Of the eight functional hits from the lsy-6(ot150) assay, seven factors were identified as ALG-1 interactors (Fig. 6d, Supplementary Table S4), consistent with the idea that ALG-1 IP perhaps precipitated proteins with broad specificities. Lack of lsy-6(ot150) phenotype modification by knockdown of let-7 and/or miR-58-associated proteins suggests that miRNA-centered complexes may be unique to the specific miRNAs, possibly due to distinct spatial or temporal expression patterns.
Several genes that modified let-7(n2853) vulval bursting in our study, cey-1, ifg-1, pab-1, and rbd-1 (Fig. 3b,c, Supplemental Table S4), were previously tested in an RNAi screen for suppressors of let-7(n2853) vulval bursting, aimed at identifying let-7 target genes55. We observed multiple differences between the results of our RNAi screen, performed at the permissive temperature of 15 °C and the previous work, performed at non-permissive 25 °C55, which eliminates let-7 activity. For example, rbd-1 knockdown enhanced vulval bursting at 15 °C (Fig. 3c, Supplementary Table S4), while it suppressed bursting at 25 °C55, suggesting that rbd-1 may have both let-7 dependent and independent functions. Knockdown of cey-1 suppressed let-7(n2853) vulval bursting at 15 °C in our study (Fig. 3c, Supplementary Table S4), however, no let-7(n2853) suppression was observed at 25 °C upon cey-1 knockdown55. These observations suggest that cey-1 may coordinate with let-7 in target mRNA regulation. Direct comparisons across RNAi studies performed under different conditions can be difficult to interpret and further explorations will be needed to understand the roles of these genes in let-7-mediated regulation of gene expression.
How could these putative physical miRNA interactors be coordinating with miRNAs to regulate gene expression? The factors identified in this study could be acting via multiple mechanisms to affect miRNA mutant phenotypes. Some of the miRNA interactors identified in this study have wide-ranging roles in regulation of gene expression. Thus, their knockdown could modify the miRNA reduction-of-function phenotypes directly through miRNA regulation and/or indirectly through regulation of mRNA lifecycle. For example, pab-1, a poly(A) binding protein and a homolog of human PABPC156, has well established roles in regulating the stability of mRNA transcripts by affecting translation initiation and mRNA stabilization and decay. PAB-1 has been previously shown to interact with miRISC19,56 and to aid miRNA-mediated deadenylation56. The enhancement of miRNA reduction-of-function phenotypes upon pab-1 knockdown may therefore be a result of miRNA-dependent and/or independent functions of pab-1, perhaps through loss of target mRNA deadenylation and subsequent mRNA stabilization.
We used the biological and molecular functions predicted by GO term analysis to consider the possible mode of action for the identified miRNA and miRISC interactors. RNA binding proteins were among the classes of genes enriched in our pulldowns (Fig. 2a–h, Supplementary Table S3). Through functional assays, we identified nine interactors with predicted and/or experimentally validated RNA binding activity as genetic interactors of miRNA mutants (Supplementary Table S4). Interestingly, knockdown of genes encoding all nine RNA binding proteins enhanced miRNA mutant phenotypes, consistent with the recent finding that 3’UTR-binding RBPs generally promote miRISC targeting57. Some of these RBPs could play a role in miRNA processing (Fig. 6e), some RBPs could potentially facilitate miRISC targeting or activity (Fig. 6f), while other RBPs could regulate localization and stability of miRNAs and/or miRNA targets, ultimately affecting gene regulation.
Translation regulators were also captured in miRNA pulldowns and ALG-1 IP (Supplementary Tables S1, S3), with two of them modifying miRNA phenotypes. Depletion of ifg-1, encoding translation initiation factor 4G (eIF4G)58, suppressed the ASEL cell fate defect of lsy-6(ot150) (Fig. 5b). Given the potential physical association of IFG-1 with ALG-119, it is possible that IFG-1 and miRNAs share common targets; with loss of IFG-1 reducing translation through loss of initiation, thereby suppressing target mRNA overexpression in miRNA reduction of function mutants. RNA helicase F33D11.10 co-precipitated in let-7 pulldown (Supplementary Table S1) and was previously identified as an interactor of ALG-119. Loss of F33D11.10 activity enhanced the ASEL cell fate defect of lsy-6(ot150) mutant (Fig. 5c). RNA helicases have been previously implicated in miRNA processing as well as miRISC activity59 and F33D11.10 may be similarly involved in either facilitating miRNA processing, miRISC activity, or both.
A surprising category of interactors identified in our study was the intermediary metabolic enzymes. RNAi depletion of metabolic enzymes DLST-1, OGDH-1 and ECH-5 modified miRNA mutant phenotypes in our study (Figs. 3c, 4d,f, Supplementary Table S4). Several reports have suggested that some metabolic enzymes possess RNA binding functions, previously unidentified due to a lack of conventional RNA binding domains60,61,62. It is possible that the metabolic enzymes identified in our study possess similar dual roles. For instance, ech-5 encodes a homolog of human AU RNA binding methylglutaconyl-CoA hydratase (AUH)63. In humans, AUH plays a dual role as a hydratase and as an RBP, binding AU-rich elements in the 3′ UTR of mRNAs64. Other ARE-binding proteins have been previously shown to aid in rapid degradation through deadenylation65. In our study, ECH-5 co-precipitated with let-7 miRNAs and ech-5 depletion enhanced let-7(n2853) vulval bursting phenotype (Fig. 3c, Supplementary Table S1). Thus, we might speculate ECH-5 could bridge miRISC complex interaction with deadenylation machinery, with loss of ech-5 exacerbating the target mRNA stabilization in miRNA mutant backgrounds (Fig. 6g). How other metabolic genes such as dlst-1 and ogdh-1, key players of TCA cycle66,67, influence gene regulation remains unclear. Thorough investigations into molecular mechanisms by which these factors coordinate with miRNAs in gene regulation will be needed.
Interestingly, LET-363, C. elegans mTOR was identified as an interactor of all three miRNAs in this study (Fig. 1f, Supplementary Table S1). RNAi of let-363 enhanced the vulval bursting of let-7(n2853) and suppressed the seam cell lineage defect of mir-48 mir-241(nDf51) mutant (Figs. 3c, 4f, Supplemental Table S4). While we did not test for functional let-363 requirement in our lsy-6(ot150) assay, RNAi of let-363 was previously reported to exacerbate the cell fate specification defect of lsy-6(ot150)50, consistent with a let-363 role in miRNA-mediated gene regulation. mTOR activation has been reported to downregulate miRNA biogenesis through Mdm2-mediated DROSHA degradation in mice68. However, the physical association of LET-363 with miRNAs was surprising. If confirmed, these persistent physical and functional interactions of LET-363 with miRNA-centered complexes should be further explored to establish the mechanistic connection between mTOR and miRNA-mediated gene regulatory activity.
Do miRNAs within the same family associate with same set of protein interactors? let-7 family miRNAs are well-studied in C. elegans. The four most abundant members of the let-7 family, let-7, mir-48, mir-84 and mir-241, are crucial components of the heterochronic pathway, regulating cell fates during larval development32,53. The miRNAs are thought to function semi-redundantly, with distinct targeting capabilities69. Part of their ability to target unique targets could come from discrete protein interactors adding a layer of specificity between a let-7 family miRNA and its target. Yet not much is known about the protein interacting partners of individual members of this family. By performing pulldowns with a let-7 specific oligo from wild type and mir-48 mir-241(nDf51); mir-84(n4037) background, we began the task of unraveling which interactors may be specific to let-7 itself or other family members (Fig. 1h, Supplementary Table S1). For example, ECH-5 and C04E6.11 were identified in let-7 pulldowns from both genetic backgrounds, suggesting that they most likely interact with let-7 (Supplementary Table S1). RNAi depletion of ech-5 or C04E6.11 enhanced the let-7(n2853) phenotype but showed no effect on the mir-48 mir-241(nDf51) associated phenotypes (Figs. 3c, 4d). Thus, it is possible that ECH-5 and C04E6.11 specifically interact with, and provide functional support for, let-7 itself. CEY-1 was captured in let-7 pulldowns from both wild type and mir-48 mir-241; mir-84 mutant background, but depletion of cey-1 suppressed let-7(n2853) vulval bursting and enhanced mir-48 mir-241(nDf51) mutant phenotypes (Figs. 3c, 4d). This suggests that cey-1 may functionally interact with multiple members of the let-7 family, potentially through distinct mechanisms. Previously, miR-241 complementary oligo pulldown captured CEY-144, although it remains difficult to assess specificity, as miR-241 oligo may capture other members of the family, similar to let-747. Overall, we cannot rule out the possibility that some of the factors precipitating in miRNA pulldowns are non-specifically interacting with the precipitating oligo, rather than with the miRNA-centered complexes. It is also possible that let-7 interactions may be altered in the mir-48 mir-241(nDf51); mir-84(n4037) background. Similarly, the severe developmental timing defect of mir-48 mir-241(nDf51); mir-84(n4037) animals could hinder identification of bona fide interactors of let-7 in that background. However, the high rate of functional relevance of these factors for miRNA-mediated gene regulation suggests that this approach captures auxiliary factors that may coordinate with miRNAs mediated gene regulation.
References
Ma, F., Zhang, X. & Yin, K.-J. MicroRNAs in central nervous system diseases: A prospective role in regulating blood–brain barrier integrity. Exp. Neurol. 323, 113094 (2019).
Ebrahimi, S. O., Reiisi, S. & Shareef, S. miRNAs, oxidative stress, and cancer: A comprehensive and updated review. J. Cell Physiol. 235, 8812–8825 (2020).
Bagga, S. et al. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell 122, 553–563 (2005).
Mayya, V. K. et al. microRNA-mediated translation repression through GYF-1 and IFE-4 in C. elegans development. Nucleic Acids Res. 49, gkab162 (2021).
Lee, Y. et al. The nuclear RNase III Drosha initiates microRNA processing. Nature 425, 415–419 (2003).
Han, J. et al. Molecular basis for the recognition of primary microRNAs by the drosha-DGCR8 complex. Cell 125, 887–901 (2006).
Hutvágner, G. et al. A cellular function for the RNA-interference enzyme dicer in the maturation of the let-7 small temporal RNA. Science 293, 834–838 (2001).
MacRae, I. J., Zhou, K. & Doudna, J. A. Structural determinants of RNA recognition and cleavage by Dicer. Nat. Struct. Mol. Biol. 14, 934–940 (2007).
Kwak, P. B. & Tomari, Y. The N domain of Argonaute drives duplex unwinding during RISC assembly. Nat. Struct. Mol. Biol. 19, 145–151 (2012).
Meijer, H. A., Smith, E. M. & Bushell, M. Regulation of miRNA strand selection: Follow the leader?. Biochem. Soc. Trans. 42, 1135–1140 (2014).
Kim, K. K., Yang, Y., Zhu, J., Adelstein, R. S. & Kawamoto, S. Rbfox3 controls the biogenesis of a subset of microRNAs. Nat. Struct. Mol. Biol. 21, 901–910 (2014).
Guil, S. & Cáceres, J. F. The multifunctional RNA-binding protein hnRNP A1 is required for processing of miR-18a. Nat. Struct. Mol. Biol. 14, 591–596 (2007).
Hammell, C. M., Lubin, I., Boag, P. R., Blackwell, T. K. & Ambros, V. nhl-2 Modulates microRNA activity in Caenorhabditis elegans. Cell 136, 926–938 (2009).
Ren, Z., Veksler-Lublinsky, I., Morrissey, D. & Ambros, V. Staufen negatively modulates microRNA activity in Caenorhabditis elegans. G3 Genes Genomes Genet. 6, 1227–1237 (2016).
Srikantan, S., Tominaga, K. & Gorospe, M. Functional Interplay between RNA-binding protein HuR and microRNAs. Curr. Protein Pept. Sci. 13, 372–379 (2012).
Kundu, P., Fabian, M. R., Sonenberg, N., Bhattacharyya, S. N. & Filipowicz, W. HuR protein attenuates miRNA-mediated repression by promoting miRISC dissociation from the target RNA. Nucleic Acids Res. 40, 5088–5100 (2012).
Landthaler, M. et al. Molecular characterization of human Argonaute-containing ribonucleoprotein complexes and their bound target mRNAs. RNA 14, 2580–2596 (2008).
Frohn, A. et al. Dicer-dependent and -independent Argonaute2 protein interaction networks in mammalian cells*. Mol. Cell Proteomics 11, 1442–1456 (2012).
Zinovyeva, A. Y., Veksler-Lublinsky, I., Vashisht, A. A., Wohlschlegel, J. A. & Ambros, V. R. Caenorhabditis elegans ALG-1 antimorphic mutations uncover functions for Argonaute in microRNA guide strand selection and passenger strand disposal. Proc. Natl. Acad. Sci. 112, E5271–E5280 (2015).
Wu, E. et al. A continuum of mRNP complexes in embryonic microRNA-mediated silencing. Nucleic Acids Res. 45, 2081–2098 (2017).
Iwasaki, S. et al. Hsc70/Hsp90 chaperone machinery mediates ATP-dependent RISC loading of small RNA duplexes. Mol. Cell 39, 292–299 (2010).
Nishihara, T., Zekri, L., Braun, J. E. & Izaurralde, E. miRISC recruits decapping factors to miRNA targets to enhance their degradation. Nucleic Acids Res. 41, 8692–8705 (2013).
Kamath, R. S. & Ahringer, J. Genome-wide RNAi screening in Caenorhabditis elegans. Methods 30, 313–321 (2003).
Li, L. & Zinovyeva, A. Y. Protein extract preparation and Co-immunoprecipitation from Caenorhabditis elegans. J. Vis. Exp. https://doi.org/10.3791/61243 (2020).
Jannot, G., Vasquez-Rifo, A. & Simard, M. J. Argonaute proteins, methods and protocols. Methods Mol. Biol. 725, 233–249 (2011).
Tabb, D. L., McDonald, W. H. & Yates, J. R. DTASelect and contrast: Tools for assembling and comparing protein identifications from shotgun proteomics. J. Proteome Res. 1, 21–26 (2002).
Huang, D. W., Sherman, B. T. & Lempicki, R. A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 (2009).
Snel, B., Lehmann, G., Bork, P. & Huynen, M. A. STRING: A web-server to retrieve and display the repeatedly occurring neighbourhood of a gene. Nucleic Acids Res. 28, 3442–3444 (2000).
Parry, D. H., Xu, J. & Ruvkun, G. A whole-genome RNAi screen for C. elegans miRNA pathway genes. Curr. Biol. 17, 2013–2022 (2007).
Porta-de-la-Riva, M., Fontrodona, L., Villanueva, A. & Cerón, J. Basic Caenorhabditis elegans methods: Synchronization and observation. J. Vis. Exp. https://doi.org/10.3791/4019 (2012).
Pasquinelli, A. E. et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature 408, 86–89 (2000).
Reinhart, B. J. et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 403, 901–906 (2000).
Jan, C. H., Friedman, R. C., Ruby, J. G. & Bartel, D. P. Formation, regulation and evolution of Caenorhabditis elegans 3′UTRs. Nature 469, 97–101 (2011).
de Lucas, M. P., Sáez, A. G. & Lozano, E. miR-58 family and TGF-β pathways regulate each other in Caenorhabditis elegans. Nucleic Acids Res. 43, 9978–9993 (2015).
Subasic, D. et al. Cooperative target mRNA destabilization and translation inhibition by miR-58 microRNA family in C. elegans. Genome Res. 25, 1680–1691 (2015).
Marco, A., Hooks, K. & Griffiths-Jones, S. Evolution and function of the extended miR-2 microRNA family. RNA Biol. 9, 242–248 (2012).
O’Hern, P. J. et al. Decreased microRNA levels lead to deleterious increases in neuronal M2 muscarinic receptors in Spinal Muscular Atrophy models. Elife 6, e20752 (2017).
Grishok, A. et al. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell 106, 23–34 (2001).
Ding, L., Spencer, A., Morita, K. & Han, M. The developmental timing regulator AIN-1 interacts with miRISCs and may target the argonaute protein ALG-1 to cytoplasmic P bodies in C. elegans. Mol. Cell 19, 437–447 (2005).
Zhang, L. et al. Systematic identification of C. elegans miRISC proteins, miRNAs, and mRNA targets by their interactions with GW182 proteins AIN-1 and AIN-2. Mol. Cell 28, 598–613 (2007).
Li, L., Veksler-Lublinsky, I. & Zinovyeva, A. HRPK-1, a conserved KH-domain protein, modulates microRNA activity during Caenorhabditis elegans development. PLoS Genet. 15, e1008067 (2019).
Long, X. et al. TOR deficiency in C. elegans causes developmental arrest and intestinal atrophy by inhibition of mRNA translation. Curr. Biol. 12, 1448–1461 (2002).
Wu, E. et al. Pervasive and cooperative deadenylation of 3′UTRs by embryonic microRNA families. Mol. Cell 40, 558–570 (2010).
Dallaire, A., Frédérick, P.-M. & Simard, M. J. Somatic and germline microRNAs form distinct silencing complexes to regulate their target mRNAs differently. Dev. Cell 47, 239-247.e4 (2018).
Kim, J. K. et al. Functional genomic analysis of RNA interference in C. elegans. Science 308, 1164–1167 (2005).
Zhou, R. et al. Comparative analysis of argonaute-dependent small RNA pathways in Drosophila. Mol. Cell 32, 592–599 (2008).
Zinovyeva, A. Y., Bouasker, S., Simard, M. J., Hammell, C. M. & Ambros, V. Mutations in conserved residues of the C. elegans microRNA argonaute ALG-1 identify separable functions in ALG-1 miRISC loading and target repression. PLoS Genet. 10, e1004286 (2014).
Vella, M. C., Choi, E.-Y., Lin, S.-Y., Reinert, K. & Slack, F. J. The C. elegans microRNA let-7 binds to imperfect let-7 complementary sites from the lin-41 3′UTR. Gene Dev. 18, 132–137 (2004).
Ecsedi, M., Rausch, M. & Großhans, H. The let-7 microRNA directs vulval development through a single target. Dev. Cell 32, 335–344 (2015).
Zhang, P. & Zhang, H. Autophagy modulates miRNA-mediated gene silencing and selectively degrades AIN-1/GW182 in C. elegans. Embo Rep. 14, 568–576 (2013).
Abbott, A. L. et al. The let-7 microRNA family members mir-48, mir-84, and mir-241 function together to regulate developmental timing in Caenorhabditis elegans. Dev. Cell 9, 403–414 (2005).
Abrahante, J. E. et al. The Caenorhabditis elegans hunchback-like gene lin-57/hbl-1 controls developmental time and is regulated by microRNAs. Dev. Cell 4, 625–637 (2003).
Johnston, R. J. & Hobert, O. A microRNA controlling left/right neuronal asymmetry in Caenorhabditis elegans. Nature 426, 845–849 (2003).
Palmer, R. E., Inoue, T., Sherwood, D. R., Jiang, L. I. & Sternberg, P. W. Caenorhabditis elegans cog-1 locus encodes GTX/Nkx6.1 homeodomain proteins and regulates multiple aspects of reproductive system development. Dev. Biol. 252, 202–213 (2002).
Rausch, M., Ecsedi, M., Bartake, H., Müllner, A. & Großhans, H. A genetic interactome of the let-7 microRNA in C. elegans. Dev. Biol. 401, 276–286 (2015).
Flamand, M. N. et al. Poly(A)-binding proteins are required for microRNA-mediated silencing and to promote target deadenylation in C. elegans. Nucleic Acids Res. 44, 5924–5935 (2016).
Kim, S. et al. The regulatory impact of RNA-binding proteins on microRNA targeting. Nat. Commun. 12, 5057 (2021).
Pan, K. Z. et al. Inhibition of mRNA translation extends lifespan in Caenorhabditis elegans. Aging Cell 6, 111–119 (2007).
Chu, Y.-D., Chen, H.-K., Huang, T. & Chan, S.-P. A novel function for the DEAD-box RNA helicase DDX-23 in primary microRNA processing in Caenorhabditis elegans. Dev. Biol. 409, 459–472 (2016).
Beckmann, B. M. et al. The RNA-binding proteomes from yeast to man harbour conserved enigmRBPs. Nat. Commun. 6, 10127 (2015).
Curtis, N. J. & Jeffery, C. J. The expanding world of metabolic enzymes moonlighting as RNA binding proteins. Biochem. Soc. Trans. 49, 1099–1108 (2021).
Rodríguez-Saavedra, C. et al. Moonlighting proteins: The case of the hexokinases. Front. Mol. Biosci. 8, 701975 (2021).
Yilmaz, L. S. & Walhout, A. J. M. A Caenorhabditis elegans genome-scale metabolic network model. Cell Syst. 2, 297–311 (2016).
Kurimoto, K. et al. AU-rich RNA-binding induces changes in the quaternary structure of AUH. Proteins Struct. Funct. Bioinform. 75, 360–372 (2009).
Otsuka, H., Fukao, A., Funakami, Y., Duncan, K. E. & Fujiwara, T. Emerging evidence of translational control by AU-rich element-binding proteins. Front. Genet. 10, 332 (2019).
Zhang, Z. et al. Broadly conserved roles of TMEM131 family proteins in intracellular collagen assembly and secretory cargo trafficking. Sci. Adv. 6, eaay7667 (2020).
Castelein, N., Hoogewijs, D., Vreese, A. D., Braeckman, B. P. & Vanfleteren, J. R. Dietary restriction by growth in axenic medium induces discrete changes in the transcriptional output of genes involved in energy metabolism in Caenorhabditis elegans. Biotechnol. J. 3, 803–812 (2008).
Ye, P. et al. An mTORC1-Mdm2-Drosha axis for miRNA biogenesis in response to glucose- and amino acid-deprivation. Mol. Cell. 57, 708–720 (2015).
Broughton, J. P., Lovci, M. T., Huang, J. L., Yeo, G. W. & Pasquinelli, A. E. Pairing beyond the seed supports microRNA targeting specificity. Mol. Cell 64, 320–333 (2016).
Acknowledgements
We thank members of the Zinovyeva lab for helpful discussions and technical assistance. We are most grateful to Victor Ambros, in whose lab this work was initiated. We thank Erik Lundquist and Helge Grosshans for sharing reagents and strains. Some of the strains were provided by the CGC (Caenorhabditis Genetics Center), which is funded by the National Institutes of Health Office of Research Infrastructure Programs P40-OD010440.
Author information
Authors and Affiliations
Contributions
S.H.: Study concept and design, performing the experiments, analysis and interpretation of data, prepared figures, manuscript writing. G.P.P.: Data analysis. I.V.-L.: Data analysis and editing manuscript. J.A.W. and A.A.V.: Proteomics experiments and data analysis. A.Y.Z.: Study concept and design, performing the experiments, editing manuscript, technical and material support, study supervision.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Hebbar, S., Panzade, G., Vashisht, A.A. et al. Functional identification of microRNA-centered complexes in C. elegans. Sci Rep 12, 7133 (2022). https://doi.org/10.1038/s41598-022-10771-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-022-10771-2
- Springer Nature Limited