Abstract
Selection of appropriate biomarker to identify inflammatory skin diseases is complicated by the involvement of thousands of differentially expressed genes (DEGs) across multiple cell types and organs. This study aimed to identify combinatorial biomarkers in inflammatory skin diseases. From one gene expression microarray profiling dataset, we performed bioinformatic analyses on dataset from lesional skin biopsies of patients with inflammatory skin diseases (atopic dermatitis [AD], contact eczema [KE], lichen planus [Li], psoriasis vulgaris [Pso]) and healthy controls to identify the involved pathways, predict upstream regulators, and potential measurable extracellular biomarkers. Overall, 434, 629, 581, and 738 DEGs were mapped in AD, KE, Li, and Pso, respectively; 238 identified DEGs were shared among four different inflammatory skin diseases. Bioinformatic analysis on four inflammatory skin diseases showed significant activation of pathways with known pathogenic relevance. Common upstream regulators, with upregulated predicted activity, identified were CNR1 and BMP4. We found the following common serum biomarkers: ACR, APOE, ASIP, CRISP1, DKK1, IL12B, IL9, MANF, MDK, NRTN, PCSK5, and VEGFC. Considerable differences of gene expression changes, involved pathways, upstream regulators, and biomarkers were found in different inflammatory skin diseases. Integrated bioinformatic analysis identified 12 potential common biomarkers of inflammatory skin diseases requiring further evaluation.
Similar content being viewed by others
Introduction
Inflammatory skin diseases are complex, chronic, multifactorial disorders, which are characterized by activation of the innate and adaptive immune system via production of proinflammatory cytokines1. Environmental, genetic, and immunologic factors apparently play a role in the pathogenesis of inflammatory skin diseases2. Itchy, red, or unsightly inflammatory skin conditions, such as hives, eczema, or psoriasis, can affect the quality of life and may be associated with psychological distress for the patients. Furthermore, common chronic inflammatory skin diseases, including atopic dermatitis (AD), contact eczema (KE), lichen planus (Li), and psoriasis vulgaris (Pso), manifest a relapsing and remitting course throughout the patient’s life1,2.
The host skin-based defence system comprises a barrier, innate immunity, and acquired immunity. The pathophysiology of inflammatory skin diseases involves various inflammatory cells and the innate immune response1. A recent study suggested that epigenetic factors, a core subset of inflammation-associated differentially methylated genes, were crucially involved in the pathophysiology of inflammatory skin diseases3. As early diagnosis and monitoring are important to prevent disease progression, reliable biomarkers are needed. However, despite decades of painstaking research with genome-wide analyses, such measurable biomarkers have proven elusive. That is why current diagnostic strategies are predominantly based on clinic-pathological reports in combination with the clinical history and physical examination4,5. With bioinformatic research into the molecular network in the skin, targeted therapies and early therapeutic intervention have revolutionized management strategies in clinical practice6.
We hypothesized that novel and potentially more specific biomarkers for inflammatory skin disease could be identified through a bioinformatic analysis of existing profiling data obtained from skin tissues of patients with inflammatory skin diseases, in comparison to the data from healthy controls (HCs). Bioinformatical analysis, especially ingenuity pathway analysis (IPA), is focused on pathways and upstream regulators rather than individual genes and provides a functional overview of the complex gene expression changes in each dataset7,8,9. The analysis on pathways and upstream regulators suggested a considerable complexity causing difficulties in finding representative biomarkers. Therefore, we aimed to identify combinatorial biomarkers in inflammatory skin diseases.
Results
Selection of eligible microarray datasets
We searched the Gene Expression Omnibus (GEO) database using the terms ‘inflammatory skin disease, ‘human’, ‘healthy control’, and ‘microarray’. This resulted in the identification of one mRNA transcriptional profiling study (GSE 63741) of whole-skin biopsies from control subjects and diseased skin tissues from patients with inflammatory skin diseases, such as AD, KE, Li, and Pso, versus the HC10. This study performed an unsupervised cluster analysis of gene expression profiles in 30 Pso patients and other inflammatory skin diseases (30 AD, 30 Li, 30 KE, and 30 HC). The study was performed using the platform, PIQOR (TM) Skin 2.0 Microarray, human, antisense (591).
Identification of differentially expressed genes in skin biopsies from patients with inflammatory skin disease
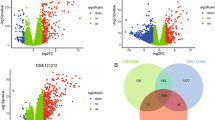
We systematically analysed the datasets for differentially expressed genes (DEGs), shared pathways, predicted upstream regulators, and biomarkers. We initially identified the DEGs from the diseased skin samples of patients with inflammatory skin diseases versus the healthy skin samples from HC. We found that AD, KE, Li, and Pso had 434, 629, 581, and 738 DEGs, respectively (Supplementary Fig. 1, Supplementary Table 1); 238 DEGs overlapped among the four datasets (Fig. 1). These genes contained several known factors that are relevant for inflammatory skin diseases, including human leukocyte antigen genes, chemokines, cytokines, and T-cell immune-regulator genes, such as TCIRG111. These analyses from both integration of gene ontology (GO) terms for DEGs and protein–protein interaction network supported the pathophysiological relevance of the shared 238 DEGs from the inflammatory skin lesions (Supplementary Fig. 2, Supplementary Table 2)12,13,14. Despite the presence of shared 238 DEGs, four different kinds of inflammatory skin disease represent different phenotypes. This led us to the current system-level analysis to examine whether there were pathway overlaps among the four different datasets.
Identification of pathways in skin biopsy specimens from patients with inflammatory skin disease
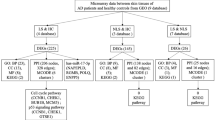
To obtain a functional overview of the complex gene expression changes in inflammatory skin diseases, we performed the IPA to identify disease-associated pathways. This resulted in the identification of a vast number of involved pathways for different inflammatory skin diseases (Supplementary Table 3). The top significant pathways (Z-score > 2) included signalling by Rho Family GTPases (2.646), white adipose tissue-browning pathway (2.236), xenobiotic metabolism general signalling pathway (2.236), cardiac hypertrophy signalling (2.236), regulation of actin-based motility by Rho (2), Tec kinase signalling (2) in AD, regulation of actin-based motility by Rho (2.236), xenobiotic metabolism aryl hydrocarbon receptor signalling pathway (2.236), Cdc42 signalling (2.236), RhoA signalling (2.236), estrogen receptor signalling (2.183), semaphorin neuronal repulsive signalling pathway (2.111), ethanol degradation II (2), agrin interactions at neuromuscular junction (2), STAT3 pathway (2) in KE, xenobiotic metabolism pregnane X receptor signalling pathway (2.449), xenobiotic metabolism AHR signalling pathway (2.236), ethanol degradation II (2), noradrenaline and adrenaline degradation (2), regulation of actin-based motility by Rho (2), Cdc42 signalling (2), serotonin degradation (2), synaptic long term potentiation (2) in LP, serotonin degradation (2.236), and synaptic long term depression (2) in Pso. Interestingly, comparative analysis of the four inflammatory skin diseases showed a similar pattern, which may explain the difficulty in clinically ascertaining the differential diagnosis in inflammatory skin diseases (Fig. 2a). These findings agreed with our previous findings that, despite the limited overlap of single genes between the gene expression profiling studies, there can be significant pathway overlap15,16. Collectively, the pathway analysis indicate a dynamic pathogenic complexity, which reflects a great challenge to the prioritization of biomarkers for inflammatory skin diseases.
Identification of upstream regulators in inflammatory skin diseases
Next, we performed IPA analysis of the DEGs to identify the predicted activated upstream regulators of those DEGs from the diseased skin samples of patients with inflammatory skin diseases. The rationale for the analysis was that, similar to the pathways, such regulators constitute higher order representations of the complex gene expression changes (Supplementary Table 4). Paralleling with the pathways, the comparative analysis of upstream regulators showed a similar pattern among the four different inflammatory diseases (Fig. 2b). We listed 5, 8, 10, and 4 predicted activated upstream regulators in AD, KE, Li, and Pso, respectively (Table 1). In AD, the predicted activated upstream regulators (Z-scores) included CNR1 (2.714), BMP4 (2.431), AMH (2.181), and CDK4/6 (2). In KE, predicted activated upstream regulators included BMP4 (2.887), CNR1 (2.496), GH1 (2.433), AMH (2.385), BMP2 (2.357), TNFSF15 (2.213), CCR2 (2.2), and EDN1 (2.152). In Li, predicted activated upstream regulators included BMP4 (2.942), IL15 (2.449), BMP2 (2.406), Jnk (2.397), CNR1 (2.35), IL7 (2.296), CDK4/6 (2.236), AMH (2.181), TNFRSF8 (2.164), and Pkc(s) (2.132). In Pso, predicted activated upstream regulators included CNR1 (2.496), BMP4 (2.3), and CAV1 (2.216). Interestingly, CNR1 and BMP4 are the two common predicted activated upstream regulators. Consistent with the fact that the Rho GTPase family signalling is most significant, the most activated predicted upstream regulator in the two studies was BMP4 (p = 1.01E−34), which can be regulated by Rho-kinase. Other important shared upstream regulators included CNR1 (p = 6.43E−33), which is an important therapeutic target in inflammatory skin diseases17.
Identification of biomarkers in inflammatory skin diseases
Finally, we used IPA to search for DEGs that encoded biomarkers. We started by screening for genes that encoded biomarkers that had been described in any disease and performed IPA to identify the potential biomarkers of those DEGs. We found 416, 605, 556, and 722 biomarkers in AD, KE, Li, and Pso, respectively (Supplementary Table 5). For clinical use, biomarkers should be measurable in the blood; therefore, we sorted potential biomarkers that were measurable in blood samples, such as the extracellular biomarkers. This identified 44, 58, 50, and 75 extracellular biomarkers in each disease (Supplementary Table 6). Next, we checked whether these extracellular biomarkers were elevated in disease conditions compared to the levels in HC, which is indicated by a positive log fold-change value. We found 12 candidate common extracellular biomarkers for all inflammatory skin diseases as follows: ACR (acrosin), APOE (apolipoprotein E), ASIP (agouti signalling protein), CRISP1 (cysteine rich secretory protein 1), DKK1 (dickkopf WNT signalling pathway inhibitor 1), IL12B (interleukin 12B), IL9 (interleukin 9), MANF (mesencephalic astrocyte derived neurotrophic factor), MDK (midkine), NRTN (neurturin), PCSK5 (proprotein convertase subtilisin/kexin type 5), and VEGFC (vascular endothelial growth factor C) (Table 2). Among the 12 candidate common biomarkers, DKK1, IL12B, and VEGFC have been targeted by known drugs in clinical settings18,19,20. Moreover, the IL12B is a known biomarker for single-nucleotide polymorphisms in psoriasis, and a monoclonal antibody against the IL-12/23 subunit is used clinically21,22.
Discussion
We found 238 shared DEGs that showed differing expressions between samples from inflammatory skin disease and HCs based on gene expression profiling data. Further bioinformatic analyses led to the identification of a wide variety of pathways and predicted upstream regulators. This complexity could explain the difficulties in identifying representative biomarkers. Although different types of biomarkers were found in each disease, we finally identified the following common biomarkers: ACR, APOE, ASIP, CRISP1, DKK1, IL12B, IL9, MANF, MDK, NRTN, PCSK5, and VEGFC, which are measurable in the serum and reflect extracellular biomarkers.
Previous studies by us and others have indicated altered expression of thousands of genes across multiple cell types locally and in blood samples from patients with inflammatory skin diseases3,4,5. This finding is consistent with the increasing recognition of the pathogenic complexity of inflammatory skin disease; however, the gene expression changes could ideally be exploited to identify highly accurate combinations of biomarkers15. From a clinical perspective, diagnostic proteins or soluble biomarkers in inflammatory skin disease are difficult to measure locally, within specimens from the diseased skin. Instead, analysis of blood samples would be more convenient. As mentioned in the introduction, there is a large collection of evidence about biomarkers or cytokine signatures in inflammatory skin diseases6. There is, however, a need for more accurate measurable biomarkers, which ideally should reflect the local disease activity. Furthermore, only few studies have tried to elucidate the inflammatory signatures and involved pathways as well as the upstream regulators in each disease, although all inflammatory skin diseases are similarly considered to be inflammatory autoimmune diseases predominantly driven by T cells. Therefore, in this study, we obtained a functional overview of the complex gene expression changes in different inflammatory diseases by focusing on pathways, upstream regulators, and measurable biomarkers rather than individual genes.
First, we checked pathways in each disease using DEGs identified from diseased patients versus HC, and comparative analysis showed quite similar patterns, which may explain the difficulties in the differential diagnosis of different inflammatory skin diseases. Despite the similar patterns observed in the comparative analysis, the rankings of individual significant pathways appeared different among each disease, reflecting the difficulty in biomarker prioritization. Next, we checked the predicted upstream regulators that affect DEGs and found that there are two common predicted activated upstream regulators—CNR1 and BMP4. Sconocchia et al. demonstrated that BMP signalling plays an important role in inflammatory Treg-cell accumulation during skin inflammation23. Moreover, psoriatic lesions are marked by constitutive high BMP7/BMPR signalling in keratinocytes, which instructs inflammatory dendritic cells to enhance Treg-cell–stimulatory activity23. Moreover, Kim et al. showed that the BMP-4 expression of epithelial cells was higher in oral Li, which suggested that the overexpression of BMP-4 was a crucial factor for the apoptosis of epithelial cells in Li24. Furthermore, the human endogenous cannabinoid system (ECS) is a complex signalling network involved in a vast number of physiological processes. The endocannabinoids and cannabinoid receptor (CB) 1, which corresponds to CNR1 gene signalling, may play a potent inhibitory role in human mast cell degranulation and activation in the airway mucosa and skin, suggesting that targeting the ECS in these tissues might well represent a novel strategy for the treatment of allergy. Martin-Fontecha et al. showed that mRNA expression level of CNR1, the gene encoding CB1 protein—the main component of the ECS—is upregulated in the tonsils and peripheral blood of patients with three different types of allergic diseases: allergic rhinitis, AD, and food allergy25. This previous finding reflected the fact that the predicted upstream regulator, CNR1, can be a potential therapeutic target that is mediated by the ECS. Collectively, our findings about the common predicted upstream regulators concur with the findings of previous studies.
Of note, the upstream regulators were predicted based on their known effects on the downstream groups of genes. Thus, if a group of genes showed coordinated changes, potential upstream regulators of those changes were identified based on previous experimental data that were accumulated in IPA26. However, for clinical purposes, the application of predicted upstream regulators based on gene expression profiles is impractical. Instead, a limited number of protein biomarkers that can be measured using routinely available methods, either in the blood or in local tissues, can be ideally used. Therefore, we ultimately checked whether extracellular biomarkers are elevated in disease states compared to the levels in HC, based on a positive log fold-change value. For all inflammatory skin diseases, we found 12 common biomarker candidates: ACR, APOE, ASIP, CRISP1, DKK1, IL12B, IL9, MANF, MDK, NRTN, PCSK5, and VEGFC. The relevance of ApoE, DKK1, IL12B, IL9, MANF, and VEGFC in specific inflammatory skin diseases was identified in several previous studies, as discussed further.
A meta-analysis of seven studies indicated that ApoE polymorphisms, especially the ε2 and ε3 alleles, are associated with an increased risk of psoriasis27. A genome-wide association study (GWAS) confirmed the association of the IL12B and IL23R genes with psoriasis22. Interestingly, robust IL12A and IL12B expression was found in patients with chronic Li, and the expression of IL-9, IFN-γ, and IL-22 was higher in cutaneous Li28 than that in oral Li29. Polymorphisms in the IL-9 and IL-9R genes have been associated with AD30, and the serum IL-9 levels were not only increased in AD patients compared with HC but were also positively correlated with the severity of AD31. Hui et. al proposed the expression of IL-9R in epidermal keratinocytes is increased by IL-432; and IL-9 was shown to induce gene expression and vascular endothelial growth factor (VEGF) secretion from mast cells by involving STAT-3 activation33. These aspects imply a pathophysiological role for IL-9/IL-9R in the skin of AD patients. Tej et al. found significantly higher levels of IL-9R in psoriatic skin than in the skin of HC, and an intradermal injection of IL-9 promoted IL-17A production; these findings suggest that IL-9 may play a role in the development of psoriatic lesions through Th17-associated inflammation and angiogenesis34. VEGF-C, a lymphangiogenesis marker in skin biopsies of psoriatic lesion35, was confirmed to be intensely expressed in Pso36 as well as in cutaneous Li37.
The treatment of ankylosing spondylitis and rheumatoid arthritis with TNF-α inhibitors was associated with a concurrent decrease in the DKK1 serum levels38,39. These abovementioned studies emphasize the role of DKK1 as a protagonist in chronic immune-mediated diseases; therefore, DKK1 may serve as a biomarker for the pathogenetic activity of these diseases. Owing to the significance of Wnt signalling in angiogenesis, Wnt antagonists, such as DKK1, have been considered as potential treatments for neovascularization-related disorders40. Gudjonsson et al. reported evidence of altered Wnt signalling in the psoriatic skin41; therefore, DKK1, a Wnt antagonist, can be a possible biomarker in inflammatory skin disease. The bioinformatics analysis of the proteome from the serum of AD patients demonstrated the altered landscape of immunological aberrations, and MANF was significantly increased in AD patients compared to HC42. These findings support the assumption that DKK1 and MANF are potential biomarkers despite the protein–gene gap.
Recent findings have established the skin as a peripheral neuroendocrine organ that is tightly networked to the central stress axes43,44,45,46,47. Specifically, epidermal and dermal cells produce, and respond to, classical stress neurotransmitters, neuropeptides, and hormones. This production is modified by ultraviolet radiation and biological, chemical, and physical factors43,44. Examples of potent epidermal products include biogenic amines (catecholamines, serotonin, and N-acetyl-serotonin), acetylcholine, melatonin and its metabolites, proopiomelanocortin-derived ACTH, β-endorphin and MSH peptides, corticotropin-releasing factor and related urocortins, corticosteroids and their precursor molecules, thyroid-related hormones, opioids and cannabinoids45,46. The production of these molecules in the skin is hierarchical, following the algorithms of classical neuroendocrine axes (e.g., hypothalamic pituitary adrenal axis, hypothalamic-thyroid axis, serotoninergic/melatoninergic, catecholaminergic and cholinergic systems). The deregulation of these systems may be involved in the etiology of skin diseases, and the control of these systems constitute novel targets through the use of specific agonists or antagonists, especially for therapy of skin diseases45,46,47. In this study, we found that 238 DEGs partially overlapped between different inflammatory skin diseases and identified 12 candidate biomarkers that overlapped, namely, ACR, APOE, ASIP, CRISP1, DKK1, IL12B, IL9, MANF, MDK, NRTN, PCSK5, and VEGFC. In the future, biomarkers might play a central role in personalized therapy by facilitating the identification of patients who will not respond to a certain treatment or those who might get adverse reactions48. Moreover, biomarkers play a very important role in deepening our understanding of the pathogenesis of psoriasis and could facilitate the development of biological therapies49. As mentioned earlier, the pathogenic and diagnostic relevance of these biomarkers is supported by not only single-nucleotide polymorphism and GWAS studies, but also through studies of the mechanism, such as Wnt signalling50. When considered with our findings, future studies are warranted to evaluate ACR, APOE, ASIP, CRISP1, DKK1, IL12B, IL9, MANF, MDK, NRTN, PCSK5, and VEGFC as candidate biomarkers in inflammatory skin diseases.
The limitations of this study include the fact that only one microarray was analysed, and that mRNA levels may not necessarily correspond to the protein expression levels. Nonetheless, the study findings are supported by those from previous studies of individual mechanisms in each inflammatory skin disease. Here, we present a systems-level overview of pathways and upstream regulators, which indicates the complexity of pathogenesis and, potentially, the relative importance of the identified mechanisms. This systems-level analyses on bulk microarray data should be assessed with data from cutting edge technologies such as single-cell RNA sequencing for reproducibility of potential biomarkers. Next, pathway analyses can be confounded by knowledge bias, inaccurate knowledge of gene interactions, or how gene interactions vary in different cell types. Despite this limitation, our pathway analyses were supported by the partially consistent results across different studies, and the findings are being agreement with the current understanding of disease mechanisms in inflammatory skin disease.
Collectively, the current system-level analysis based on profiling microarray data suggested possible combinatorial biomarkers for inflammatory skin diseases. Future studies for the identification of combinatorial biomarkers are warranted.
Methods
Identification and selection of eligible gene expression dataset
We systematically mined the Gene Expression Omnibus (GEO) database51 for expression profiling datasets. The following key words and their combinations were used: ‘inflammatory skin disease’, ‘human’, ‘microarray’, ‘gene expression dataset’, ‘tissues’, and ‘biopsy’. Specifically, the gene expression data were extracted for each diseased condition in comparison to the data of HCs. The inclusion criteria were specified and strictly followed for dataset selection: human case/control study, comparable conditions, untreated samples, and the availability of raw and processed data. In this study, we finally selected GSE 63,741 because it included data on diseased skin tissues, as well as control skin tissues, and it had enough number of patients, covering four different kinds of inflammatory skin diseases based on the same microarray platform to decrease possible technical bias.
Analysis of gene expression data
We first identified DEGs between inflammatory skin diseases and HCs using GEO2R53. The data were annotated using the National Centre for Biotechnology Information-generated platform and adjusted for multiple testing using the Benjamini–Hochberg procedure16. The data were then sorted only to include significant DEGs (q value < 0.05 based on false discovery rate) for downstream analysis16.
Identification of DEGs
For bulk microarray data, DEGs were identified using the LIMMA R package (Bioconductor, version 3.5) as described in the ‘Analysis of gene expression data’ that were reported in the Methods section of previous studies16,54,55. A negative binomial distribution was used to define the dataset with the lowest detection limit of 0.5. Genes were considered as significant DEGs if the adjusted p value was less than 0.05.
Ingenuity pathway analysis
Our bioinformatics strategy was based on finding pathways among the DEGs, and upstream regulators of the DEGs. The objective of pathway analysis is the obtain an overview of disease-associated mechanisms, while the objective of upstream regulators is to find key regulators of such mechanisms. The analyses were performed using the IPA software (Qiagen, Hilden, Germany)26. The IPA includes a global network, which is based on the manual curation of a vast body of medical literature and biomedical databases and is continuously updated56. The core analysis in IPA was used to identify pathways that were significantly enriched among the DEGs, as well as to predict upstream regulators of those DEGs, which were either activated or inhibited. The statistical analysis was performed using Fisher’s exact test (right-tailed) within the IPA software26. We performed an analysis of IPA’s upstream regulators to identify transcription factors that were predicted to be activated based on the activation Z-score26. The upstream regulators of groups of interacting DEGs were identified by Upstream Regulator Analysis (URA) that is based on the Ingenuity Knowledge Base database. For the upstream regulator analysis, we focused on seven molecule types—specifically, cytokines, complexes, groups, growth factors, G-protein-coupled receptors, ligand-dependent nuclear receptors, and transmembrane receptors16. IPA has a biomarker filter function, which identifies the most promising and relevant biomarker candidates in experimental data. Using the function of biomarker filter, we prioritized molecular biomarker candidates based on key biological characteristics and selected extracellular biomarkers that could be detected in blood.
Protein–protein network interaction
We used Cytoscape software to visualize protein–protein interaction network for common DEGs. Cytoscape is an open-source software platform for visualizing molecular interaction networks and biological pathways and integrating these networks with annotations, gene expression profiles and other state data.
Ethics declaration
This study was conducted in accordance with the Declaration of Helsinki. Using GEO2R, we identified DEGs between lesional and HC skin samples from patients with AD, KE, Li, and Pso. Skin samples of 150 donors (30 patients with AD, KE, Li, Pso, and HC, respectively) were analysed in GSE 63741. Only biopsies taken from untreated patients with typical skin lesions were included. The patients with KE included those with allergic contact dermatitis and irritant contact dermatitis. The public GEO data which can be assessable for everyone was annotated using the national centre for biotechnology information (NCBI) generated platform.
Data availability
The processed bulk data generated in this study are publicly available on GEO database (https://www.ncbi.nlm.nih.gov/geo/) with accession no. GSE63741.
References
Sa, D. C. & Festa, C. N. Inflammasomes and dermatology. An. Bras. Dermatol. 91, 566–578. https://doi.org/10.1590/abd1806-4841.20165577 (2016).
Zeng, J., Luo, S., Huang, Y. & Lu, Q. Critical role of environmental factors in the pathogenesis of psoriasis. J. Dermatol. 44, 863–872. https://doi.org/10.1111/1346-8138.13806 (2017).
Mobus, L., Weidinger, S. & Emmert, H. Epigenetic factors involved in the pathophysiology of inflammatory skin diseases. J. Allergy Clin. Immunol. 145, 1049–1060. https://doi.org/10.1016/j.jaci.2019.10.015 (2020).
Carretero, M. et al. Differential features between chronic skin inflammatory diseases revealed in skin-humanized psoriasis and atopic dermatitis mouse models. J. Invest. Dermatol. 136, 136–145. https://doi.org/10.1038/JID.2015.362 (2016).
Guttman-Yassky, E., Nograles, K. E. & Krueger, J. G. Contrasting pathogenesis of atopic dermatitis and psoriasis–part I: clinical and pathologic concepts. J. Allergy Clin. Immunol. 127, 1110–1118. https://doi.org/10.1016/j.jaci.2011.01.053 (2011).
Gilliet, M. Targeted therapies and precision medicine for inflammatory skin diseases. Eur. J. Dermatol. 29, 19–24. https://doi.org/10.1684/ejd.2019.3539 (2019).
Benson, M. et al. DNA microarray analysis of transforming growth factor-beta and related transcripts in nasal biopsies from patients with allergic rhinitis. Cytokine 18, 20–25. https://doi.org/10.1006/cyto.2002.1012 (2002).
Barrenas, F. et al. Gender differences in inflammatory proteins and pathways in seasonal allergic rhinitis. Cytokine 42, 325–329. https://doi.org/10.1016/j.cyto.2008.03.004 (2008).
Bruhn, S. et al. A generally applicable translational strategy identifies S100A4 as a candidate gene in allergy. Sci. Transl. Med. 6(218), 214. https://doi.org/10.1126/scitranslmed.3007410 (2014).
D’Erme, A. M. et al. IL-36gamma (IL-1F9) is a biomarker for psoriasis skin lesions. J Invest Dermatol 135, 1025–1032. https://doi.org/10.1038/jid.2014.532 (2015).
Coda, A. B., Icen, M., Smith, J. R. & Sinha, A. A. Global transcriptional analysis of psoriatic skin and blood confirms known disease-associated pathways and highlights novel genomic “hot spots” for differentially expressed genes. Genomics 100, 18–26. https://doi.org/10.1016/j.ygeno.2012.05.004 (2012).
Nedoszytko, B., Sokolowska-Wojdylo, M., Ruckemann-Dziurdzinska, K., Roszkiewicz, J. & Nowicki, R. J. Chemokines and cytokines network in the pathogenesis of the inflammatory skin diseases: atopic dermatitis, psoriasis and skin mastocytosis. Postepy. Dermatol. Alergol. 31, 84–91. https://doi.org/10.5114/pdia.2014.40920 (2014).
Georgescu, S. R. et al. Advances in understanding the immunological pathways in psoriasis. Int. J. Mol. Sci. https://doi.org/10.3390/ijms20030739 (2019).
Singh, S. et al. Genomic alterations driving psoriasis pathogenesis. Gene 683, 61–71. https://doi.org/10.1016/j.gene.2018.09.042 (2019).
Gawel, D. R. et al. A validated single-cell-based strategy to identify diagnostic and therapeutic targets in complex diseases. Genome Med. 11, 47. https://doi.org/10.1186/s13073-019-0657-3 (2019).
Lee, E. J. et al. Bulk and single cell transcriptomic data indicate that a dichotomy between inflammatory pathways in peripheral blood and arthritic joints complicates biomarker discovery. Cytokine 127, 154960. https://doi.org/10.1016/j.cyto.2019.154960 (2020).
Kupczyk, P., Reich, A. & Szepietowski, J. C. Cannabinoid system in the skin—A possible target for future therapies in dermatology. Exp. Dermatol. 18, 669–679. https://doi.org/10.1111/j.1600-0625.2009.00923.x (2009).
Tampellini, M., Sonetto, C. & Scagliotti, G. V. Novel anti-angiogenic therapeutic strategies in colorectal cancer. Expert Opin. Investig. Drugs 25, 507–520. https://doi.org/10.1517/13543784.2016.1161754 (2016).
Katoh, M. & Katoh, M. Molecular genetics and targeted therapy of WNT-related human diseases (Review). Int. J. Mol. Med. 40, 587–606. https://doi.org/10.3892/ijmm.2017.3071 (2017).
Armstrong, A. W. & Read, C. Pathophysiology, clinical presentation, and treatment of psoriasis: A review. JAMA 323, 1945–1960. https://doi.org/10.1001/jama.2020.4006 (2020).
Leonardi, C. L. et al. Efficacy and safety of ustekinumab, a human interleukin-12/23 monoclonal antibody, in patients with psoriasis: 76-week results from a randomised, double-blind, placebo-controlled trial (PHOENIX 1). Lancet 371, 1665–1674. https://doi.org/10.1016/S0140-6736(08)60725-4 (2008).
Liu, Y. et al. A genome-wide association study of psoriasis and psoriatic arthritis identifies new disease loci. PLoS Genet. 4, e1000041. https://doi.org/10.1371/journal.pgen.1000041 (2008).
Sconocchia, T. et al. Bone morphogenetic protein signaling regulates skin inflammation via modulating dendritic cell function. J. Allergy Clin. Immunol. 147, 1810-1822 e1819. https://doi.org/10.1016/j.jaci.2020.09.038 (2021).
Kim, S. G. et al. Apoptosis of oral epithelial cells in oral lichen planus caused by upregulation of BMP-4. J. Oral. Pathol. Med. 35, 37–45. https://doi.org/10.1111/j.1600-0714.2005.00373.x (2006).
Martin-Fontecha, M. et al. The expression of cannabinoid receptor 1 is significantly increased in atopic patients. J. Allergy Clin. Immunol. 133, 926-929 e922. https://doi.org/10.1016/j.jaci.2013.12.1032 (2014).
Kramer, A., Green, J., Pollard, J. Jr. & Tugendreich, S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 30, 523–530. https://doi.org/10.1093/bioinformatics/btt703 (2014).
Han, Y., Liu, T. & Lu, L. Apolipoprotein E gene polymorphism in psoriasis: A meta-analysis. Arch. Med. Res. 44, 46–53. https://doi.org/10.1016/j.arcmed.2012.10.009 (2013).
Pietschke, K. et al. The inflammation in cutaneous lichen planus is dominated by IFN-Upsilon and IL-21-A basis for therapeutic JAK1 inhibition. Exp. Dermatol. 30, 262–270. https://doi.org/10.1111/exd.14226 (2021).
Weber, B. et al. Distinct interferon-gamma and interleukin-9 expression in cutaneous and oral lichen planus. J. Eur. Acad. Dermatol. Venereol. 31, 880–886. https://doi.org/10.1111/jdv.13989 (2017).
Namkung, J. H. et al. An association between IL-9 and IL-9 receptor gene polymorphisms and atopic dermatitis in a Korean population. J. Dermatol. Sci. 62, 16–21. https://doi.org/10.1016/j.jdermsci.2011.01.007 (2011).
Ciprandi, G., De Amici, M., Giunta, V., Marseglia, A. & Marseglia, G. Serum interleukin-9 levels are associated with clinical severity in children with atopic dermatitis. Pediatr. Dermatol. 30, 222–225. https://doi.org/10.1111/j.1525-1470.2012.01766.x (2013).
Hong, C. H., Chang, K. L., Wang, H. J., Yu, H. S. & Lee, C. H. IL-9 induces IL-8 production via STIM1 activation and ERK phosphorylation in epidermal keratinocytes: A plausible mechanism of IL-9R in atopic dermatitis. J. Dermatol. Sci. 78, 206–214. https://doi.org/10.1016/j.jdermsci.2015.03.004 (2015).
Sismanopoulos, N. et al. IL-9 induces VEGF secretion from human mast cells and IL-9/IL-9 receptor genes are overexpressed in atopic dermatitis. PLoS ONE 7, e33271. https://doi.org/10.1371/journal.pone.0033271 (2012).
Singh, T. P. et al. Involvement of IL-9 in Th17-associated inflammation and angiogenesis of psoriasis. PLoS ONE 8, e51752. https://doi.org/10.1371/journal.pone.0051752 (2013).
Henno, A. et al. Histological and transcriptional study of angiogenesis and lymphangiogenesis in uninvolved skin, acute pinpoint lesions and established psoriasis plaques: an approach of vascular development chronology in psoriasis. J Dermatol. Sci. 57, 162–169. https://doi.org/10.1016/j.jdermsci.2009.12.006 (2010).
Liew, S. C. et al. Differential expression of the angiogenesis growth factors in psoriasis vulgaris. BMC Res. Notes 5, 201. https://doi.org/10.1186/1756-0500-5-201 (2012).
Vybohova, D., Mellova, Y., Adamicova, K., Adamkov, M. & Heskova, G. Quantitative comparison of angiogenesis and lymphangiogenesis in cutaneous lichen planus and psoriasis: Immunohistochemical assessment. Acta Histochem. 117, 20–28. https://doi.org/10.1016/j.acthis.2014.10.008 (2015).
Wang, S. Y. et al. Circulating Dickkopf-1 is correlated with bone erosion and inflammation in rheumatoid arthritis. J. Rheumatol. 38, 821–827. https://doi.org/10.3899/jrheum.100089 (2011).
Hu, Z. et al. Adalimumab significantly reduces inflammation and serum DKK-1 level but increases fatty deposition in lumbar spine in active ankylosing spondylitis. Int. J. Rheum. Dis. 15, 358–365. https://doi.org/10.1111/j.1756-185X.2012.01734.x (2012).
Choi, H. J., Park, H., Lee, H. W. & Kwon, Y. G. The Wnt pathway and the roles for its antagonists, DKKS, in angiogenesis. IUBMB Life 64, 724–731. https://doi.org/10.1002/iub.1062 (2012).
Gudjonsson, J. E. et al. Evidence for altered Wnt signaling in psoriatic skin. J. Invest. Dermatol. 130, 1849–1859. https://doi.org/10.1038/jid.2010.67 (2010).
Hou, T. et al. Skewed inflammation is associated with aberrant interleukin-37 signaling pathway in atopic dermatitis. Allergy 76, 2102–2114. https://doi.org/10.1111/all.14769 (2021).
Slominski, A., Gomez-Sanchez, C. E., Foecking, M. F. & Wortsman, J. Active steroidogenesis in the normal rat skin. Biochim. Biophys. Acta 1474, 1–4. https://doi.org/10.1016/s0304-4165(99)00215-9 (2000).
Slominski, A. T. et al. Key role of CRF in the skin stress response system. Endocr. Rev. 34, 827–884. https://doi.org/10.1210/er.2012-1092 (2013).
Slominski, A. & Wortsman, J. Neuroendocrinology of the skin. Endocr. Rev. 21, 457–487. https://doi.org/10.1210/edrv.21.5.0410 (2000).
Slominski, A. T. et al. Sensing the environment: regulation of local and global homeostasis by the skin’s neuroendocrine system. Adv. Anat. Embryol. Cell Biol. 212, 1–115. https://doi.org/10.1007/978-3-642-19683-6_1 (2012).
Slominski, A., Wortsman, J., Luger, T., Paus, R. & Solomon, S. Corticotropin releasing hormone and proopiomelanocortin involvement in the cutaneous response to stress. Physiol. Rev. 80, 979–1020. https://doi.org/10.1152/physrev.2000.80.3.979 (2000).
Landeck, L., Kneip, C., Reischl, J. & Asadullah, K. Biomarkers and personalized medicine: Current status and further perspectives with special focus on dermatology. Exp. Dermatol. 25, 333–339. https://doi.org/10.1111/exd.12948 (2016).
Tampa, M. et al. The pathophysiological mechanisms and the quest for biomarkers in psoriasis, a stress-related skin disease. Dis. Markers 2018, 5823684. https://doi.org/10.1155/2018/5823684 (2018).
Shi, J. et al. Emerging role and therapeutic implication of Wnt signaling pathways in autoimmune diseases. J. Immunol. Res. 2016, 9392132. https://doi.org/10.1155/2016/9392132 (2016).
National Center for Biotechnology Information, Home - GEO - NCBI, (2015). https://www.ncbi.nlm.nih.gov/geo/ (accessed June, 2021).
NCBI, About GEO2R - GEO - NCBI, (n.d.). https://www.ncbi.nlm.nih.gov/geo/info/geo2r.html).
Smyth, G. K. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. https://doi.org/10.2202/1544-6115.1027 (2004).
Qiu, X. et al. Single-cell mRNA quantification and differential analysis with Census. Nat Methods 14, 309–315. https://doi.org/10.1038/nmeth.4150 (2017).
Trapnell, C. et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 32, 381–386. https://doi.org/10.1038/nbt.2859 (2014).
FAQ - Ingenuity Knowledge Base, (n.d.). http://qiagen.force.com/KnowledgeBase/KnowledgeIPAPage?id=kA1D0000000PIoaKAG.
Funding
This work was supported by a faculty research grant of Yonsei University Wonju College of Medicine for 2021-52-0055 (granted to E.J.L.) and the Basic Science Research Program through the National Research Foundation of Korea (NRF) grant funded by the Korea government (Ministry of Science and ICT) (NRF-2021R1A2C1010082, granted to E.J.L.).
Author information
Authors and Affiliations
Contributions
E.J.L. and J.E.K. designed the study, managed the data extraction, and interpreted the data. H.B. and E.J.L. performed bioinformatics analyses. H.B., J.E.K., and E.J.L. wrote the manuscript. H.B. and E.J.L. formatted the manuscript. H.S.L., S.M.P., and D.-J.P. interpreted the data and reviewed the manuscript. E.J.L. initiated and supervised the study. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Bang, H., Kim, J.E., Lee, H.S. et al. Integrated bioinformatic analysis of gene expression profiling data to identify combinatorial biomarkers in inflammatory skin disease. Sci Rep 12, 5889 (2022). https://doi.org/10.1038/s41598-022-09840-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-022-09840-3
- Springer Nature Limited