Abstract
Breast cancers expressing high levels of Ki67 are associated with poor outcomes. Oncotype DX test was designed for ER+/HER2− early-stage breast cancers to help adjuvant chemotherapy decision by providing a Recurrent Score (RS). RS measures the expression of 21 specific genes from tumor tissue, including Ki67. The primary aim of this study was to assess the agreement between Ki67RNA obtained with Oncotype DX RS and Ki67IHC. Other objectives were to analyze the association between the event free survival (EFS) and the expression level of Ki67RNA; and association between RS and Ki67RNA. Herein, we report a low agreement of 0.288 by Pearson correlation coefficient test between Ki67IHC and Ki67RNA in a cohort of 98 patients with early ER+/HER2− breast cancers. Moreover, Ki67RNAhigh tumors were significantly associated with the occurrence of events (p = 0.03). On the other hand, we did not find any association between Ki67IHC and EFS (p = 0.26). We observed a low agreement between expression level of Ki67RNA and Ki67 protein labelling by IHC. Unlike Ki67IHC and independently of the RS, Ki67RNA could have a prognostic value. It would be interesting to better assess the prognosis and predictive value of Ki67RNA measured by qRT-PCR. The Ki67RNA in medical routine could be a good support in countries where Oncotype DX is not accessible.
Similar content being viewed by others
Introduction
In terms of incidence, prevalence and mortality, breast cancer is in first place worldwide in women1. The vast majority of patients with breast cancer does not show detectable distant metastasis on diagnosis. Treatment of early breast cancer is based on surgical tumor resection with conditional lymph node dissection. Adjuvant systemic treatments including endocrine or anti-Hormonal therapy (HT) and chemotherapy (CT) aim at the reduction of the distant recurrence rate and improvement of breast cancer specific survival. The decision of adjuvant therapeutic modalities is taken according to several prognostic and/or predictive factors including patient age, tumor size, histological type and grade, lymph node involvement and expression on the tumor of hormone receptors for estrogen (ER) progesterone (PR), and the human epidermal growth factor receptor 2 (HER2) as well as the percentage of tumor cells expressing the nuclear proliferation marker Ki672.
Genomic signatures were designed to give prognostic and predictive information to streamline adjuvant chemotherapy decision in ER-positive, HER2-negative breast cancer patients. Oncotype DX Breast (ODX) is the most widely used molecular signature in this setting and is included in treatment guidelines for estimating both the risk of distant recurrence and predicting adjuvant chemotherapy benefit. ODX measures the RNA of 21 genes (16 cancer-associated genes and 5 housekeeping genes) and uses the expression pattern to calculate a recurrence score (RS) that ranges between 0 and 1003. The RS result provides two types of information on tumor biology: (i) prognosis information: an estimate of the individual risk of distant cancer recurrence within 10 years, (ii) predictive information: an estimate of the likelihood of a benefit from chemotherapy4,5,6.
Interestingly in breast cancer, Ki67 RNA (Ki67RNA) is a parameter analyzed by several molecular signatures such as PAM50 and ODX7. Furthermore, Ki67 is an interesting biomarker in early breast cancer and breast cancers expressing high levels of Ki67 are associated with poor outcomes8,9,10. To provide individualized patient care in the concept of precision medicine, reliability of prognostic and predictive information deriving from Ki67 value is essential11,12. Some studies13,14 indicate that lowering in Ki67 expression after neoadjuvant endocrine treatment may predict long-term outcome. Nevertheless, substantial variability in Ki67 staining of breast cancer tissue by immunohistochemistry (IHC) and interpretation was found between 30 routine pathology labs. Clinical use of Ki67 staining for therapeutic decisions should be considered with caution and only fully aware of lab-specific reference values15. Ki67 staining lacks scoring standardization; various studies have focused on assessment methodology standardization16, interobserver reproducibility17 and digital image analyses of Ki67 staining18,19. However, little is known about variability in IHC Ki67-labelling results between routine pathology labs20,21 and its potential influence on interpretation of Ki67 levels in breast cancer. When using Ki67 assessment by IHC in order to consider an indication of adjuvant chemotherapy22, clinicians should be aware of the low reproducibility of Ki67 scoring and its questionable analytical validity.
In the present study, we analyzed 98 patients tested by the Oncotype DX Breast from June 2012 to April 2014. For this cohort, Ki67RNA level obtained in patients Oncotype DX signatures were available. The primary aim of this study was to assess the agreement between Ki67RNA and Ki67 staining by IHC (Ki67IHC). The other objectives were to analyze the association between the event free survival (EFS) and the expression level of Ki67RNA in ODX signature; and association between RS and Ki67RNA.
Results
Characteristics of the patient population (Table 1)
Complete data sets from 98 breast cancer patients who underwent RS testing were provided from 4 public treatment centers (public hospitals and university hospitals). The included patients were exclusively female and showed a wide age distribution (from 31–81 years) with a mean age of 57 years. The predominant tumor characteristics were no special type (NST) (91%), N0 or Nmic (71%), grade 1 (62%), and tumor size pT1c (1–2 cm) (58%). All patients had ER positive/ HER2 negative tumors. Table 1 shows the patient and disease characteristics of the full population. The RS values were < 18 in 38% (n = 37), 18–30 in 51% (n = 50) and > 30 in 11% (n = 11) of the patients. After surgery and collegial decision, all patients have received a treatment according to the result of ODX test (HT alone or CT-HT) in adjuvant situation.
During the follow-up (57 months), we observed 19 events (19%): three local relapses, ten metastatic relapses (bone, lung, liver and pancreas), three other cancers (contralateral breast, colorectal carcinoma and pancreatic carcinoma) and three deaths.
Agreement between Ki67IHC and Ki67RNA
The Ki67IHC positivity rate of > 20%23 was used to define for the “high-risk” tumor group. We showed by ROC curve analysis an optimal threshold for Ki67RNA at 6.35 with a specificity of 48% and a sensitivity of 84%. With this cut off, the Ki67RNAhigh were ≥ 6.35 and Ki67RNAlow were < 6.35 (Fig. 1). A correlation between Ki67RNA expression and Ki67IHC score was found (R = 0.288; p = 0.0047) (Supplementary data 2). In the cohort, 29 tumors were Ki67high and 29 were Ki67low by the two methods, which represent a match between the results in 58 patients (61%) (Table 2). Discordances were observed in 37 patients (39%): 10 samples were Ki67IHClow/Ki67RNAhigh and 27 samples were Ki67IHChigh/Ki67RNAlow.
Threshold determination, specificity and sensitivity of Ki67RNA. Threshold of Ki67RNA was determined using package pROC version 1.16.144. An optimal threshold of Ki67RNA at 6.35 units was found with a specificity of 48% and sensitivity of 84%.
Events-free survival among all patients according RS groups and Ki67 status
A significant relationship was observed between EFS and the level of Ki67RNA expression (p = 0.03) (Fig. 2A) unlike Ki67IHC (p = 0.26) (Fig. 2B). In univariate Cox analysis, we observed a hazard ratio of 3.92 [1.14–13.49] for Ki67RNA and 0.57 [0.263–1.48] for Ki67IHC. The same difference was observed in multivariate analysis (Table 3). However, we did not observe any significant difference on EFS according to the three RS groups risk (p = 0.4) (Fig. 3).
Events-free survival according Ki67 status by ODX test and by immunochemistry. The survival follow-up was analyzing by the package survival version 3.1-845,46. The events include the local or metastatic relapse, other cancers occurrence and death with a mean follow-up of 57 months. (A) Ki67RNA were analyzed according to two groups using the 6.35 units’ threshold. (B) The Ki67IHC were analyzed according to two groups using the rate of > 20%. Statistical analysis were obtained by a Cox univariate test.
Events-free survival among all patients according RS groups. The survival follow-up was analyzed by the package survival version 3.1-845,46. The events include the local or metastatic relapse, other cancers occurred and death with a mean follow-up of 57 months. The RS are interpreted in three categories (low risk: RS < 18; intermediate risk: RS = 18–30; high risk: RS > 30). Statistical analysis were obtained by a Cox univariate test.
Using the age categories from the TAILORx study24, the association between EFS and Ki67RNA status, was found only for women under 50 years (p = 0.01) and not for women over 50 years (p = 0.4) (Supplementary data 3). No association between Ki67IHC and EFS was observed whatever the patient age (p > 0.20).
Ki67RNA association with RS
As expected, Ki67RNA levels were significantly associated with RS (p = 5 × 10−4) (Fig. 4A). To a lesser extent, an association between ODX test with Ki67IHC status was also observed (p = 0.013) (Fig. 4B). Supplementary data 4 shows the distribution of Ki67IHC score and Ki67RNA level according to patient age, tumor grade and size, PR status, nodal status, Recurrence score, treatment and events.
Discussion
In this study, we focused on the Ki67RNA expression level of 98 ER-positive, HER2-negative breast cancer with Oncotype DX signatures. The primary aim of this study was to assess the agreement between Ki67RNA and Ki67 staining by IHC (Ki67IHC). Outcomes and tumors characteristics were also assessed.
Study population
The majority of our data are consistent with the PONDx real life study25 which found a female population over 50 years (70%) with NST tumors (78%) ranging between 2–5 cm (89%), grade II (68%) and without lymph node involvement in 79% of cases. The PONDx cohort included more grade II than in our population (68% vs 29%) with a higher contingent of lobular carcinoma (13% vs 7%). In PONDx, RS results by prognostic category used between 2012–2014 were: < 18: 54%, 18–30: 36%; > 30: 10%. Compared to this real-life study, the local cohort shows an inversion of the proportions between populations at low risk (38% vs 54%) and those at intermediate risk (51% vs 36%).
The few differences observed in the distribution of the risk groups established by the ODX test can be explained by an over-representation of grade II tumors in the PONDx study. However, our cohort remains comparable to this larger population for most other demographic parameters.
Differences between Ki67RNA and Ki67IHC
Comparisons of Ki67 status showed a discrepancy between the two different types of evaluation. Indeed, half of Ki67RNAhigh (> 6.35) were found Ki67IHClow (Table 2). The observed correlation between the two techniques, is therefore weak even if significant (R = 0.288, p = 0.0047) (Supplementary data 2). Technically, IHC shows greater variability20,21 and is less reproducible than the quantification of mRNAs. Another study comparing microarray data with IHC confirm in a series of 520 samples26. These results are in contradiction with those observed by Finsterbusch et al.27. These authors found a strong correlation between Ki67 in IHC and mRNA level of Ki67 obtained by Mammatyper in 101 patients. In this retrospective study, their method of assessing Ki67 protein expression was different from ours. On the other hand, all the contradictory data around its interpretation highlight the lack of reproducibility and the variation between observers.
The prognostic value of Ki67 in IHC is now no more debated because it was demonstrated in numerous publications10,28. These two meta-analyses of Stuart-Harris et al. and Azambuja et al. have confirmed the prognostic information on 44 and 46 studies but the threshold were comprising between 0–34% of Ki67 positive cells by anti-Ki67 or anti-MIB-I antibodies. Currently, there is no standardization for the interpretation of the Ki67 status by IHC16. As a consequence, it generates inter-site and inter-observer variability within the same site17. Recently, Nielsen et al. have updated the recommendations from the International Ki67 in Breast Cancer Working Group29. These authors provide solutions for a pre-analytical standardization of Ki67IHC. However, they specify that the Ki67IHC should be used to drive patient care only when 5% or less or 30% or more cells are positive due to poor reproducibility between observers29,30. A single-center prospective study with double-blind reading seems more suitable in order to avoid interpretation bias. In contrast, the Oncotype DX test which is a molecular based and standardized test provides objective results that are independent of the observer.
Furthermore, the ODX test analyzes the level of Ki67 messenger RNAs in the tumor sample. This analysis therefore considers the transcriptional status of the Ki67 gene level not only in tumor cells but also in associated cells such as lymphocytes, stromal or endothelial cells. Therefore, it is an overall estimate of Ki67 expression. By IHC, the protein expression is scored only on tumor cells. The two methods do not provide the same types of information on Ki67 status. These two types of evaluation of Ki67 are probably complementary because the Ki67IHC gives informations about the tumor specific proliferation index and the Ki67RNA would reflect the proliferation of the tumor and its microenvironment. For all the reasons mentioned above, the association between RS and Ki67IHC status is lower but remains significant (p = 0.013).
Events free survival analyses
On the one hand in this study, adaptation of treatment based on RS permit to obtain equivalent EFS in populations with low and high risk of recurrence. Indeed, the lack of association between RS and EFS can be explained by the treatment (HT alone or CT-HT) performed according the RS interpretation based on Paiks et al. retrospective study31. Indeed, during the inclusion period, we used the Paiks et al. criteria for treatment as follow: low risk (RS < 18) patients were treated by HT alone and high risk (RS > 30) patients by CT-HT. The treatment for intermediate risk (RS = 18–30) patients was assigned on a case-by-case basis. The clinical validity and utility of the RS has been demonstrated prospectively across multiple studies in breast cancer patients worldwide including multiple validation studies as well as long-term prospective studies (TAILORx24, WSG Plan B32 and analyses from a prospective epidemiological database33). Based on level Ia evidence, the Oncotype DX test has been incorporated in leading internationally-accepted clinical guidelines on the treatment of early breast cancer (St. Gallen34, ESMO35, and ASCO36). This therapeutic adaptation will therefore smooth the differences between the categories of the ODX test and at the same time demonstrates the value of the information provided by its score.
On the other hand, we were able to show that Ki67RNAhigh was significantly associated with the occurrence of events (p = 0.03) but we did not find any association between Ki67IHC and EFS (p = 0.26).
In univariate analysis of Ki67RNA, we obtained a hazard ratio of 3.92 [1.14–13.49] and performed a power calculation under the Cox proportional-hazards model and found a power of 0.82. The significant of our study is near the decisional threshold for clinical studies. Further investigations will be required to confirm these results.
Correlation between Ki67RNA and RS
As expected in our cohort, the RS obtained is strongly associated with the Ki67RNA (p = 5 × 10−4) which is one of the components of the ODX test (Fig. 4A). To optimize RS interpretation, the TAILORx subgroup study shows a benefit of CT in women ≤ 50 years old with an RS between 16 and 25 (p = 0.004). CT could be avoided in women over 50 years old with a RS < 26, in women 50 years old or less with an RS < 1637. Recently at San Antonio Breast cancer symposium, the results of RxPONDER study were presented38. Now, postmenopausal women with 1–3 positive nodes and RS 0–25 can safely forego adjuvant CT without compromising invasive disease-free survival. The premenopausal women with positive nodes and RS 0–25 likely significantly benefit from chemotherapy38.
Oncotype DX Breast enables relevant net reductions in chemotherapy use, sparing patients from serious toxicities7. On this other side, its clinical impact and pharmacoeconomic benefit in routine care have been shown in 20 decision-impact studies39,40,41. However, in many places, the oncotype score remains inaccessible or not reimbursed7,25,41. This is why the evaluation of Ki67RNA level by molecular biology techniques42,43 could be a low-cost prognostic alternative in some countries. However, this hypothesis will have to be validated during a prospective translational study.
In conclusion, we observed of low agreement between Ki67RNA tumor level measured by qRT-PCR and Ki67 protein labelling by IHC. Substantial variabilities in Ki67IHC of breast cancer tissue and interpretation have been shown and widely published. The clinical use of Ki67 labelling should be cautious and limited by the low reproducibility of Ki67 scoring and its questionable analytical validity.
Unlike Ki67IHC and independently of the RS, Ki67RNA could have a prognostic value. The assessment of Ki67RNA by qRT-PCR on breast cancer tumor would be feasible and cost effective. It would be of great clinical utility to better assess the prognosis and predictive value of Ki67RNA. Presentation of Ki67 status in Genomic Health report could also be helpful for therapeutic decisions on borderline situation.
Methods
Ethics statement
All procedures performed in studies involving human participants were in accordance with the ethical standards of the national research committee and with the 1964 Helsinki Declaration and its later amendments. In France, this search is considered like a non-interventional study according to European legislation. All patients were individually informed that their data should be used for scientific research. Informed and written consents were obtained from all subjects, or if subject are deceased, from a parent and/or legal guardian. All experimental protocols were approved by the « comité de protection des personnes » (CCP) of Besançon, France.
Patients and tumors characteristics
Our study was an observational multicenter retrospective study collecting data on the real-life use of Oncotype DX Breast Recurrence Score test by physicians in clinical practice settings in France. Patients eligibility criteria for this analysis correspond to the population for which the Oncotype DX Breast Recurrence Score test is validated, i.e. adult patients with a recent first diagnosis of a single early invasive breast tumor with ER+/HER2− status, plus available documentation of lymph node involvement as either N0 (node negative), Nmic (micrometastatic) or N1 (1–3 positive nodes). The following data were documented: patient age and sex, conventional clinical and pathological disease characteristics including histologic type, tumor size and grade, nodal status, receptor status including ER, PR, and HER2, and the Ki67 proliferation marker by IHC. Between 2012 and 2014, the RS results were interpreted in three categories (low risk; intermediate risk; high risk with two cut-offs: 18 and 30). The event free survival (EFS) includes the local or metastatic relapse, other cancers and death with a mean follow-up of 57 months. Following the guidelines, the Oncotype DX Breast Recurrence Score test was realized after surgery and adjuvant treatments were adapted according to the RS. The interpretation of RS values based on Paiks et al. study31 and the associated treatment (hormonal therapy plus/minus chemotherapy, or other modalities) were also collected.
Immunohistochemistry analyses
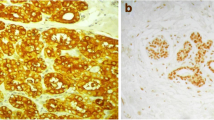
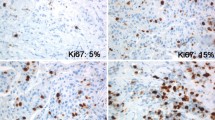
The hormonal receptor status (ER and PR) was considered positive according to standardized European guidelines using a cut-off of ≥ 10% stained tumor cell nuclei. The Ki67IHC evaluation was realized in 2 publics and 2 private pathological departments. In these structures, the Ki67IHC was assessed with monoclonal antibody MIB1, based on the recommendations from the International Ki67 in Breast Cancer Working Group16. Indeed, Ki67IHC is a nuclear staining and is evaluated regardless the staining intensity. Ki67IHC was defined as the percentage of positively stained tumor cells among the total number of tumor cells scored. For each cases, at least 3 randomly selected high power (40 × objective) fields were scored16. The Ki67IHChigh corresponding to Ki67 > 20% were defined “high-risk” subpopulations of tumors according to the recommendations established by St Gallen in 201523. In 2019, the ESMO clinical practice guidelines for diagnosis, treatment and follow-up in early-stage breast cancer validated this threshold, while presenting its limits30.
Ki67RNA threshold determination and events statistical analyses
Descriptive statistics were used to summarize clinicopathological characteristics. Variables were described by the size and rate. For each of the 98 patients of our cohort, Genomic Health, Inc. provided normalized unit values of Ki67RNA expression. Based on 5 housekeeping genes expressions (GAPDH, RPLPO, GUS, TFRC, β-actin), the Ki67 RNA expression is normalized in Oncotype DX test. These values were distributed according to a Gaussian curve in our cohort (Supplementary data 1).
We have determined the optimal threshold using ROC curves approach with the point maximizing the area under ROC curve with R package pROC version 1.16.144. The quantitative values of IHC and RNA were turned in clinically used qualitative variables. The statistical comparison between clinically used qualitative IHC and RNA was realized by Pearson coefficient. The statistical comparison between quantitative IHC and RNA was realized by the Pearson correlation coefficient. Statistical analysis were performed using R version 3.6.245. The survival follow-up was analyzed with the Cox test on Kaplan–Meier estimates using the package survival version 3.1-846.
Data availability
The datasets generated and analyzed during this study (birthdate, admission date, discharge date, date of death…), are available from the corresponding author on reasonable request.
References
Erratum: Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA. Cancer J. Clin. 70, 313–313 (2020).
Curigliano, G. et al. De-escalating and escalating treatments for early-stage breast cancer: The St. Gallen International Expert Consensus Conference on the Primary Therapy of Early Breast Cancer 2017. Ann. Oncol. 28, 1700–1712 (2017).
Gluz, O. et al. Prospective WSG phase III Plan B trial: Clinical outcome at 5 year follow up and impact of 21 Gene Recurrence Score result, central/local-pathological review of grade, ER, PR and Ki67 in HR+/HER2-high risk node-negative and -positive breast cancer. Eur. J. Cancer 57, S6 (2016).
Curtit, E., Mansi, L., Maisonnette-Escot, Y., Sautière, J.-L. & Pivot, X. Prognostic and predictive indicators in early-stage breast cancer and the role of genomic profiling: Focus on the Oncotype DX® Breast Recurrence Score Assay. Eur. J. Surg. Oncol. J. Eur. Soc. Surg. Oncol. Br. Assoc. Surg. Oncol. 43, 921–930 (2017).
Pivot, X. et al. In the era of genomics, should tumor size be reconsidered as a criterion for neoadjuvant chemotherapy?. Oncologist 20, 344–350 (2015).
McVeigh, T. P. et al. The impact of Oncotype DX testing on breast cancer management and chemotherapy prescribing patterns in a tertiary referral centre. Eur. J. Cancer Oxf. Engl. 1990(50), 2763–2770 (2014).
Gligorov, J. et al. Prospective clinical utility study of the use of the 21-gene assay in adjuvant clinical decision making in women with estrogen receptor-positive early invasive breast cancer: Results from the SWITCH study. Oncologist 20, 873–879 (2015).
Domagala, W., Markiewski, M., Harezga, B., Dukowicz, A. & Osborn, M. Prognostic significance of tumor cell proliferation rate as determined by the MIB-1 antibody in breast carcinoma: Its relationship with vimentin and p53 protein. Clin. Cancer Res. 2, 147–154 (1996).
Trihia, H. et al. Ki-67 expression in breast carcinoma. Cancer 97, 1321–1331 (2003).
de Azambuja, E. et al. Ki-67 as prognostic marker in early breast cancer: A meta-analysis of published studies involving 12 155 patients. Br. J. Cancer 96, 1504–1513 (2007).
Hayes, D. F. Biomarker validation and testing. Mol. Oncol. 9, 960–966 (2015).
Duffy, M. J. et al. Clinical use of biomarkers in breast cancer: Updated guidelines from the European Group on Tumor Markers (EGTM). Eur. J. Cancer 75, 284–298 (2017).
Ellis, M. J. et al. Letrozole inhibits tumor proliferation more effectively than tamoxifen independent of HER1/2 expression status. Cancer Res. 63, 6523–6531 (2003).
Dowsett, M. et al. Prognostic value of Ki67 expression after short-term presurgical endocrine therapy for primary breast cancer. JNCI J. Natl. Cancer Inst. 99, 167–170 (2007).
Focke, C. M. et al. Interlaboratory variability of Ki67 staining in breast cancer. Eur. J. Cancer 84, 219–227 (2017).
Dowsett, M. et al. Assessment of Ki67 in Breast Cancer: Recommendations from the International Ki67 in Breast Cancer Working Group. JNCI J. Natl. Cancer Inst. 103, 1656–1664 (2011).
Polley, M.-Y.C. et al. An international study to increase concordance in Ki67 scoring. Mod. Pathol. 28, 778–786 (2015).
Stålhammar, G. et al. Digital image analysis outperforms manual biomarker assessment in breast cancer. Mod. Pathol. 29, 318–329 (2016).
Gudlaugsson, E. et al. Comparison of the effect of different techniques for measurement of Ki67 proliferation on reproducibility and prognosis prediction accuracy in breast cancer. Histopathology 61, 1134–1144 (2012).
Mengel, M. et al. Inter-laboratory and inter-observer reproducibility of immunohistochemical assessment of the Ki-67 labelling index in a large multi-centre trial. J. Pathol. 198, 292–299 (2002).
Ekholm, M. et al. Highly reproducible results of breast cancer biomarkers when analysed in accordance with national guidelines—A Swedish survey with central re-assessment. Acta Oncol. 54, 1040–1048 (2015).
Sonnenblick, A. et al. Final 10-year results of the Breast International Group 2–98 phase III trial and the role of Ki67 in predicting benefit of adjuvant docetaxel in patients with oestrogen receptor positive breast cancer. Eur. J. Cancer 51, 1481–1489 (2015).
Focke, C. M., van Diest, P. J. & Decker, T. St Gallen 2015 subtyping of luminal breast cancers: Impact of different Ki67-based proliferation assessment methods. Breast Cancer Res. Treat. 159, 257–263 (2016).
Sparano, J. A. et al. Prospective validation of a 21-gene expression assay in breast cancer. N. Engl. J. Med. 373, 2005–2014 (2015).
Curtit, E. et al. Results of PONDx, a prospective multicenter study of the Oncotype DX® breast cancer assay: Real-life utilization and decision impact in French clinical practice. The Breast 44, 39–45 (2019).
Rossing, M. et al. Molecular subtyping of breast cancer improves identification of both high and low risk patients. Acta Oncol. 57, 58–66 (2018).
Finsterbusch, K., Decker, T., van Diest, P. J. & Focke, C. M. Luminal A versus luminal B breast cancer: MammaTyper mRNA versus immunohistochemical subtyping with an emphasis on standardised Ki67 labelling-based or mitotic activity index-based proliferation assessment. Histopathology 76, 650–660 (2020).
Stuart-Harris, R., Caldas, C., Pinder, S. E. & Pharoah, P. Proliferation markers and survival in early breast cancer: A systematic review and meta-analysis of 85 studies in 32,825 patients. The Breast 17, 323–334 (2008).
Nielsen, T. O. et al. Assessment of Ki67 in breast cancer: Updated recommendations from the International Ki67 in Breast Cancer Working Group. JNCI J. Natl. Cancer Inst. 113, 808–819 (2020).
Cardoso, F. et al. Early breast cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up†. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 30, 1194–1220 (2019).
Paik, S. et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N. Engl. J. Med. 351, 2817–2826 (2004).
Gluz, O. et al. West German Study Group Phase III PlanB Trial: First prospective outcome data for the 21-gene recurrence score assay and concordance of prognostic markers by central and local pathology assessment. J. Clin. Oncol. https://doi.org/10.1200/JCO.2015.63.5383 (2016).
Roberts, M. C., Miller, D. P., Shak, S. & Petkov, V. I. Breast cancer-specific survival in patients with lymph node-positive hormone receptor-positive invasive breast cancer and Oncotype DX Recurrence Score results in the SEER database. Breast Cancer Res. Treat. 163, 303–310 (2017).
Gnant, M., Harbeck, N. & Thomssen, C. St. Gallen/Vienna 2017: A brief summary of the consensus discussion about escalation and de-escalation of primary breast cancer treatment. Breast Care 12, 102–107 (2017).
Senkus, E. et al. Primary breast cancer: ESMO clinical practice guidelines for diagnosis, treatment and follow-up†. Ann. Oncol. 26, v8–v30 (2015).
Krop, I., Ismaila, N. & Stearns, V. Use of biomarkers to guide decisions on adjuvant systemic therapy for women with early-stage invasive breast cancer: American Society of Clinical Oncology clinical practice focused update guideline summary. J. Oncol. Pract. 13, 763–766 (2017).
Sparano, J. A. et al. Adjuvant chemotherapy guided by a 21-gene expression assay in breast cancer. N. Engl. J. Med. 379, 111–121 (2018).
Kalinsky, K. et al. RxPONDER: A Clinical Trial Rx for Positive Node, Endocrine Responsive Breast Cancer. First results from a phase III randomized clinical trial of standard adjuvant endocrine therapy +/− chemotherapy in patients (pts) with 1–3 positive nodes, hormone receptor-positive (HR+) and HER2-negative breast cancer with recurrence score of 25 or less: SWOG S1007 (2020).
Davidson, J. A. et al. A prospective clinical utility and pharmacoeconomic study of the impact of the 21-gene Recurrence Score® assay in oestrogen receptor positive node negative breast cancer. Eur. J. Cancer 49, 2469–2475 (2013).
Holt, S. et al. A decision impact, decision conflict and economic assessment of routine Oncotype DX testing of 146 women with node-negative or pNImi, ER-positive breast cancer in the UK. Br. J. Cancer 108, 2250–2258 (2013).
Nerich, V. et al. Economic assessment of the routine use of Oncotype DX® assay for early breast cancer in Franche-Comte region. Bull. Cancer (Paris) 101, 681–689 (2014).
Ellis, M. J. et al. Ki67 Proliferation Index as a tool for chemotherapy decisions during and after neoadjuvant aromatase inhibitor treatment of breast cancer: Results from the American College of Surgeons Oncology Group Z1031 Trial (Alliance). J. Clin. Oncol. 35, 1061–1069 (2017).
Issac, M. S. M., Yousef, E., Tahir, M. R. & Gaboury, L. A. MCM2, MCM4, and MCM6 in breast cancer: Clinical utility in diagnosis and prognosis. Neoplasia N. Y. N 21, 1015–1035 (2019).
Robin, X. et al. pROC: An open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinform. 12, 77 (2011).
R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing. https://www.R-project.org/ (2019).
Therneau T. A Package for Survival Analysis in S_. version 2.38 (2015).
Acknowledgements
We thank all patients who agreed to participate in this observational study. Authors also thank Genomic Health, Inc. (now Exact sciences) and Mahi Y. for the Ki67RNA data. The authors are grateful to all participating physicians for their cooperation and for private and public anatomopathology departments (CH de Trevenans, Besançon Pathologie and Dr Chargeboeuf M.) for their participation in IHC analyses.
Author information
Authors and Affiliations
Contributions
Conception and design: E.C., Z.S. Development of methodology: Z.S., E.C. Acquisition of data: E.C., Z.S., F.B., E.D., L.C., L.M., N.M., M.J.P., G.M., J.V., X.P. Analysis and interpretation of data: Z.S., A.O., C.M., J.L.P., J.P.F. Writing, review, and/or revision of the manuscript: Z.S., C.M., J.L.P., E.C. Administrative, technical, or material support: E.C., Z.S., J.L.P., M.P.A., J.P.F., C.B. Study supervision: E.C., Z.S.
Corresponding author
Ethics declarations
Competing interests
ZS: Honoraria from Pfizer, Genomic Health EC: Honoraria from Novartis, Pfizer, Daiichi, Genomic Health FB: Honoraria from Roche, Novartis, Pfizer, Astra-Zeneca, Clovis, Daiichi. CB: advisory board from Bayer, BMS, msd, servier and research grant from Roche NM, JV, CM, AO, JLP, JPF, ED, LM, MPA, LC, MJP, GM, XP: have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Selmani, Z., Molimard, C., Overs, A. et al. Low correlation between Ki67 assessed by qRT-PCR in Oncotype Dx score and Ki67 assessed by Immunohistochemistry. Sci Rep 12, 3617 (2022). https://doi.org/10.1038/s41598-022-07593-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-022-07593-7
- Springer Nature Limited