Abstract
Estrogen receptor alpha (ERα, encoded by ESR1) is a well-characterized transcription factor expressed in more than 75% of breast tumors and is the key biomarker to direct endocrine therapies. On the other hand, much less is known about estrogen receptor beta (ERβ, encoded by ESR2) and its importance in cancer. Previous studies had some disagreement, however most reports suggested a more favorable prognosis for patients with high ESR2 expression. To add further clarity to ESR2 in breast cancer, we interrogated a large population-based cohort of primary breast tumors (n = 3207) from the SCAN-B study. RNA-seq shows ESR2 is expressed at low levels overall with a slight inverse correlation to ESR1 expression (Spearman R = −0.18, p = 2.2e−16), and highest ESR2 expression in the basal- and normal-like PAM50 subtypes. ESR2-high tumors had favorable overall survival (p = 0.006), particularly in subgroups receiving endocrine therapy (p = 0.03) and in triple-negative breast cancer (p = 0.01). These results were generally robust in multivariable analyses accounting for patient age, tumor size, node status, and grade. Gene modules consistent with immune response were associated to ESR2-high tumors. Taken together, our results indicate that ESR2 is generally expressed at low levels in breast cancer but associated with improved overall survival and may be related to immune response modulation.
Similar content being viewed by others
Introduction
Breast cancer (BC) is the most frequently diagnosed cancer in women worldwide1 and although the 5-year prognosis is good, it remains a public health issue on a global scale as it has overtaken lung cancer as the most commonly diagnosed cancer in the world according to recent global cancer estimates2.
Three quarters of all breast cancers are positive for expression of estrogen receptor alpha (ERα), encoded by the ESR1 gene3, making ER signaling the most important target of clinical treatments in ERα-positive BC. The effects of estrogen are also mediated by estrogen receptor beta (ERβ) encoded by ESR24. The mechanisms of ERα signaling in BC has been well studied over the past decades, with high expression being a potent driver of dysregulated endocrine signaling at multiple levels in BC5,6,7,8. While the role of ERα/ESR1 is largely established9,10,11, the potential therapeutic role and the extent of involvement of ERβ/ESR2 in treatment, progression and prognosis of BC remains uncertain12,13,14,15,16.
ERβ has been found to be expressed in normal breast epithelial cells as well as in various other tissues such as uterus, ovary, prostrate and brain, as well as in breast cancer cell lines17,18,19,20. The role of ERβ in breast cancer has been studied in various in vivo and in vitro models, suggesting its contribution in inhibiting BC tumor progression and its potential role as tumor suppressor. In cell models, ERβ has been found to enhance the response to tamoxifen21,22 and ERβ selective agonists reduce anti-apoptotic signaling23. ERβ activation increases cell autophagy21,24 and the generation of reactive oxygen species22 which may be part of the explanation for these results. Conversely, ERβ has been found to decrease the response to cytotoxic agents such as cisplatin, paclitaxel and doxorubicin21 and in triple-negative cell lines, enhances the antiproliferative effects of raloxifene25,26 and increases sensitivity to anti-androgens27.
Reports on the prognostic value of ERβ are conflicting. On one hand, some studies showed that high ERβ expression, irrespective of the ERα status, is a treatment response marker for BC patients receiving chemotherapy14,28,29 and endocrine therapy29,30,31,32. On the other hand, some report the opposite, where increased expression of ERβ in patients receiving endocrine therapy predicted poor prognosis and significantly reduced median tumor-free survival time33 as well as lower disease-free survival (DFS) in postmenopausal primary BC patients34. The association to poor prognosis was reported in particular for patients with triple-negative breast cancer (TNBC)35,36,37, but even in this subgroup, some studies have indicated a favorable prognosis38,39. Other studies reported no remarkable association between ERβ expression and patient outcome40,41.
Part of the reason for the conflicting results may be the lack of standardized methods for detecting ERβ in immunohistochemical (IHC) analysis and variable performance of the antibodies utilized across the many studies. Andersson et al. applied rigorous methods for validating commonly used ERβ antibodies and found that only one out of thirteen was specific for ERβ and that expression levels in human tissues were accordingly lower than previously reported42. Inadequate validity and poor specificity of ERβ antibodies has been an issue in much of the literature on ERβ protein expression, highlighted by the finding that neither of the well-studied purported ERβ-positive cancer cell lines, MCF-7 (breast) and LNCaP (prostate), expressed any ERβ when using validated antibodies and independent mass spectrometry-based approaches43. To address these issues, there have been efforts to standardize IHC protocols19 which can serve as a reference for future antibody-based ERβ studies.
Hence, the involvement and importance of ERβ (ESR2) in breast cancer remains controversial; moreover, most studies have been focused on measuring ERβ protein levels. In this study, we set out to characterize ESR2 mRNA expression levels and investigate its association to clinicopathological features and patient outcomes. To accomplish this, we analyzed gene expression in a large, population-based cohort of 3207 primary invasive breast tumors using RNA-sequencing (RNA-seq).
Results
SCAN-B cohort
SCAN-B (ClinicalTrials.gov identifier NCT02306096) is an ongoing, large, population-based breast cancer study started in 2010 and now enrolling patients at nine hospitals in Sweden, wherein all newly diagnosed patients are offered to participate44,45. From this cohort, we analyzed RNA-seq data from 3207 patients with longer follow-up (diagnosed between 1 September 2010 and 31 March 2015). The present cohort is a subset of the previously described cohort of 3217 patients46,47,48, which has been reduced to 3207 samples due to additional quality controls. The clinical characteristics are presented in Table 1 and are in concordance with the typical clinicopathological properties of breast cancer patients in Sweden. RNA-seq-based gene expression data was used to determine the PAM50 molecular subtypes of the tumors: 48% were classified as luminal A, 28% luminal B, 8.7% HER2-enriched, 9.9% basal-like, and 3.5% normal-like. Endocrine treatment was administered to 78.0% (n = 2502) of the patients in the cohort, out of which 218 patients also received chemotherapy.
ESR1 and ESR2 mRNA expression in SCAN-B breast tumor tissues
Quantification of ESR2 mRNA levels in transcripts per million mapped reads (TPM) across the entire SCAN-B dataset revealed a generally low expression. The median expressi on wa s 0.05 log2(TPM + 0.1), and in 1027 samples ESR2 was not expressed at all. We stratified the cohort into two groups based on ESR2 expression levels (upper tertile: “ESR2-high”; lower two tertiles: “ESR2-low”) and performed statistical two-group comparisons for relevant clinical factors between the ESR2-high and ESR2-low groups. Twice as many samples of basal subtype and ERα-negative status could be found in the ESR2-high group as compared to the ESR2 - low group. Tumor grade and Ki67 status did not differ between the ESR2-high vs -low subgroups. Median age and median tumor size differed between the ESR2-high vs -low subgroup s (p < 0.05) (Table 1).
We investigated the expression patterns of ESR1 and ESR2 according to clinical ERα status (positive or negative). ESR1 followed the well-known bimodal distribution pattern49,50, whereas the expression of ESR2 was very low and exhibited a left-skewed distribution (Fig. 1A). The two genes showed a weak inverse correlation (Spearman rank correlation test R = −0.18, p = 2e−10), consistent with a prior report49. Of the 475 ERα-negative tumors, 46.9% (n = 223) were classified as ESR2-high, and among the 2715 ERα-positive cases, 30.9% (n = 838) were classified ESR2-high.
ESR1 and ESR2 mRNA expression in SCAN-B and TCGA data sets. (A) Scatterplot of ESR1 and ESR2 mRNA expression (log2TPM). ESR1 and ESR2 data points are colored by density using 2D kernel density estimation function from the MASS R package. Adjacent to the scatterplot, histograms are shown indicating the frequency of expression values and color-coded according to ERα clinical status. (B) Expression of ESR1 and ESR2 is shown for the TCGA dataset as in panel A.
We compared the relative expression of the estrogen receptor genes across molecular subtypes. Within the SCAN-B data set, median ESR2 expression followed the trend: normal-like > basal-like > HER2-enriched > luminal A > luminal B, being highest in normal-like tumors (Fig. 2A). Conversely, the median ESR1 expression was highest in luminal B and lowest in the basal-like subtype (luminal B > luminal A > normal-like > HER2-enriched > basal-like). We also analyzed the expression levels across patients stratified by age at diagnosis (Fig. 2C). Median ESR1 expression increased with increasing patient age at diagnosis (Spearman correlation R = + 0.28, p = 2.2e−16), whereas median ESR2 mRNA quantities remained largely stable across age groups (Spearman correlation R = −0.078, p = 1.1e−05).
High ESR2 expression is associated with better prognosis for patients receiving endocrine therapy and in triple-negative disease
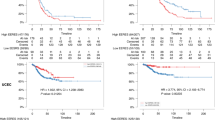
We analyzed patient outcome regarding overall survival (OS) and relapse-free interval (RFI). The median follow-up time was 6.2 years. We found that high ESR2 expression was not associated with improved RFI (Fig. 3A), however was associated with improved OS (logrank test p = 0.006; Fig. 3B). These results are in concordance with another study where higher levels of ERβ were found to be associated with favorable OS in inflammatory breast cancer patients51.
We also analyzed outcome for the sub-group of patients that received endocrine therapy (ET) with or without other systemic therapies (n = 2502) and for the patients receiving chemotherapy with or without other systemic therapy (n = 1258). ESR2 expression was not associated with RFI outcome (Fig. 3C), but higher ESR2 expression was associated with better OS in the endocrine-treated group (logrank test p = 0.03; Fig. 3D). No significant associations to RFI and OS were found in the patients who received chemotherapy (Fig. 3E, 3F). Furthermore, patients were stratified based on clinical groups: ERα-positive/HER2-negative (n = 2308), ERα-positive/HER2-positive (n = 287), ERα-negative/HER2-positive (n = 124) and triple-negative breast cancer (TNBC, n = 320). We found that low ESR2 expression was associated with poor OS in TNBC (logrank p = 0.01), but not in the other clinical sub-groups (Fig. 4).
ESR2 expression and association to overall survival (OS) and relapse-free interval (RFI) in the SCAN-B clinical groups. (A,B) Patients with ERα-positive, HER2-negative breast cancer; (C,D) ERα-positive, HER2-positive breast cancer; (E,F) Triple-negative breast cancer (TNBC); and (G,H) ERα-negative, HER2-positive breast cancer.
To further examine our findings that high ESR2 expression is associated with improved OS, we performed Cox regression multivariable analysis, adjusting for age, tumor size, lymph node status, and grade (Fig. 5). In the full cohort, ESR2 expression remained a significant prognostic factor with hazard ratio (HR) 1.34 (95% CI 1.06–1.32; p = 0.01). For patients receiving endocrine therapy, low ESR2 expression carried an HR of 1.24 (95% CI 0.96–1.61; p = 0.1, not significant). In triple-negative tumors, low expression of ESR2 exhibited an increased HR of 2.0 (95% CI 1.25–3.23; p = 0.004).
Validation of findings in the TCGA dataset
We set out to validate our results in the TCGA breast cancer cohort52. Albeit not population-based, and as previously reported, having a bias towards larger tumors with higher grade and stage53, this dataset represents a comparably large tumor collection with publicly available RNA-seq data. Generally, the SCAN-B results were confirmed in TCGA. ESR2 mRNA overall showed low expression across TCGA. As expected, in TCGA ESR1 followed a bimodal distribution pattern in the histogram (Fig. 1B). Moreover, ESR1 and ESR2 levels showed a weak inverse correlation (Fig. 1B, Spearman correlation R = −0.20, p = 3.5e−12). The ESR2-high group was comprised of 29% of the ERα-positive tumors (231/800), as compared to 48% ERα-negative tumors (115/239). Expression of ESR2 in TCGA followed the same trend as in SCAN-B with higher expression in normal-like, basal-like, and HER2-enriched groups and lower expression in the luminal subtypes (Fig. 2B). While there were small but significant differences in ESR2 expression across molecular subtypes in SCAN-B we did not find significant differences in TCGA (Supplementary Table S1). ESR1 and ESR2 expression patterns in the TCGA cohort followed same trend as in SCAN-B, where ESR1 expression increased with patient age at diagnosis but ESR2 expression was largely stable across age groups (Fig. 2D).
OS and RFI for the TCGA breast tumors were analyzed using patient survival at 10 years, after which all events were censored, for comparison with the SCAN-B cohort. The median follow-up time was 2.3 years. As with the SCAN-B dataset, patients were subdivided based on treatment received; endocrine therapy with or without other systemic treatment (n = 524) and chemotherapy with or without other treatment (n = 576). Within these groups we could not find any association with outcome for OS, as we had seen in the SCAN-B cohort (Supplementary Figure S1). However, when analyzing outcome in the clinical subgroups, we found that, in contrast to our findings in the SCAN-B cohort, OS and RFI were significantly improved for ESR2-high patients in the ERα-negative HER2-positive subgroup (RFI, p = 0.02, OS, p = 0.03; Supplementary Figure S2).
Differential gene expression and GSEA analysis of ESR2 high vs low groups
To shed light on the potential biology behind the differences in outcome of ESR2 expression groups, we next performed differential gene expression (DGE) analysis for tumors with ESR2-high versus ESR2-low expression to determine the genes co-modulated with ESR2 within the SCAN-B cohort. To remove the influence of ERα effects, which are known to have a strong impact on global gene expression patterns, we performed separate DGE analyses within the ERα-positive and -negative sub-groups. We applied a false discovery rate (FDR) cut-off of ≤ 0.05 and identified up- and down-regulated genes according to the log2 fold change (log2FC) with the criteria log2FC ≥ 1 .5 for up-regulated genes and log2FC ≤ − 1.5 for down-regulated genes. Within the ERα-positive subgroup, a total of 64 genes were found to be upregulated in ESR2-high vs -low tumors, and 6 genes were downregulated. In the ERα-negative subgroup, 199 genes were upregulated and 22 genes were downregulated in ESR2-high vs -low cases.
Of all the genes identified in the DGE analyses, a total of 42 up-regulated genes (BANK1, BLK, CCL19, CD19, CD79, IGLL5, IRF4, JCHAIN, PAX5, TCL1A, TNFRSF17, VPREB3 and others) were found to be upregulated in ESR2-high tumors in both the ERα-positive and -negative subgroups, and two down-regulated genes (COL11A1, EEF1A2) were found to be common between these subgroups (Supplementary Table S2; common genes highlighted). Up-regulated genes were found to be involved in processes such as immune response, B-cells signature (CCL19, JCHAIN, VPREB3, IGLL5, CD19, BLK, IGHD, CD79A), chromosomal rearrangement (TCL1A, TNFRSF17, IRF4, PAX5) as well as proto-oncogenes such as TCL1A, PAX5, which have been shown to be potent regulators of malignant processes in breast cancer54,55,56,57.
Next, we performed gene set enrichment analysis (GSEA) to find the statistically significant, concordant gene sets that differed between ESR2-high vs -low in both ERα-positive and -negative tumors. The log2FC ranked gene expression values were analyzed for enrichment within Gene Ontology (GO) category ‘non-redundant biological processes’. Most of the GO categories enriched within ERα-positive and ERα-negative subgroup analyses were found to be shared, with a common theme related to immune system modulation including the positively enriched GO categories immune responses, B cell activation and proliferation, response to chemokine, and cellular defense (Fig. 6). The genes involved in these positively enriched GO categories within the ERα-positive and -negative subgroups were also upregulated in the ERα-positive and -negative DGE list (Supplementary Tables S3 and S4). Genes involved in negatively-enriched GO categories such as NADH dehydrogenase complex assembly were also found to be common within the ERα-positive and -negative subgroups. Other GO categories negatively enriched in ESR2-high were unique within the ERα-positive subgroup analysis (such as base-excision repair, DNA damage response, protein localization to chromosome, microtubule bundle formation, kinetochore organization, and DNA strand elongation) or within the ERα-negative subgroup analysis (cell aggregation) (Fig. 6).
Gene Set Enrichment Analysis (GSEA; GO category: Biological Process) of genes ranked by fold change (log2FC) and p-value < 0.05, associated with ESR2-high vs -low in SCAN-B. GSEA analysis based on ESR2-high vs -low was performed separately for the ERα-positive (A) and ERα-negative (B) subgroups. Categories found enriched in both subgroup analyses are indicated by red text.
Importantly, the common positively enriched categories between the two ERα groups were associated with a myriad of immune response processes such as adaptive immune response, cellular defense response, lymphocyte mediated immunity, leukocyte cell–cell adhesion and proliferation, B and T cell activation, production of interleukins, cell killing, cellular defense response, and regulation of inflammatory response. Together, this raises the hypothesis that the improved survival for ESR2-high tumors may be partly associated with the local and systemic immune response.
Discussion
In this study we have characterized the expression of ESR2 mRNA using RNA-seq analysis of a large cohort of breast cancer samples from SCAN-B. Our analyses revealed that ESR2 transcripts are generally much less abundant than ESR1 across all breast cancers. Within this general low expression, ESR2 expression was highest in the ERα-negative subtypes (normal-like, basal-like, and HER2-enriched) and lower in the ERα-positive subtypes (luminal A and luminal B). The relatively higher expression in basal-like subtype may be of clinical interest, since some studies report that ERβ expression in ERα-negative tumors may be a predictor for response to endocrine therapy in these patients30; our results support this conclusion.
We also found that higher expression of ESR2 was associated with better OS for patients treated with endocrine therapy, although the effect did not remain significant when adjusted for other clinical variables. Interestingly, the clinical subgroup analyses revealed that the overall survival effect was most pronounced in the TNBC subgroup. In the SCAN-B cohort, we could not observe any association of ESR2 expression with RFI, which may be an effect of shorter follow-up times compared to OS.
Analysis of the TCGA breast tumors confirmed that ESR2 was generally expressed at low levels, but higher in ERα-negative PAM50 subtypes. For association of ESR2 expression and improved OS, the results within the entire SCAN-B cohort and TCGA cohort showed a similar trend, with SCAN-B showing a significant association whereas in TCGA, the survival curves had a later separation that did not reach statistical significance. The SCAN-B results on patient OS following endocrine treatment or in TNBC were not reproduced. There could be several reasons for these discrepancies. First, due to smaller sample size in TCGA, a potential survival association may not be as readily detectable. Furthermore, TCGA is not a population-based cohort, but rather spans samples collected from large number of clinical sites, varying timeframes of diagnosis, various treatment regimens of diverse countries, and furthermore is biased towards more advanced tumors52. It may be that this heterogeneity affects the analysis for patient outcome within TCGA. To note, we did find that ESR2-high patients had a significantly improved OS and RFI in TCGA HER2-positive patients, which we did not observe in the SCAN-B cohort. Eighty percent of HER2-positive patients in SCAN-B received anti-HER2 therapy, compared to 28% in TCGA (note, 37% of HER2-positive patients were missing treatment information) and this may have improved the overall outcome for the SCAN-B group. A potential weakness of our study is that it relies on the quantities of mRNA rather than protein. The global concordance of mRNA to protein is expected to be high, with a commonly stated correlation of 0.658, but it does not completely explain the variance in protein levels, which are also affected by translation, post-translational modifications, and regulation of the rate of protein decay. Consequently, our study must be interpreted in the context of the biological phenotype related to high ESR2 mRNA expression. On the other hand, our approach allows us to circumvent the problematic use of ERβ antibodies, which have been shown to be exceedingly unreliable to date42,43.
Another possible limitation of our study is its reliance on RNA-seq of bulk tumor tissue samples. Since bulk RNA-seq mainly reflects the averaged gene expression across thousands of cells at different transcriptomic states or even different cell types within the same tissues (for example, infiltrating immune cells or normal cells in tumor samples), it is not possible to determine from which compartment in the tumor or tumor microenvironment the gene expression signals originate. It is possible that, along with BC cells, immune cells such as lymphocytes may be contributing to ESR2 expression, which has also been shown in previous studies59,60,61. Indeed, our DGE analyses demonstrate the enrichment of lymphocytic markers in the upregulated gene lists within ERα-positive and -negative subgroups. This may suggest that ESR2 is co-expressed within the immune cell compartment, or that ESR2 is expressed in the tumor compartment and is associated to signatures of immune cell infiltration. Additional studies at the protein level, or using approaches such as single-cell sequencing, will be needed to further decipher the origin of the ESR2 expression signature.
GSEA analysis revealed co-expressed genes, many of which were enriched in immune response biological processes and pathways in both ERα-positive and ERα-negative tumors. This robust result may suggest that the improved survival seen in patients with ESR2-high expression could be related to the local and systemic immune response. In this respect, ESR2 may be an active participant or be an associated biomarker for immune cell activity.
TNBC accounts for approximately 10–15% of all breast cancers62, which lacks expression of ERα, PR, HER263,64. TNBCs are associated with aggressive features, do not benefit from treatments with targeted therapies currently used, and have poorer prognosis65. Our analysis showed that ESR2-high tumors had favorable OS (p = 0.006), ESR2 expression was high in the basal-like tumors, associated with better OS in TNBC (p = 0.01), and associated to immune response in GSEA analysis. Taken together, these results indicate that ERβ could be an interesting biomarker for more favorable-prognosis TNBC, a target for re-activation, possibly providing alternative therapeutic options for patients with TNBC.
In conclusion, we have characterized the expression of ESR2 across the largest population-based breast cancer cohort to date, and described its association to clinicopathological parameters and patient outcomes. We found that ESR2 mRNA is not abundantly expressed in primary breast cancer, but that higher ESR2 expression is found particularly within ERα-negative breast cancer subtypes and that ESR2-high has a significant association to survival in endocrine-treated patients as well as patients with TNBC. Our study brings further clarity to the ERβ/ESR2 field of research and sets the stage for further exploration of this poorly understood receptor.
Materials and methods
Patient enrollment and study design
The study was conducted in accordance with the Declaration of Helsinki and was approved by the Regional Ethics Review Board of Lund at Lund University (diary numbers 2007/155, 2009/658, 2009/659, 2010/383, 2012/58, 2013/459), the county governmental biobank center, and the Swedish Data Inspection group (diary number 364–2010). Trained health professionals provided the written information and all patients gave written informed consent.
Clinical/medical records were retrieved from the Swedish National Cancer Registry (NKBC). The median overall follow-up time for the early BC patients in the SC AN-B cohort was 6.2 years (IQR = 2.2). Hormone receptor positive early breast tumors were defined as cases expressing estrogen (ERα) or progesterone (PR) receptors using an immunohistochemical staining cutoff ≥ 10% of neoplastic/BC cells as indicated by Swedish guidelines and HER2 status was assessed according to standard recommendations66.
Tumor processing and RNA-seq gene expression measurements
SCAN-B tissue collection, tumor sample processing, preservation in RNA-later, mRNA enrichment by poly-A selection, mRNA-sequencing and read processing were performed as described previously44,46,47.
In brief, the RNA-seq data was processed through an automated multistep analysis pipeline implemented in BASE67,68 with extension package Reggie69. Picard toolkit70 v2.22.3 was used for demultiplexing raw sequencing read data using tools ExtractIlluminaBarcodes and IlluminaBasecallsToFastq with default parameters except –INCLUDE_NON_PF_READS = false. Trimmomatic71 v0.33 with the recommended parameters for PE reads was used to remove adaptor sequences and poor-quality reads (ILLUMINACLIP:TruSeq3-PE-2.fa:2:30:12:1:true; MINLEN:20; MAXINFO:40:0.9 and MINLEN:20). Each data set was filtered to remove reads that align (using Bowtie272 v2.2.9 with default parameters except -k 1 –phred33 –local) to ribosomal RNA/DNA (GenBank loci NR_023363.1, NR_003285.2, NR_003286.2, NR_003287.2, X12811.1, U13369.1), phiX174 Illumina control (NC_001422.1), and sequences contained in the UCSC hg38 RepeatMasker track.
Reads were aligned using HISAT273 v2.1.0 to the human genome reference GRCh38/hg38 using the GENCODE release 27 transcriptome model, with default parameters except –no-unal –non-deterministic –novel-splicesite-outfile ${SPLICEFILE} –rna-strandness RF. HISAT2 indexes were created using the –snp parameter and dbSNP build 150. StringTie74 v1.3.3b was used to calculate expression levels as fragments per kilobase of transcript per million mapped reads (FPKM), with default parameters including –rf -e using protein coding transcripts from GENCODE release 27 as transcriptome model. Novel transcripts were discarded. An FPKM gene expression matrix was generated from .ctab files using tximport75 and subsequently transformed to TPM values. TPM values were log2 transformed. To avoid zero values and large negative values in log2 transformation, a fixed pseudo-count of 0.1 was added to all transcripts in the TPM matrix prior to transformation. Molecular subtyping using the PAM50 gene list was performed as described previously44. All data are available from the NCBI Gene Expression Omnibus (Accession No. GSE96058).
Validation using TCGA-BRCA cohort
TCGA clinical and expression data was obtained from the GDC Legacy Archive (https://portal.gdc.cancer.gov) and accessed using TCGABiolinks76. The TCGA BRCA samples were filtered for distinct barcodes (n = 1222), only primary tumor samples (n = 1102), and female gender (n = 1089). Gene expression data was obtained as FPKM, converted to TPM, and transformed using log2(TPM + 0.1) for use in gene expression analysis.
Statistical analysis
All analyses were performed using R 3.6.1. P values of ≤ 0.05 were considered significant. Spearman rank correlation was used to determine correlations between expression of ESR1 and ESR2. Since the data was not normally distributed, Kruskal–Wallis non-parametric test (for significant difference between groups) as well as Wilcoxon rank sum test (for multiple pairwise comparisons between groups) were used to compare and plot expression of the ESR1 and ESR2 genes in various clinical groups such as PAM50 subtype and age groups in both the SCAN-B and TCGA cohorts. To evaluate significant differences in the clinicopathological variables for the ESR2-high and ESR2-low groups, Mann Whitney U test (for continuous variables) and Fisher’s exact test (for categorical data) were used. DGE was performed using the limma-voom package77 in R. GSEA was performed using the fgsea package78,79 in R as well as WebGestalt80,81,82.
Patients were sub-grouped according to the treatment received (endocrine therapy with or without other treatments and chemotherapy with or without other treatments) for both SCAN-B and TCGA cohorts. Patients were also subdivided based on receptor status for ER, PR and HER2, resulting in four clinical groups: (1) ERα-positive, HER2-negative (PR-positive or -negative), (2) ERα-positive, HER2-positive (PR-positive or -negative), (3) ERα-negative, HER2-positive (PR-positive or -negative) and (4) triple-negative (TNBC): ERα-negative, PR-negative, and HER2-negative.
Survival analysis
For SCAN-B, overall survival (OS) outcome was defined as death from any cause and the relapse-free interval (RFI) endpoint as locoregional or distant recurrence. For TCGA, OS and RFI were calculated as described earlier83 with a modification to RFI calculation. For patients having new tumor event, only local recurrence and distant metastasis were taken into account as endpoints. Survival analysis was performed by Kaplan–Meier and Cox regression survival analyses. Transformed ESR2 expression data was divided into tertiles, with the first tertile defined as ESR2-high, and the bottom two tertiles at ESR2-low.
Proportional hazards assumptions were checked graphically by Schoenfeld residual plots. One of the variables in the multivariable model for the full SCAN-B cohort, Nottingham Histological Grade (NHG), showed a time varying effect. Therefore, three models were fitted to estimate the adjusted effect of ESR2 on outcome, a model with adjustment for NHG despite its non-proportional effect on outcome, a model with stratification for NHG, and finally a model with interaction between NHG and follow-up time allowing for a time dependent effect of NHG on outcome. The estimated HRs for ESR2 status were essentially the same in these three models (range 1.30–1.32).
References
Bray, F. et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 68, 394–424 (2018).
Sung, H. et al. Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 71, 209–249 (2021).
Green, S. et al. Human oestrogen receptor cDNA: Sequence, expression and homology to v-erb-A. Nature 320, 134–139 (1986).
Kuiper, G. G., Enmark, E., Pelto-Huikko, M., Nilsson, S. & Gustafsson, J. A. Cloning of a novel receptor expressed in rat prostate and ovary. Proc. Natl. Acad. Sci. U. S. A. 93, 5925–5930 (1996).
Jensen, E. V. On the mechanism of estrogen action. Perspect. Biol. Med. 6, 47–59 (1962).
Jensen, E. V., Block, G. E., Smith, S., Kyser, K. & DeSombre, E. R. Estrogen receptors and breast cancer response to adrenalectomy. Natl. Cancer Inst. Monogr. 34, 55–70 (1971).
Bundred, N. J. Prognostic and predictive factors in breast cancer. Cancer Treat. Rev. 27, 137–142 (2001).
Cleator, S. & Ashworth, A. Molecular profiling of breast cancer: Clinical implications. Br. J. Cancer 90, 1120–1124 (2004).
Park, Y. R. et al. Absence of estrogen receptor is associated with worse oncologic outcome in patients who were received neoadjuvant chemotherapy for breast cancer. Asian J. Surg. 43, 467–475 (2020).
Kumar, M., Salem, K., Tevaarwerk, A. J., Strigel, R. M. & Fowler, A. M. Recent advances in imaging steroid hormone receptors in breast cancer. J. Nucl. Med. 61, 172–176 (2020).
van Kruchten, M. et al. PET imaging of estrogen receptors as a diagnostic tool for breast cancer patients presenting with a clinical dilemma. J. Nucl. Med. 53, 182–190 (2012).
Oueslati, M. et al. ERα and ERβ co-expression: An indicator of aggressive tumors and hormonal sensitivity. Oncol. Lett. 14, 1675–1682 (2017).
Mirza, S. et al. Clinical significance of promoter hypermethylation of ERβ and RARβ2 in tumor and serum DNA in Indian breast cancer patients. Ann. Surg. Oncol. 19, 3107–3115 (2012).
Elebro, K. et al. High estrogen receptor β expression is prognostic among adjuvant chemotherapy-treated patients-results from a population-based breast cancer cohort. Clin. Cancer Res. 23, 766–777 (2017).
Girault, I. et al. Altered expression pattern of alternatively spliced estrogen receptor beta transcripts in breast carcinoma. Cancer Lett. 215, 101–112 (2004).
Zhou, Y. & Liu, X. The role of estrogen receptor beta in breast cancer. Biomark. Res. 8, 39 (2020).
Crandall, D. L., Busler, D. E., Novak, T. J., Weber, R. V. & Kral, J. G. Identification of estrogen receptor beta RNA in human breast and abdominal subcutaneous adipose tissue. Biochem. Biophys. Res. Commun. 248, 523–526 (1998).
Kuiper, G. G., Shughrue, P. J., Merchenthaler, I. & Gustafsson, J. A. The estrogen receptor beta subtype: A novel mediator of estrogen action in neuroendocrine systems. Front. Neuroendocrinol. 19, 253–286 (1998).
Hawse, J. R. et al. Optimized immunohistochemical detection of estrogen receptor beta using two validated monoclonal antibodies confirms its expression in normal and malignant breast tissues. Breast Cancer Res. Treat. 179, 241–249 (2020).
Omoto, Y. & Iwase, H. Clinical significance of estrogen receptor β in breast and prostate cancer from biological aspects. Cancer Sci. 106, 337–343 (2015).
Pons, D. G. et al. The presence of estrogen receptor β modulates the response of breast cancer cells to therapeutic agents. Int. J. Biochem. Cell Biol. 66, 85–94 (2015).
Razandi, M., Pedram, A., Jordan, V. C., Fuqua, S. & Levin, E. R. Tamoxifen regulates cell fate through mitochondrial estrogen receptor beta in breast cancer. Oncogene 32, 3274–3285 (2013).
Ruddy, S. C. et al. Preferential estrogen receptor β ligands reduce Bcl-2 expression in hormone-resistant breast cancer cells to increase autophagy. Mol. Cancer Ther. 13, 1882–1893 (2014).
Song, P. et al. Estrogen receptor β inhibits breast cancer cells migration and invasion through CLDN6-mediated autophagy. J. Exp. Clin. Cancer Res. CR 38, 354 (2019).
Greish, K., Nehoff, H., Bahman, F., Pritchard, T. & Taurin, S. Raloxifene nano-micelles effect on triple-negative breast cancer is mediated through estrogen receptor-β and epidermal growth factor receptor. J. Drug Target. 27, 903–916 (2019).
Tu, Z. et al. The enhanced antiproliferative response to combined treatment of trichostatin A with raloxifene in MCF-7 breast cancer cells and its relevance to estrogen receptor β expression. Mol. Cell. Biochem. 366, 111–122 (2012).
Anestis, A. et al. Estrogen receptor beta increases sensitivity to enzalutamide in androgen receptor-positive triple-negative breast cancer. J. Cancer Res. Clin. Oncol. 145, 1221–1233 (2019).
Chang, J. et al. Expression of ERβ gene in breast carcinoma and the relevance in neoadjuvant therapy. Oncol. Lett. 13, 1641–1646 (2017).
Yan, M., Rayoo, M., Takano, E. A., kConFab Investigators & Fox, S. B. Nuclear and cytoplasmic expressions of ERβ1 and ERβ2 are predictive of response to therapy and alters prognosis in familial breast cancers. Breast Cancer Res. Treat. 126, 395–405 (2011).
Gruvberger-Saal, S. K. et al. Estrogen receptor beta expression is associated with tamoxifen response in ERalpha-negative breast carcinoma. Clin. Cancer Res. 13, 1987–1994 (2007).
Hopp, T. A. et al. Low levels of estrogen receptor beta protein predict resistance to tamoxifen therapy in breast cancer. Clin. Cancer Res. 10, 7490–7499 (2004).
Hamilton-Burke, W. et al. Phosphorylation of estrogen receptor beta at serine 105 is associated with good prognosis in breast cancer. Am. J. Pathol. 177, 1079–1086 (2010).
Guo, L., Zhang, Y., Zhang, W. & Yilamu, D. Correlation between estrogen receptor β expression and the curative effect of endocrine therapy in breast cancer patients. Exp. Ther. Med. 7, 1568–1572 (2014).
Guo, L., Zhang, Y. U., Yilamu, D., Liu, S. & Guo, C. ERβ overexpression results in endocrine therapy resistance and poor prognosis in postmenopausal ERα-positive breast cancer patients. Oncol. Lett. 11, 1531–1536 (2016).
Austin, D. et al. Estrogen receptor-beta is a potential target for triple negative breast cancer treatment. Oncotarget 9, 33912–33930 (2018).
Hamilton, N. et al. Biologic roles of estrogen receptor-β and insulin-like growth factor-2 in triple-negative breast cancer. BioMed Res. Int. 2015, 925703 (2015).
Guo, L. et al. Expression and prognostic value of estrogen receptor β in patients with triple-negative and triple-positive breast cancer. Exp. Ther. Med. 9, 2147–2150 (2015).
Wang, J. et al. ERβ1 inversely correlates with PTEN/PI3K/AKT pathway and predicts a favorable prognosis in triple-negative breast cancer. Breast Cancer Res. Treat. 152, 255–269 (2015).
Honma, N. et al. Clinical importance of estrogen receptor-beta evaluation in breast cancer patients treated with adjuvant tamoxifen therapy. J. Clin. Oncol. 26, 3727–3734 (2008).
de Cremoux, P. et al. Quantitation of estradiol receptors alpha and beta and progesterone receptors in human breast tumors by real-time reverse transcription-polymerase chain reaction. Correlation with protein assays. Biochem. Pharmacol. 64, 507–515 (2002).
Stefanou, D., Batistatou, A., Briasoulis, E., Arkoumani, E. & Agnantis, N. J. Estrogen receptor beta (ERbeta) expression in breast carcinomas is not correlated with estrogen receptor alpha (ERalpha) and prognosis: The Greek experience. Eur. J. Gynaecol. Oncol. 25, 457–461 (2004).
Andersson, S. et al. Insufficient antibody validation challenges oestrogen receptor beta research. Nat. Commun. 8, 15840 (2017).
Nelson, A. W. et al. Comprehensive assessment of estrogen receptor beta antibodies in cancer cell line models and tissue reveals critical limitations in reagent specificity. Mol. Cell. Endocrinol. 440, 138–150 (2017).
Saal, L. H. et al. The Sweden Cancerome Analysis Network - Breast (SCAN-B) Initiative: A large-scale multicenter infrastructure towards implementation of breast cancer genomic analyses in the clinical routine. Genome Med. 7, 20 (2015).
Rydén, L. et al. Minimizing inequality in access to precision medicine in breast cancer by real-time population-based molecular analysis in the SCAN-B initiative. Br. J. Surg. 105, e158–e168 (2018).
Brueffer, C. et al. Clinical value of RNA sequencing-based classifiers for prediction of the five conventional breast cancer biomarkers: A report from the population-based multicenter Sweden cancerome analysis network-breast initiative. JCO Precis. Oncol. 2, 1–18 (2018).
Brueffer, C. et al. The mutational landscape of the SCAN-B real-world primary breast cancer transcriptome. EMBO Mol. Med. 12, 12118 (2020).
Dahlgren, M. et al. Preexisting somatic mutations of estrogen receptor alpha (ESR1) in early-stage primary breast cancer. JNCI Cancer Spectr. 5, 028 (2021).
Bièche, I. et al. Quantification of estrogen receptor α and β expression in sporadic breast cancer. Oncogene 20, 8109–8115 (2001).
Collins, L., Botero, M. & Schnitt, S. Bimodal frequency distribution of estrogen receptor immunohistochemical staining results in breast cancer. Am. J. Clin. Pathol. 123, 16–20 (2005).
Thomas, C. et al. Estrogen receptor β-mediated inhibition of actin-based cell migration suppresses metastasis of inflammatory breast cancer. Cancer Res. 81, 2399–2414 (2021).
Koboldt, D. C. et al. Comprehensive molecular portraits of human breast tumours. Nature 490, 61–70 (2012).
Xie, Y. et al. Breast cancer gene expression datasets do not reflect the disease at the population level. NPJ Breast Cancer 6, 39 (2020).
Benzina, S. et al. Pax-5 is a potent regulator of E-cadherin and breast cancer malignant processes. Oncotarget 8, 12052–12066 (2017).
Leblanc, N., Harquail, J., Crapoulet, N., Ouellette, R. J. & Robichaud, G. A. Pax-5 inhibits breast cancer proliferation through miR-215 up-regulation. Anticancer Res. 38, 5013–5026 (2018).
Garaud, S. et al. Tumor-infiltrating B cells signal functional humoral immune responses in breast cancer. JCI Insight 4, 129641 (2019).
Shen, M., Wang, J. & Ren, X. New insights into tumor-infiltrating B lymphocytes in breast cancer: Clinical impacts and regulatory mechanisms. Front. Immunol. 9, 470 (2018).
Larsson, O., Tian, B. & Sonenberg, N. Toward a genome-wide landscape of translational control. Cold Spring Harb. Perspect. Biol. 5, a012302 (2013).
Yakimchuk, K., Jondal, M. & Okret, S. Estrogen receptor α and β in the normal immune system and in lymphoid malignancies. Mol. Cell. Endocrinol. 375, 121–129 (2013).
Kovats, S. Estrogen receptors regulate innate immune cells and signaling pathways. Cell. Immunol. 294, 63–69 (2015).
Khan, D. & Ansar Ahmed, S. The immune system is a natural target for estrogen action: Opposing effects of estrogen in two prototypical autoimmune diseases. Front. Immunol. 6, 635 (2016).
National Cancer Institute. Surveillance, Epidemiology, and End Results (SEER) program website. http://www.seer.cancer.gov. Published 2018. Accessed August 10, 2021.
Nielsen, T. O. et al. Immunohistochemical and clinical characterization of the basal-like subtype of invasive breast carcinoma. Clin. Cancer Res. 10, 5367–5374 (2004).
Dent, R. et al. Triple-negative breast cancer: Clinical features and patterns of recurrence. Clin. Cancer Res. 13, 4429–4434 (2007).
Bianchini, G., Balko, J. M., Mayer, I. A., Sanders, M. E. & Gianni, L. Triple-negative breast cancer: Challenges and opportunities of a heterogeneous disease. Nat. Rev. Clin. Oncol. 13, 674–690 (2016).
Wolff, A. C. et al. Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. J. Clin. Oncol. 31, 3997–4013 (2013).
Saal, L. H. et al. BioArray Software Environment (BASE): A platform for comprehensive management and analysis of microarray data. Genome Biol. 3, SOFTWARE0003 (2002).
Vallon-Christersson, J., Nordborg, N., Svensson, M. & Häkkinen, J. BASE - 2nd generation software for microarray data management and analysis. BMC Bioinformatics 10, 330 (2009).
Häkkinen, J., Nordborg, N., Månsson, O. & Vallon-Christersson, J. Implementation of an open source software solution for laboratory information management and automated rnaseq data analysis in a large-scale cancer genomics initiative using base with extension package reggie. bioRxiv 038976 (2016) https://doi.org/10.1101/038976.
Picard toolkit. Broad Institute, GitHub repository (2019).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinforma. Oxf. Engl. 30, 2114–2120 (2014).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Kim, D., Paggi, J. M., Park, C., Bennett, C. & Salzberg, S. L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 37, 907–915 (2019).
Pertea, M. et al. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 33, 290–295 (2015).
Soneson, C., Love, M. & Robinson, M. Differential analyses for RNA-seq: Transcript-level estimates improve gene-level inferences. F1000Research 4, 1521 (2016).
Colaprico, A. et al. TCGAbiolinks: An R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res. 44, e71–e71 (2016).
Ritchie, M. E. et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015).
Sergushichev, A. A. An algorithm for fast preranked gene set enrichment analysis using cumulative statistic calculation. bioRxiv 060012 (2016). https://doi.org/10.1101/060012.
Korotkevich, G. et al. Fast gene set enrichment analysis. bioRxiv 060012 (2021). https://doi.org/10.1101/060012.
Wang, J., Duncan, D., Shi, Z. & Zhang, B. WEB-based GEne SeT AnaLysis Toolkit (WebGestalt): Update 2013. Nucleic Acids Res. 41, W77–W83 (2013).
Zhang, B., Kirov, S. & Snoddy, J. WebGestalt: An integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res. 33, W741–748 (2005).
Wang, J., Vasaikar, S., Shi, Z., Greer, M. & Zhang, B. WebGestalt 2017: A more comprehensive, powerful, flexible and interactive gene set enrichment analysis toolkit. Nucleic Acids Res. 45, W130–W137 (2017).
Liu, J. et al. An integrated TCGA pan-cancer clinical data resource to drive high-quality survival outcome analytics. Cell 173, 400-416.e11 (2018).
Acknowledgements
The authors thank the patients, clinicians, and hospital staff participating in the SCAN-B study, the staff at the central SCAN-B laboratory at Lund University, the Swedish National Breast Cancer Quality Registry (NKBC), Regional Cancer Center South, the South Swedish Breast Cancer Group (SSBCG), and the members of the Translational Oncogenomics Unit, Division of Oncology, Lund University. This work was supported by the European Commission Horizon 2020 MSCA-COFUND-2016 754299 CanFaster programme; Mrs. Berta Kamprad Foundation; Swedish Research Council; Swedish Cancer Society; Governmental Funding of Clinical Research within National Health Service; Lund University Medical Faculty; Gunnar Nilsson Cancer Foundation; and Skåne University Hospital Foundation.
Funding
Open access funding provided by Lund University.
Author information
Authors and Affiliations
Contributions
H.D., M.D., S.K.G.-S., and L.H.S. conceived the study. H.D., M.D., S.G., C.B., S.K.G.-S., and L.H.S. analyzed data. H.D. and M.D. prepared all figures and tables. L.H.S. supervised the project, and H.D. and M.D. wrote the report with assistance from all authors. All authors discussed, critically revised, and approved the final version of the report for publication.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Dalal, H., Dahlgren, M., Gladchuk, S. et al. Clinical associations of ESR2 (estrogen receptor beta) expression across thousands of primary breast tumors. Sci Rep 12, 4696 (2022). https://doi.org/10.1038/s41598-022-08210-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-022-08210-3
- Springer Nature Limited
This article is cited by
-
Digital PCR quantification of ultrahigh ERBB2 copy number identifies poor breast cancer survival after trastuzumab
npj Breast Cancer (2024)
-
Exploring Prognostic Gene Factors in Breast Cancer via Machine Learning
Biochemical Genetics (2024)
-
Elevated galectin-3 levels detected in women with hyperglycemia during early and mid-pregnancy antagonizes high glucose − induced trophoblast cells apoptosis via galectin-3/foxc1 pathway
Molecular Medicine (2023)
-
A comparative analysis of clinicopathological features and survival between pre and postmenopausal breast cancer from an Indian cohort
Scientific Reports (2023)
-
Toxic PARP trapping upon cAMP-induced DNA damage reinstates the efficacy of endocrine therapy and CDK4/6 inhibitors in treatment-refractory ER+ breast cancer
Nature Communications (2023)