Abstract
Pancreatic adenocarcinoma (PAAD) is one of the most lethal malignant tumors in the world. The GSE55643 and GSE15471 microarray datasets were downloaded to screen the diagnostic and prognostic biomarkers for PAAD. 143 downregulated genes and 118 upregulated genes were obtained. Next, we performed gene ontology (GO) and The Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis on these genes and constructed a protein–protein interaction (PPI) network. We screened out two important clusters of genes, including 13 upregulated and 5 downregulated genes. After the survival analysis, 3 downregulated genes and 10 upregulated genes were identified as the selected key genes. The KEGG analysis on 13 selected genes showed that GNG7 and ADCY1 enriched in the Pathway in Cancer. Next, the diagnostic and prognostic value of GNG7 and ADCY1 was investigated using independent cohort of the Cancer Genome Atlas (TCGA), GSE84129 and GSE62452. We observed that the expression of the GNG7 and ADCY1 was decreased in PAAD. The diagnostic receiver operating characteristic (ROC) analysis indicated that the GNG7 and ADCY1 could serve as sensitive diagnostic markers in PAAD. Survival analysis suggested that expression of GNG7, ADCY1 were significantly associated with PAAD overall survival (OS). The multivariate cox regression analysis showed that the expression of GNG7, ADCY1 were independent risk factors for PAAD OS. Our study indicated GNG7 and ADCY1 may be potential diagnostic and prognostic biomarkers in patients with PAAD.
Similar content being viewed by others
Introduction
Pancreatic adenocarcinoma (PAAD) is one of the most malignant cancers. It was reported that PAAD accounts for more than 0.33 million deaths per year1. The 5-year survival rate of PAAD patients is still less than 5%, and the survival time of patients after multi-drug chemotherapy is generally still less than 1 year2. The cause of pancreatic adenocarcinoma is currently unknown. Studies have found that the occurrence of chronic pancreatitis is one of an important risk factor for PAAD3,4. Previous studies have also found that the occurrence of PAAD is obviously related to the intestinal flora, especially the types of oral microorganisms5,6. Early diagnosis and treatment of pancreatic cancer are of great importance to prolong the survival time of patients.
By reason of the lack of sensitive early diagnosis methods, it is challenging to develop effective treatments to delay and reverse the progression of PAAD. Serum CA19-9 is of great value for the diagnosis of PAAD and for assessing tumor progression, but it is not recommended for general screening because it is elevated in non-tumor diseases such as chronic pancreatitis and acute cholangitis7,8. However, CA19-9 is still considered to be the most helpful tumor marker for the diagnosis and prognosis for PAAD. In recent years, plenty of researches was devoted to identifying the diagnostic and prognostic biomarkers for PAAD. Lv et, al. reported that high TXLNA expression predicts favorable outcomes for PAAD patients9. Zheng and his colleagues identified COL11A1 as an immune infiltrate correlated prognosticator in PAAD through integrated bioinformatics analysis10. Nevertheless, biomarkers for the diagnosis and prognosis for PAAD are still lacking due to its heterogeneity11.
This present study aimed to screen out the genes that has diagnostic and prognostic significance for PAAD. We downloaded the GSE55643 and GSE15471 mRNA expression profiles from the Gene Expression Omnibus (GEO) database and then used built-in packages such as limma12 (version 3.22.1; Fred Hutchinson Cancer Research Center) with R software13 to perform differentially expressed genes (DEGs) analysis between the primary tumor tissue of PAAD patients and the paired adjacent non-malignant tissue. Next, we used Venn plots to query the overlapping DEGs obtained from GSE55643 and GSE15471. Then, the Database for Annotation, Visualization and Integrated Discovery (DAVID) database was used to perform the gene ontology (GO) and the Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis on the overlapping DEGs14. We constructed the protein–protein interaction (PPI) network of DEGs via the Search Tool for the Retrieval of Interacting Genes (STRING) database to identify the highly interconnected protein molecules and visualized by Cytoscape software15,16,17,18. Finally, we investigated the association between the selected gene expression with clinical outcomes using independent PAAD patient data obtained from the Cancer Genome Atlas (TCGA) database, GSE84129, and GSE62452 datasets19,20. Through integrated bioinformatics approaches, we screened out two potentially valuable biomarkers for the diagnosis and prognosis of PAAD.
Methods
Acquisition and processing of microarray data
mRNA expression data from microarray files GSE5564321 and GSE1547122 were downloaded from the GEO database23,24. GSE55643 contains 45 primary and 8 adjacent non-malignant tissue samples from PAAD patients. GSE15471 contains 78 pairs of primary and para-cancerous tissue samples from PAAD patients. We downloaded the normalized log2 ratio representing tumor/normal of the GSE55643 and GSE15471 dataset and converted the probe identification numbers to gene symbols using the Whole Human Genome Microarray 4 · 44 K G4112F (Probe Name Version) and the Affymetrix Human Genome U133 Plus 2.0 Array, respectively.
Identification of DEGs
We utilized the R software and built-in limma packages (version 3.22.1; Fred Hutchinson Cancer Research Center) to perform DEGs analysis between the primary tumor tissue of PAAD patients and the paired adjacent non-malignant tissue. DEGs must meet our two important criteria to be selected for the next stage of analysis: adjusted P value < 0.05, |log2 fold change (FC)|> 1.225.
KEGG and GO enrichment analysis of DEGs
The GO clarifies the functions of genes from three aspects: cellular component (CC), molecular function (MF), and biological process (BP)26. The KEGG provides data resources for understanding high-level functions and utilities of the biological metabolic pathways27. DAVID, a network server that provides a comprehensive set of functional annotation tools to determine the biological meaning of a large list of genes, was used for GO function annotation and KEGG pathway enrichment analysis14. P value < 0.05 was considered statistically significant.
Construction of PPI networks and identification of gene clusters
STRING database (version11.0), containing more than 2 billion interactions of 24.6 million proteins involving more than 5,000 organisms, was used to search for DEGs’ encoded protein and PPI network information. We uploaded the overlapping DEGs from the microarray data GSE55643 and GSE15471 into the STRING database and the significance threshold was set to an interaction score of 0.900 (highest confidence). Then, we visualized the PPI network in Cytoscape software. Next, we used built-in Molecular Complex Detection (MCODE) of Cytoscape to screen out highly interconnected protein molecules as molecular clusters.
Survival and expression analysis of gene clusters and KEGG enrichment analysis
The Kaplan Meier plotter is capable to assess the effect of 54 k genes (mRNA, miRNA, protein) on survival in 21 cancer types. Sources for the databases include GEO, EGA, and TCGA19. But for PAAD, the expression and survival data are all sources from the TCGA database (n = 177). The TNMplot database contains 56,938 unique multilevel quality-controlled samples of normal tissues, tumor tissues, and metastatic tissues from GEO, GTex/TCGA, and TARGET databases. We performed the Kaplan–Meier curve and log-rank test analyses to investigate the prognostic value of gene clusters in PAAD tissues. The TNMplot database (https://www.tnmplot.com/) was used for the comparison of gene expression in normal, tumor and metastatic tissues of PAAD patients. Then, the member of gene clusters validated by the Kaplan–Meier plotters and TNMplot database were screened as selected genes. Finally, the DAVID was used to perform the KEGG enrichment analysis on the selected genes to explore the pathway that may regulate the occurrence and development of PAAD.
Diagnostic and prognostic significance of selected gene in TCGA and GEO database
We analyzed the expression of the selected genes in PAAD tissues and para carcinoma tissues from patients in the TCGA database and GEO database (GSE84129, and GSE62452 datasets). We performed the Kaplan–Meier curve and log-rank test and ROC analyses as well as multivariate Cox regression analysis to investigate the relationship between the expression of selected genes and the clinical outcome of PAAD patients.
Statistical analysis
SPSS 21 (SPSS Inc., Chicago, IL, USA) was used to perform statistical analysis. GraphPad Prism 5.0 (GraphPad Software, Inc., San Diego, CA, USA) was used to draw figures. Pearson’s chi-square test was used to compare the categorical variables. The student’s t-test was used to analyze normally distributed continuous variables. Kaplan–Meier plots with the log-rank test were used to estimate survival differences. The diagnostic significance of selected genes for PAAD was evaluated by the ROC curve. P < 0.05 was considered statistically significant.
Results
Identification of DEGs in PAAD
We screened DEGs from 123 cases of primary tumor tissues using limma packages in R language, compared with 86 cases paired para-cancerous nonmalignant tissues. Using both adjusted P value < 0.05 and logFC (fold change) > 1.2 criteria, a total of 1228 genes were identified in GSE55643, including 529 up-regulated genes and 699 down-regulated genes. A total of 2280 genes were screened in GSE15471, including 1289 up-regulated genes and 991 down-regulated genes. Then, the overlapping DEGs in the microarray data GSE55643 and GSE15471 were analyzed by Venn plot (Fig. 1), and finally, 261 DEGs were obtained, including 118 up-regulated genes and 143 down-regulated genes (Supplementary Table 1).
KEGG and GO enrichment analysis of DEGs
To further explore the function of the identified DEGs in PAAD, the online biological classification software DAVID was applied to perform GO and KEGG pathway analyses on PAAD. As shown in Supplementary Fig. 1, we list the top 6 significant terms for the BP, CC, MF, and KEGG pathways of DEGs, respectively (Supplementary Fig. 1). We list the annotation of the up-regulated genes and down-regulated genes. The up-regulated DEGs were mainly enriched for genes involved in the type I interferon signaling pathway, response to virus and extracellular matrix organization in the BP group. The down-regulated DEGs were mainly enriched genes in the reactive oxygen species metabolic process, regulation of cell adhesion and multicellular organism aging. In the CC group, the up-regulated DEGs were mainly enriched for genes associated with the extracellular exosome, extracellular space and extracellular region. The down-regulated DEGs were mainly enriched in the integral component of plasma membrane, extracellular exosome and integral component of membrane. In the MF group, the up-regulated DEGs were mainly enriched from genes associated with chemokine activity, laminin binding and calcium ion binding. The down-regulated DEGs were enriched in the lipid binding, phospholipid binding and serine-type endopeptidase inhibitor activity in the MF group. In addition, 3 significant KEGG pathways for up-regulated genes, such as ECM-receptor interaction, PI3K-Akt signaling pathway, focal adhesion. Moreover, 3 significant KEGG pathways for down-regulated genes, such as fat digestion and absorption, protein digestion and absorption, vitamin digestion and absorption (Supplementary Table 2).
Construction of PPI networks and identification of gene clusters
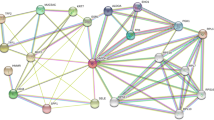
The 118 up‑ and 143 downregulated genes were input into the STRING database to identify the significant interactions between proteins and then visualized by Cytoscape software. 186 nodes with 348 edges were selected to construct the PPI networks with a confidence score of > 0.900 (highest confidence). 2 gene clusters both including 9 nodes with 36 edges were identified by MCODE which was built in the Cytoscape (Fig. 2). Gene cluster 1 includes five down-regulated genes (C5, ADCY1, GNG7, CXCL12, LPAR3) and four up-regulated genes (CXCL3, CXCL5, CCL20, NMU). All genes of gene cluster 2 were up-regulated genes (IFIT2, IRF4, BST2, XAF1, OAS1, OAS2, IFI6, IFI27, RSAD2).
PPI networks of DEGs. Based on the STRING online database, 186 genes/nodes were filtered into the DEG PPI network and visualized by Cytoscape software (version 3.7.2). The two highlighted circle areas are the most significant modules. upregulated DEGs are represented by orange circles and downregulated DEGs are represented by red circles.
Survival and expression analysis of cluster of genes in the Kaplan–Meier plotters and the TNMplot database
We performed the Kaplan–Meier curve and log-rank test analyses to investigate the prognosis of gene clusters in PAAD tissues. The 177 PAAD specimens were divided into high expression group and low expression group taking the median expression as the critical point. The results showed that 13 genes expression including 10 up-regulated genes (XAF1, IFI27, OAS1, OAS2, IFIT2, RSAD2, BST2, CCL20, NMU, CXCL5, Fig. 3A) and 3 down-regulated genes (C5, ADCY1, GNG7, Fig. 3B) were associated with significantly poorer overall survival in patients with PAAD, whereas 5 genes including 3 up-regulated genes (IRF4, IFI6 and CXCL3) and 2 down-regulated genes (CXCL12, LPAR3) showed no significant correlations19. We also used the TNMplot database for the comparison of these genes expression validated by Kaplan–Meier curve and log-rank test analyses in normal, tumor, and metastatic tissues of PAAD patients. Compared with normal samples, 10 genes were significantly overexpressed (Fig. 4A) and 3 genes were significantly down expressed (Fig. 4B) in PAAD samples, which was consistent with the original datasets. In addition, OAS1, CXCL5 and BST2 expression levels were exhibited higher in PAAD metastatic samples than no metastatic samples. The down-regulated genes, C5, GNG7, and ADCY1 were exhibited significant lower expression in metastatic samples than no metastatic samples.
Overall survival of the genes with expression in accordance with that in Kaplan–Meier Plotter database (http://kmplot.com/analysis/). (A) High expression of 10 up-regulated genes was associated with poor overall survival in PAAD (a-j). (B) Low expression of 3 down-regulated genes was associated with poor overall expression in PAAD (a-c).
The comparison of the cluster of genes expression in normal, tumor, and metastatic tissues of PAAD patients in the TNMplot database (https://www.tnmplot.com/). The 177 PAAD specimens were divided into high expression group and low expression group taking the median expression as the critical point. (A) 10 up-regulated genes significantly overexpressed in PAAD samples compared with normal tissues (a–j). The mRNA expression of OAS1, CXCL5, and BST2 in metastatic tissues was higher than tumor tissues (a, f, g). (B)The mRNA expression of 3 down-regulated genes was incrementally downregulated in the normal, tumor, and metastatic tissues (a–c).
KEGG enrichment analysis of 13 selected Genes by DAVID.
We performed KEGG analysis on 13 selected genes in DAVID to explore the potential pathway and mechanism that 13 genes may regulate the occurrence and development of PAAD. The result showed 6 significant KEGG pathways for 13 validated genes, including Chemokine signaling pathway, Cytokine-cytokine receptor interaction, Rheumatoid arthritis, Pathways in cancer, Influenza A, and Herpes simplex infection (Table 1). We observed that two genes (GNG7, ADCY1) were meaningfully enriched in the Pathways in Cancer (P = 0.016, Fig. 5) (https://www.kegg.jp/kegg/kegg1.html).
Reanalysis of 13 validated genes via KEGG pathway enrichment. two genes (GNG7, ADCY1) were markedly enriched in the Pathways in Cancer (https://www.kegg.jpkegg/kegg1.html). AC means ADCY1. βy means GNG7.
Diagnostic value and clinical outcomes of GNG7 and ADCY1 in TCGA database.
We downloaded the PAAD data set from the TCGA database to analyze the relationship between GNG7, ADCY1 mRNA expression and the clinical outcome. The mRNA expression of GNG7 and ADCY1 in the PAAD tissues was significantly lower than in the normal tissues. In addition, the GNG7 and ADCY1 mRNA expression was incrementally downregulated with increasing neoplasm histology grades as well as tumor stages (Fig. 6A–D). The low expression of GNG7 mRNA was related to the neoplasm histology grades (P = 0.045) and survival (P = 0.007) in PAAD patients. The low expression of ADCY1 mRNA was related to gender (P = 0.035), neoplasm histology grades (P = 0.008), and survival (P = 0.036) (Table 2). The Multivariate Cox Regression analysis showed that the GNG7 (HR (95%CI): 2.106 (1.357–3.269); P = 0.010) and ADCY1 (HR (95%CI): 1.881 (1.221–2.898); P = 0.004) low expression, neoplasm histology grades 3/4 (HR (95%CI): 0.637 (0.408–0.995); P = 0.047), preoperative pharmaceutical history (HR (95%CI): 0.452 (0.281–0.725); P = 0.001) were independent risk factors for overall survival of PAAD (Table 3). And we constructed a combined expression model of GNG7 and ADCY1 to evaluate the diagnostic value for PAAD. The Combined expression score = 0.651* GNG7 + 0.691*ADCY1. The ROC analysis showed that combined expression of GNG7 and ADCY1 have diagnostic value in distinguishing between PAAD and normal pancreas tissue (AUC = 0.937, P = 0.0028, Fig. 6E). Finally, we performed the Pearman correlation test analysis in the LinkedOmics database28 to investigate the correlation of the mRNA expression between GNG7 and ADCY1 in the PAAD samples. The result exhibited a significantly positive correlation (r = 0.622, P = 0.000, Fig. 6F).
Analysis of GNG7 AND ADCY1 expression and diagnostic significance of 177 cases of PAAD patients in the TCGA database. (A, B) The mRNA expression of GNG7 in the PAAD tissues was significantly lower than in the adjacent normal tissues. The GNG7 mRNA expression was incrementally downregulated with increasing neoplasm histology grades (A) as well as tumor stages (B). (C–D) The mRNA expression of ADCY1 in the PAAD tissues was significantly lower than in the adjacent normal tissues. The ADCY1 mRNA expression was incrementally downregulated with increasing neoplasm histology grades (C) as well as tumor stages (D). (E) The ROC curve showed that the combined expression of GNG7 and ADCY1 has diagnostic value for PAAD patients (AUC = 0.937, P = 0.0028). (F) The Pearman correlation test analysis exhibited a significantly positive correlation between GNG7 and ADCY1 (r = 0.622, P = 0.000). This figure was drawn by GraphPad Prism 5.0 (GraphPad Software, Inc., San Diego, CA, USA; https://www.graphpad.com/).
Diagnostic and prognostic value of GNG7 and ADCY1 was investigated in the GSE84129 and GSE62452 datasets.
We downloaded two independent PAAD datasets from the GEO database (GSE84129 and GSE62452 datasets) to analyze the diagnostic and prognostic value of GNG7 and ADCY1. In the GSE84129 dataset, PAAD patients with high survival time (more than 24 months) have significantly higher mRNA expression of GNG7 and ADCY1 than patients with short survival time (no more than 24 months) (Fig. 7A,B). In addition, the ROC analysis showed that combined expression of GNG7 and ADCY1 have diagnostic value in distinguishing between PAAD and normal pancreas tissue (AUC = 0.863, P = 0.0008, Fig. 7C). In the independent GSE62452 dataset, the GNG7 and ADCY1 mRNA expression was incrementally downregulated with increasing tumor stages as well as neoplasm histology grades (Fig. 7D–G). Furthermore, PAAD patients with high expression of GNG7 and ADCY1 have a longer survival time than patients with low GNG7 and ADCY1 expression (Fig. 7H,I). We also performed the ROC analysis showed that combined expression of GNG7 and ADCY1 have diagnostic value for HCC (AUC = 0.798, P < 0.0001, Fig. 7J). Moreover, the Multivariate Cox Regression analysis and the results showed that low expression of GNG7 and ADCY1 were independent risk factors for OS of PAAD patients (Fig. 7K). All these results demonstrated that the GNG7 and ADCY1 has diagnostic and prognostic value for PAAD.
Diagnostic and prognostic value of GNG7 and ADCY1 in the GSE84129 and GSE62452 datasets. (A, B) PAAD patients with high survival time have significantly higher mRNA expression of GNG7 (A) and ADCY1 (B) than patients with short survival time. (C) The ROC curve showed that the combined expression of GNG7 and ADCY1 has diagnostic value for PAAD in the GSE84129 dataset (AUC = 0.863, P = 0.0008). (D–E) The GNG7 (D) and ADCY1 (E) mRNA expression was incrementally downregulated with increasing tumor stages. (F, G) The GNG7 (F) and ADCY1 (G) mRNA expression was incrementally downregulated with increasing neoplasm histology grades. (H, I) PAAD patients with high expression of GNG7 (H) and ADCY1 (I) have a longer survival time than patients with low expression. (J) The ROC curve showed that the combined expression of GNG7 and ADCY1 has diagnostic value for PAAD in the GSE62452 dataset (AUC = 0.798, P < 0.0001). (K) Low expression of GNG7 and ADCY1 were independent risk factors for OS of PAAD patients.
Discussion
PAAD is one of the most malignant cancers. Due to the lack of sensitive early diagnosis biomarker, it is challenging to develop effective treatments to delay and reverse the progression of PAAD. The 5-year survival percentage of PAAD patients for all stages was only 9%, based on data in 2019 released by the American Cancer Society29. Therefore, more meaningful biomarkers should be discovered to improve the diagnostic and prognostic efficiency of PAAD patients30,31. At present, a variety of biomarkers with prognosis and prognosis for PAAD has been reported32,33,34. Jiang et al. suggest that CDK1 and CCNA2 may be potential diagnostic and prognostic biomarkers in patients with PAAD35. In addition, CDK1 and CCNA2 has been reported promoting progression of PAAD via regulating the P53 and cell cycle signaling pathway, which were subpathways of Pathway in cancer35,36,37. Georgiadou et al. reported that overexpression of VEGF and Id-1 was associated with high microvessel density and emerged as prognostic factors in patient survival in PAAD38. Additionally, the VEGF signaling pathways take an important position in Pathway in cancer (Fig. 6).
This present study aimed to screen out the genes that has diagnostic and prognostic significance for PAAD by comparing the tumor tissues with adjacent non-malignant tissues. 118 up-regulated and 143 down-regulated overlapping DEGs were screened out from the microarray data GSE55643 and GSE15471. Then, we performed the GO and KEGG enrichment analysis for overlapping DEGs using the DAVID database. Subsequently, we constructed a PPI network with 186 nodes/genes and 348 edges in the STRING database and visualized it by Cytoscape software. From the PPI network, we screened out the two most core gene clusters, which including 13 up-regulated and 5 down-regulated genes. We analyzed the mRNA expression and the prognosis of survival of these genes in the TNMplot and Kaplan–Meier plotters database, respectively. By this process, 10 up-regulated and 3 down-regulated genes were screened. Next, we performed KEGG analysis on 13 selected genes in DAVID to explore the potential pathway and mechanism that 13 genes may regulate the occurrence and development of PAAD. We observed that two genes (GNG7, ADCY1) meaningfully enriched in the Pathways in Cancer. Additionally, we performed diagnostic and prognostic analysis of GNG7 and ADCY1 in the independent TCGA cohort, GSE84129 and GSE62452 datasets. The result showed that the mRNA expression of GNG7 and ADCY1 in the PAAD tissues was significantly lower than in the normal tissues. The Multivariate Cox Regression analysis showed that the low expression of GNG7 (HR (95%CI): 2.106 (1.357–3.269); P = 0.010) and ADCY1 (HR (95%CI): 1.881 (1.221–2.898); P = 0.004) were independent risk factors for overall survival of PAAD (Table 3). The ROC analysis showed that low combined expression of GNG7 and ADCY1 have diagnostic value in distinguishing between PAAD and normal pancreas tissue. Meanwhile, the Pearman correlation test analysis in the LinkedOmics database exhibited a significantly positive correlation between GNG7 and ADCY1 (r = 0.622, P = 0.000). We observed that the expression of GNG7 and ADCY1 in the TCGA database has no significant difference between adjacent normal tissue and tumors with early stages (stage 1) or tumor grades (grade 1). This lack of significance may relate to the small normal sample size (n = 4). After all, we notice a significantly different expression in the GSE62452 dataset between adjacent normal tissue and tumors with early stages or grades. However, the diagnostic efficiency of GNG7 and ADCY1 for early stage PAAD indeed requires further investigations. Therefore, all our results indicate that GNG7 and ADCY1 may as diagnostic and prognostic biomarker for PAAD.
GNG7 are involved as a modulator or transducer in various transmembrane signaling systems. The beta and gamma chains are required for the GTPase activity, for replacement of GDP by GTP, and for G protein-effector interaction. Plays a role in the regulation of adenylyl cyclase signaling in certain regions of the brain39. A previous study showed that GNG7 was down-regulated in clear cell renal cell carcinoma tissues due to promoter methylation and frequent gene mutations, and negatively associated with overall survival40. Additionally, previous studies showed that reduced expression of GNG7 was associated with breast cancer, lung carcinogenesis, head and neck cancer and esophageal cancer41,42,43,44,45. However, rarely the previous study has addressed the role of GNG7 in PAAD and its prognostic value. This study is the first systematic investigation of diagnostic value, clinical significance of GNG7 in PAAD.
Adenylate cyclase 1 (ADCY1) encodes a member of the adenylate cyclase gene family that is primarily expressed in the brain46,47. This protein is regulated by calcium/calmodulin concentration and may be involved in brain development. ADCY1 can regulate drug resistance in lung cancer through participating in cAMP signaling pathways and was of great significance to be a novel prognostic biomarker48. It is worth noting that the cAMP signaling pathways was the subpathways of Pathway in cancer. Additionally, the upregulation of ADCY1 by regulating microRNA-127-3p exerts anti-tumor effects on colon cancer49. Y Li et al. showed that the expression of ADCY1 was downregulated in osteosarcoma compared with benign bone tumors, suggesting that ADCY1 may be potential biomarkers for osteosarcoma tumorigenesis and therapeutics50. Furthermore, previous studies reported that ADCY1 was a key candidate gene in melanoma and rectal adenocarcinoma metastasis51,52. However, Ma M et al. reported that ADCY1 was regulated by miR-23a-3p and plays a cancer‐promoting role in mucosal melanoma53.
In summary, this study screened out 2 potential genes that has diagnostic and prognostic significance for PAAD through the systematic bioinformatic-based analyses. Of course, all of the DEG candidate genes we have screened out related to PAAD should be confirmed through molecular biology and cytology experiments.
Conclusion
The mRNA expression level of GNG7, ADCY1 was significantly down-regulated in PAAD tissues compared with adjacent normal tissues. Low GNG7, ADCY1 mRNA expression level was associated with poor clinical outcomes of PAAD patients. GNG7 and ADCY1 may be an oncogene that regulated tumorigenesis and development through pathways in cancer and could be used as a biomarker of the diagnostic and prognostic value of PAAD.
Data availability
All datasets generated for this study are available within the article.
References
Ilic, M. & Ilic, I. Epidemiology of pancreatic cancer. World J. Gastroenterol. 22, 9694–9705. https://doi.org/10.3748/wjg.v22.i44.9694 (2016).
McGuigan, A. et al. Pancreatic cancer: a review of clinical diagnosis, epidemiology, treatment and outcomes. World J. Gastroenterol. 24, 4846–4861. https://doi.org/10.3748/wjg.v24.i43.4846 (2018).
Navaneethan, U. et al. Volatile organic compounds in bile can distinguish pancreatic cancer from chronic pancreatitis: a prospective observational study. Eur. J. Phys. 41, doi:https://doi.org/10.1055/a-1255-9169 (2020).
Grassia, R. et al. Eus-guided tissue acquisition in chronic pancreatitis: differential diagnosis between pancreatic cancer and pseudotumoral masses using eus-fine needle aspiration or core biopsy. Digest Liver Dis. 52, S6–S7 (2020).
Ren, Z. G. et al. Gut microbial profile analysis by MiSeq sequencing of pancreatic carcinoma patients in China. Oncotarget. 8, 95176–95191. https://doi.org/10.18632/oncotarget.18820 (2017).
Thomas, R. M. et al. Intestinal microbiota enhances pancreatic carcinogenesis in preclinical models. Carcinogenesis 39, 1068–1078. https://doi.org/10.1093/carcin/bgy073 (2018).
Duffy, M. J. et al. Tumor markers in pancreatic cancer: a European Group on Tumor Markers (EGTM) status report. Ann. Oncol. 21, 441–447. https://doi.org/10.1093/annonc/mdp332 (2010).
Poruk, K. E., Pawlik, T. M. & Weiss, M. J. Perioperative management of hilar cholangiocarcinoma. J. Gastrointest. Surg. 19, 1889–1899. https://doi.org/10.1007/s11605-015-2854-8 (2015).
Lv, S. et al. High TXLNA expression predicts favourable outcome for pancreatic adenocarcinoma patients. Biomed. Res. Int. 2020, 2585862. https://doi.org/10.1155/2020/2585862 (2020).
Zheng, X., Liu, X., Zheng, H., Wang, H. & Hong, D. Integrated bioinformatics analysis identified COL11A1 as an immune infiltrates correlated prognosticator in pancreatic adenocarcinoma. Int. Immunopharmacol. 90, 106982. https://doi.org/10.1016/j.intimp.2020.106982 (2021).
Samuel, N. & Hudson, T. J. The molecular and cellular heterogeneity of pancreatic ductal adenocarcinoma. Nat. Rev. Gastroenterol. Hepatol. 9, 77–87. https://doi.org/10.1038/nrgastro.2011.215 (2011).
Gentleman, R., Carey, V. J., Huber, W., Irizarry, R. A. & Dudoit, S. [Statistics for Biology and Health] bioinformatics and computational biology solutions using R and bioconductor. Limma. Linear Models Microarray Data. https://doi.org/10.1007/0-387-29362-0, 397–420 (2005).
Dessau, R. B. & Pipper, C. B. “R”–project for statistical computing. Ugeskr Laeger 170, 328–330 (2008).
Dennis, G. Jr. et al. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol 4, P3 (2003).
Franceschini, A. et al. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res 41, D808-D815, doi:https://doi.org/10.1093/nar/gks1094 (2013).
Kohl, M., Wiese, S. & Warscheid, B. J. M. Cytoscape: software for visualization and analysis of biological networks. Methods Mol. Biol. 696, 291–303 (2011).
Zhang, C. et al. The identification of key genes and pathways in hepatocellular carcinoma by bioinformatics analysis of high-throughput data. Med. Oncol. 34, 101. https://doi.org/10.1007/s12032-017-0963-9 (2017).
Rezaei-Tavirani, M., Rezaei-Taviran, S., Mansouri, M., Rostami-Nejad, M. & Rezaei-Tavirani, M. Protein-protein interaction network analysis for a biomarker panel related to human esophageal adenocarcinoma. Asian Pac. J. Cancer Prev. 18, 3357–3363. https://doi.org/10.22034/APJCP.2017.18.12.3357 (2017).
Ádám, N., András, L., Otília, M. & Reports, G. R. B. J. S. Validation of miRNA prognostic power in hepatocellular carcinoma using expression data of independent datasets. Sci Rep 8, 9227 (2018).
Weinstein, J. N. et al. The Cancer Genome Atlas Pan-Cancer analysis project. Nature Genetics (2013).
Lunardi, S. et al. IP-10/CXCL10 induction in human pancreatic cancer stroma influences lymphocytes recruitment and correlates with poor survival. Oncotarget 5, 11064–11080. https://doi.org/10.18632/oncotarget.2519 (2014).
Badea, L., Herlea, V., Dima, S. O., Dumitrascu, T. & Popescu, I. Combined gene expression analysis of whole-tissue and microdissected pancreatic ductal adenocarcinoma identifies genes specifically overexpressed in tumor epithelia. Hepatogastroenterology 55, 2016–2027 (2008).
Barrett, T. et al. NCBI GEO: archive for functional genomics data sets–update. Nucleic Acids Res. 41, D991-995. https://doi.org/10.1093/nar/gks1193 (2013).
Edgar, R., Domrachev, M. & Lash, A. E. Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30, 207–210. https://doi.org/10.1093/nar/30.1.207 (2002).
Benjamini, Y. & Hochberg, Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. Roy. Stat. Soc. 57, 289–300 (1995).
Ashburner, M. et al. Gene ontology: tool for the unification of biology the gene ontology consortium. Nat. Genet. 25, 25–29. https://doi.org/10.1038/75556 (2000).
Kanehisa, M., Goto, S., Sato, Y., Furumichi, M. & Tanabe, M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 40, D109-114. https://doi.org/10.1093/nar/gkr988 (2012).
Vasaikar, S. V., Straub, P., Wang, J. & Zhang, B. LinkedOmics: analyzing multi-omics data within and across 32 cancer types. Nucleic Acids Res. 46, D956–D963. https://doi.org/10.1093/nar/gkx1090 (2018).
Siegel, R. L., Miller, K. D. & Jemal, A. Cancer statistics, 2019. CA Cancer J. Clin. 69, 7–34. https://doi.org/10.3322/caac.21551 (2019).
Khomiak, A. et al. Recent discoveries of diagnostic, prognostic and predictive biomarkers for pancreatic cancer. Cancers (Basel) https://doi.org/10.3390/cancers12113234 (2020).
Turanli, B., Yildirim, E., Gulfidan, G., Arga, K. Y. & Sinha, R. Current state of “Omics” biomarkers in pancreatic cancer. J. Pers. Med. https://doi.org/10.3390/jpm11020127 (2021).
Li, H. et al. SRPX2 and RAB31 are effective prognostic biomarkers in pancreatic cancer. J. Cancer 10, 2670–2678. https://doi.org/10.7150/jca.32072 (2019).
Chen, H. et al. Three hypomethylated genes were associated with poor overall survival in pancreatic cancer patients. Aging (Albany NY) 11, 885–897. https://doi.org/10.18632/aging.101785 (2019).
Yu, Y., Feng, X. & Cang, S. A two-microRNA signature as a diagnostic and prognostic marker of pancreatic adenocarcinoma. Cancer Manag. Res. 10, 1507–1515. https://doi.org/10.2147/CMAR.S158712 (2018).
Jiang, P., Zhang, M., Gui, L. & Zhang, K. Expression patterns and prognostic values of the cyclin-dependent kinase 1 and cyclin A2 gene cluster in pancreatic adenocarcinoma. J. Int. Med. Res. 48, 300060520930113. https://doi.org/10.1177/0300060520930113 (2020).
Chen, L. Y. et al. Overexpression of CBX3 in Pancreatic adenocarcinoma promotes cell cycle transition-associated tumor progression. Int. J. Mol. Sci. https://doi.org/10.3390/ijms19061768 (2018).
Hou, X. et al. KDM1A and KDM3A promote tumor growth by upregulating cell cycle-associated genes in pancreatic cancer. Exp. Biol. Med. (Maywood) https://doi.org/10.1177/15353702211023473 (2021).
Georgiadou, D. et al. VEGF and Id-1 in pancreatic adenocarcinoma: prognostic significance and impact on angiogenesis. Eur. J. Surg. Oncol. 40, 1331–1337. https://doi.org/10.1016/j.ejso.2014.01.004 (2014).
Schwindinger, W. F. et al. Loss of G protein gamma 7 alters behavior and reduces striatal alpha(olf) level and cAMP production. J. Biol. Chem. 278, 6575–6579. https://doi.org/10.1074/jbc.M211132200 (2003).
Liu, J. et al. G protein gamma subunit 7 induces autophagy and inhibits cell division. Oncotarget 7, 24832–24847. https://doi.org/10.18632/oncotarget.8559 (2016).
Ohta, M. et al. Clinical significance of the reduced expression of G protein gamma 7 (GNG7) in oesophageal cancer. Br. J. Cancer 98, 410–417. https://doi.org/10.1038/sj.bjc.6604124 (2008).
Sheng, H., Li, X. & Xu, Y. Knockdown of FOXP1 promotes the development of lung adenocarcinoma. Cancer Biol. Ther. 20, 537–545. https://doi.org/10.1080/15384047.2018.1537999 (2019).
Demokan, S. et al. Identification of guanine nucleotide-binding protein gamma-7 as an epigenetically silenced gene in head and neck cancer by gene expression profiling. Int. J. Oncol. 42, 1427–1436. https://doi.org/10.3892/ijo.2013.1808 (2013).
Mei, J., Wang, T., Zhao, S. & Zhang, Y. Osthole inhibits breast cancer progression through upregulating tumor suppressor GNG7. J. Oncol. 2021, 6610511. https://doi.org/10.1155/2021/6610511 (2021).
Hartmann, S. et al. Loss of protein expression and recurrent DNA hypermethylation of the GNG7 gene in squamous cell carcinoma of the head and neck. J. Appl. Genet. 53, 167–174. https://doi.org/10.1007/s13353-011-0079-4 (2012).
Lubker, C. et al. Membranous adenylyl cyclase 1 activation is regulated by oxidation of N- and C-terminal methionine residues in calmodulin. Biochem. Pharmacol. 93, 196–209. https://doi.org/10.1016/j.bcp.2014.11.007 (2015).
Villacres, E. C. et al. Cloning, chromosomal mapping, and expression of human fetal brain type I adenylyl cyclase. Genomics 16, 473–478. https://doi.org/10.1006/geno.1993.1213 (1993).
Zou, T. et al. A perspective profile of ADCY1 in cAMP signaling with drug-resistance in lung cancer. J. Cancer 10, 6848–6857. https://doi.org/10.7150/jca.36614 (2019).
Liu, G. et al. Long non-coding RNA DPP10-AS1 exerts anti-tumor effects on colon cancer via the upregulation of ADCY1 by regulating microRNA-127–3p. Aging (Albany NY) https://doi.org/10.18632/aging.202729 (2021).
Li, Y. et al. Comparative proteomics analysis of human osteosarcomas and benign tumor of bone. Cancer Genet. Cytogenet. 198, 97–106. https://doi.org/10.1016/j.cancergencyto.2010.01.003 (2010).
Chen, J. et al. Identification of key candidate genes involved in melanoma metastasis. Mol. Med. Rep. 20, 903–914. https://doi.org/10.3892/mmr.2019.10314 (2019).
Hua, Y. et al. Identification of the potential biomarkers for the metastasis of rectal adenocarcinoma. APMIS 125, 93–100. https://doi.org/10.1111/apm.12633 (2017).
Ma, M. et al. MicroRNA-23a-3p Inhibits Mucosal Melanoma Growth and Progression through Targeting Adenylate Cyclase 1 and Attenuating cAMP and MAPK Pathways. Theranostics 9, 945–960. https://doi.org/10.7150/thno.30516 (2019).
Acknowledgements
We thank the contributors of the GSE15471 and GSE55643 for sharing the PAAD expression profile data set on open access. Our deepest gratitude goes to the editors and anonymous reviewers for their careful work and thoughtful suggestions that have helped to improve this paper substantially.
Author information
Authors and Affiliations
Contributions
Conceived and designed the study: Youfu Zhang, Xinchang Li. Wrote the paper: Youfu Zhang, Jinran Yang, Xuyang wang, Xinchang Li. Analyzed the data: Youfu Zhang, Xinchang Li. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zhang, Y., Yang, J., Wang, X. et al. GNG7 and ADCY1 as diagnostic and prognostic biomarkers for pancreatic adenocarcinoma through bioinformatic-based analyses. Sci Rep 11, 20441 (2021). https://doi.org/10.1038/s41598-021-99544-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-99544-x
- Springer Nature Limited
This article is cited by
-
An integrated ensemble learning technique for gene expression classification and biomarker identification from RNA-seq data for pancreatic cancer prognosis
International Journal of Information Technology (2024)
-
MALAT1-regulated gene expression profiling in lung cancer cell lines
BMC Cancer (2023)
-
VIVA1: a more invasive subclone of MDA-MB-134VI invasive lobular carcinoma cells with increased metastatic potential in xenograft models
British Journal of Cancer (2022)
-
CUX2 prevents the malignant progression of gliomas by enhancing ADCY1 transcription
Experimental Brain Research (2022)