Abstract
The NAC (NAM, ATAF1/2, and CUC2), is a large family of plant-specific transcription factors (TFs) that exert crucial regulatory roles in various physiological processes and abiotic stresses. There is scanty information on the role of the NAC family in sunflower (Helianthus annuus L.). In this study, we conducted a genome-wide survey and expression analysis of the NAC family in sunflower. A total of 150 HaNACs were identified in sunflower. Phylogenetic analysis to compare HaNACs with Arabidopsis NACs generated 15 clusters. Among them, eight membrane-bound NAC TFs with transmembrane helixes were found (designated as NTLs), which were suggested to be localized in the membrane and transferred to the nucleus through proteolysis. Notably, 12 HaNACs were potentially regulated via miR164 cleavage or translational inhibition. By analyzing RNA-seq data from Sequence Read Archive (SRA), the expression of HaNACs showed tissue specificity and strong response to drought stress. Additionally, phylogenetic analysis of 150 HaNACs with the previously reported NACs related to abiotic stress revealed that 75% of the abiotic stress-related NACs were clustered into the SNAC (abiotic stress-related NAC) group, and only 25% were in the Non-SNAC group. qRT-PCR further demonstrated that about 75% of the HaNACs in the SNAC subgroup were induced by salt and drought stress, and the expression of some HaNACs showed tissue specificity. These findings provide valuable information that can deepen the understanding of how NAC TFs in sunflower respond to abiotic stress.
Similar content being viewed by others
Introduction
Environmental stresses are the main limiting factors for the growth, productivity, and distribution of plants. One-third of the Earth's surface is classified as either arid or semi-arid, and nearly 40% of the world's land is characterized by potentially high salinity levels1. For sunflower, poor weather and soil conditions are among the major obstacles to high yield production2, and soil salinity is particularly the major constraint3. High salt concentration can lower the seed germination rate, limit the water absorption of plant roots, delay plant growth, and cause nutritional imbalance4. In addition, secondary pressures, such as oxidative damage, can exacerbate the influence of the above outcomes5. Drought stress is the most prevalent environmental factor limiting crop productivity6, and global climate change is increasing the frequency of severe drought conditions7. Drought stress causes the transpiration rate to exceed the water uptake, and the growth of plants will encounter water shortage. Water deficiency can cause the changes of solute concentration in cells, which leads to the disruption of water potential gradients, changes of cell volume, denaturation of protein, and disruption of membrane integrity, and eventually the plants are damaged6.
Globally, sunflower is the fourth most important oil crop after palm, soybean, and rapeseed, and second in Europe. A growing trend for sunflower cooking oil is attributed to the rise in sunflower production, worldwide8. Sunflower is considered a moderately salt-tolerant crop and is widely cultivated in saline-alkali areas around the world9. Reconstruction of crops can enhance their resistance to abiotic stress. However, it is still challenging to grow plants in extreme environments. A few crops can grow in highly salinized soils. There is enormous potential to develop saline-alkali characteristic crops for sunflowers based on its well-known moderate salt tolerance, compared with non-resistant crops. Limited rainfall or shortage of water for irrigation during the growing season constraints its seed yield with significant reductions10. Therefore, the breeding of drought tolerant cultivars will contribute to more stable sunflower production. Several studies have established sunflower regeneration and genetic modification methods, which makes it possible to use genetic engineering to cultivate salt and drought-tolerant sunflower11,12,13,14. However, there is also a need to find genes to endow the salt and drought resistance of plants.
Abiotic stress resistance requires the production of important functional proteins, such as osmoprotectants and regulatory proteins, that play roles in signal transduction pathways, including kinases and transcription factors (TFs)15. NAC (NAM, ATAF1/2, and CUC2) family is a plant-specific transcription factor firstly discovered in Petunia hybrida16, and involved in plant salt and drought stress response (multiple abiotic-stress responses)17,18,19. They possess a conserved NAC domain (about 150 amino acids in the N-terminal region), which usually exists in five subdomains (A, B, C, D and E). The NAC domain is responsible for DNA binding and dimer formation, whereas different C-terminal regions participate in transcriptional regulation19,20.
NACs have been identified from a variety of plants, such as banana21, Fragaria vesca22, Fragaria × ananassa23, Oxytropis ochrocephala24, Sesamum indicum25, Capsicum annuum26. More than 100 genes from this family have been identified in Arabidopsis, rice and soybean20,27,28. Data on sunflower genome sequences were published and released in 201729, which was a breakthrough in the molecular biological research process of sunflowers. Since then, a few studies on NACs in Helianthus annuus are ongoing. Of note, only a study by Yuce et al.30 has so far reported that sunflower NAC TFs responded to chromium stress. Elsewhere, Moschen et al. found some NACs were associated with leaf senescence31, and a sunflower NAC was induced under drought stress32. However, the functions of most sunflower NAC members are unknown. This study gap prompted us to comprehensively explore the sunflower NAC family by identification, protein property, phylogenetic analysis, transmembrane helix, miRNA164 target site, tissue distribution, and analysis of abiotic stress expression profiles. This work will lay a foundation for further study on the function and regulation mechanism of NAC TFs in sunflower.
Results
Identification and annotation of NAC members
Initially, a total of 157 non-redundant putative NACs were identified via HMM (Hidden Markov Model) search. First, through the identification of CCD and pfam, one protein was deleted, because it lacked the NAC domain. Then, six proteins among them with incomplete NAC subdomains were also removed, after which we obtained a final total of 150 NACs with intact NAC domains from the sunflower. The number was slightly higher than that of plants such as tomato (104 NACs)33, Arabidopsis (106)34, sesame (87)25, and soybean (101)35. It was speculated that the presence of more NACs in the sunflower highly reflects their participation in the complex transcriptional regulation of sunflower. This phenomenon also appears to be caused by multiple gene duplication events.
Moreover, the characteristics of NAC protein sequences greatly varied in sunflower. The sequence length of 150 HaNACs ranged from 139 to 636 aa, the molecular weight from 16.16 to 72.13 kDa and the isoelectric point from 4.17 to 10.44. Detailed information on these data was shown in Table S1.
Due to the lack of comprehensive standard annotations, we mapped the 150 HaNACs to all 17 chromosomes and named them from HaNAC1-HaNAC150 based on their position on the chromosome. Among them, chromosome 13 had a maximum of 18 HaNACs (~ 12%), whereas chromosome 6 had only 3 HaNACs (~ 2%) (Fig. 1). The uneven distribution of HaNACs on the chromosomes reflected the diversity and complexity of the HaNAC gene family.
Phylogenetic analysis of sunflower NACs
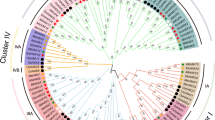
In 2003, Ooka et al. established a classification system for the NAC family20, which has so far been applied in various plants, such as sorghum36, pepper26, and rice37, among others. Herein, to reveal the phylogenetic relationship and potential functional characteristics of the HaNAC family members, a phylogenetic tree was constructed using 255 full-length protein sequences from sunflower and Arabidopsis using MEGA X, Neighbor-joining (NJ) and Maximum Likelihood (ML). Since the trees produced by the two methods were largely similar (data not shown), only the result generated via the NJ method was shown in Fig. 2. We divided the 150 HaNACs into two groups and 15 subgroups. The largest clade was the ONAC003 subgroup containing 25 HaNACs, while the NAC1 subgroup constituted the smallest clade with 5 HaNACs and this clade only contained NACs from Arabidopsis thaliana. Remarkably, Arabidopsis NACs with the same function were clustered to the same subgroup. For instance, the AtNAC3, NAP, and ATAF subgroups contained several well-known stress-responsive NACs, including ANAC019, ANAC056 (AtNAC2), ANAC055 (AtNAC3), ANAC002 (ATAF1), ANAC081 (ATAF2), and ANAC072 (RD26). In addition, we examined other evidence to support the reliability of subgroup classification, such as gene structure and motifs, as described below.
A phylogenetic tree of sunflower and Arabidopsis NAC proteins. The amino acid sequences of NAC proteins were aligned using the ClustalW of MEGA X, and a phylogenetic tree was generated using the neighbor-joining method with 1000 bootstrap replicates in MEGA X. Red and black fonts denoted sunflower and Arabidopsis NACs, respectively. All NACs were classified into two groups: Group I (red) and Group II (blue), and 15 subgroups (different colors for each clade).
Gene structure and conserved motifs of sunflower NACs
Multiple sequence alignment of the sunflower NACs demonstrated that the N-terminus of all HaNACs have a conserved NAC domain, and five subdomains (Fig. S1). To better understand the relationship between the structure and function of these HaNAC proteins, the gene structure, and conserved motifs were analyzed. The exon numbers of sunflower NAC genes ranged from 2 to 7, among which, most genes had three exons (Table S2). Generally, members from the same subgroup were characterized by similar exon/intron structure and gene length. For example, subgroups i and j contained three exons, whereas subgroups a, h and k contained 2 or 3 exons (Fig. 3C).
Phylogenetic tree, motif, and genetic structure of 150 HaNAC TFs. (A) The phylogenetic tree was constructed using the Neighbor-Joining (NJ) method and 1000 bootstrap tests. (B) Showing the positions of 15 motifs in the proteins. Different colors indicated different motifs. (C) Gene structures, exons and untranslated regions (UTR) were indicated by green and yellow boxes, and black lines indicated introns. The ruler at the bottom was used to estimate their length.
A total of 15 conservative motifs of HaNAC proteins in sunflower were shown in Fig. S2. As expected, the most closely related members of the same subfamily shared a common motif composition, which also meant that they had similar functions (Fig. 3B). Most of the predicted motifs were at the N-terminus, which was more conservative. Motif5 and motif2 were present in most HaNACs, and only specific proteins in the subgroup c did not have motif1. A phylogenetic tree of 150 sunflower NACs was constructed using the above method (Fig. 3A). It was worth noting that several special motifs (10, 13, 14, and 15) at the C-terminus were clustered together. These results not only proved the reliability of the classification, but also revealed a correlation between the subfamily and motif.
Syntenic and evolutionary analysis of HaNAC gene family
Duplication events are related to plant evolution patterns, while tandem duplication and segmental duplication are sources of gene family expansion and genomic complexity. The duplication events of HaNACs were identified using blastp and MCscan. Segment duplication analysis revealed segment duplications in 17 HaNAC gene pairs such as HaNAC19–HaNAC63, and HaNAC40–HaNAC60 (Fig. 4 and Table S3). Tandem duplication (when two genes were located in a 100 kb region on the same chromosome and separated by five or fewer genes), was found in 10 HaNAC gene pairs, such as in HaNAC3–HaNAC4, and HaNAC28–HaNAC29 (Table S3). To evaluate the selection limits for duplicated HaNAC genes, we estimated ratios of the number of non-synonymous substitutions per non-synonymous site (Ka) to the number of synonymous substitutions per synonymous site (Ks). Of the 27 duplication gene pairs, 26 pairs evolved under purified selection (Ka/Ks < 1), whereas one pair evolved under positive selection (HaNAC112-HaNAC113, Ka/Ks = 1.15). The divergence time of the HaNAC genes indicated that the duplication event could be potentially traced back to 83.51 million years ago (Mya) and continued to 0.46 Mya. (Table S3).
Membrane-bound HaNACs and miR164s target site prediction
Subcellular localization prediction indicated that most of the 150 NAC proteins were localized in the nucleus (Table S1), which was consistent with the characteristics of NAC as a transcription factor. Numerous MTFs (membrane-bound transcription factors) had been identified in the NAC family and were named NTL (NAC with transmembrane motif1-like)38. They were, in most cases bound to the membrane, and possibly released from the membrane into the nucleus by proteolysis39. Using the TMHMM2.0, eight NAC proteins containing a transmembrane helix were predicted, which were designated as HaNTL1-HaNTL8 (Table 1 and Fig. S3). A phylogenetic tree of NTLs from sunflower, Arabidopsis, and rice was generated, from which sunflower NTLs were closely related to Arabidopsis NTLs, indicating a similar function of these genes in the two species (Fig. S4).
Based on the current understanding, microRNAs (small non-coding RNAs of about 22 nucleotides in length) can regulate target gene expression40. Researchers have revealed that NAC family genes are regulated by miRNA164s41,42,43. For instance, a study by Badouin et al. in 2017 sequenced small RNAs in the sunflower genome and found four exceptionally reliable miRNA164s29. The regulatory relationship between these miRNA164s and HaNACs was predicted using the psRNATarget online tool44, which was a total of 12 HaNACs were probably regulated by these miRNA164s. Among them, ten could be regulated at the transcriptional level by mRNA cleavage, whereas the other two could be regulated at the translational level via inhibition of mRNA translation. Notably, these target sites were distributed in the 5′ untranslated regions, 3′ untranslated regions, different coding sequences, but not in the conserved NAC domains (Fig. 5). This signified the complexity in the regulation of HaNACs by miRNA164s.
Cis-element analysis
To explore the potential functions of HaNACs, 24,425 cis-elements were detected in the promoter region (2000 bp upstream of the start codon of the gene) using PlantCare. Excluding conventional cis-elements (such as TATA-box and CAAT-box), the cis-elements with functional annotation were divided into 20 groups, including light-, methyl jasmonate-, and anaerobic-responsive elements, among others. (Tables S4 and S5). Among them, light-related cis-elements accounted for the largest portion (45.97%), followed by methyl jasmonate (12.11%), anaerobic (10.91%), abscisic acid (9.93%), and low temperature (3.11%). Since previous studies revealed that both methyl jasmonate and abscisic acid can play various roles in plant stress and defense45,46,47, it can be speculated that HaNACs were closely related to the photoperiod, stress, and defense of sunflower. In addition, there were some cis-element groups related to special tissue, such as these groups of meristem and endosperm expression, and differentiation of the palisade mesophyll cells (Table S5). Additional information on cis-elements was provided in Fig. S5, and Table S4.
HaNAC gene expression profiles in different organs and under PEG stress
To infer the role of HaNACs in different tissues, the RNA-seq data of sunflower from the Sequence Read Archive (SRA) database were searched, downloaded, and analyzed. In total, 80 genes were expressed in the pistil, stamen, ligule, mature leaves, roots, and seeds. HaNAC20, HaNAC115, HaNAC119, and HaNAC138 genes were uniquely expressed in pistil; HaNAC2, HaNAC31, HaNAC44, HaNAC74, HaNAC97, HaNAC123, and HaNAC145 were only expressed in stamen; HaNAC1, HaNAC21 in ligule; HaNAC14, HaNAC45, HaNAC56, HaNAC57, HaNAC85, and HaNAC101 were only expressed in the root. Higher expressions of HaNAC83, HaNAC129, and HaNAC132 were observed in mature leaf, but could also be found in other organs. Except for HaNAC23 and HaNAC105, all genes were lowly expressed in seeds. This result could be attributed to the dormant state of seeds and low gene expression (Fig. 6A). Overall, these findings implied that different HaNACs might play different roles in various tissues.
The expression profiles of HaNAC genes in different organs and under PEG stress based on transcriptome data from SRA. The legend represented a logarithmic normalized TPM. (A) HaNAC gene expression profiles in different organs. (B, C) HaNAC gene expression profiles in leaves and roots after 15% PEG stress treatment, respectively.
To assess the function of the HaNAC gene under drought stress, RNA-seq data of leaves and roots treated under 15% PEG stress for 24 h with sunflower seedlings at four-leaf stage from the SRA database showed that 67 HaNAC genes were present in all samples, among them, 47 HaNAC genes were detected in leaves and 58 in roots, respectively (Fig. 6B,C). Using p-value < 0.05 as the standard for differentially expressed genes (DEGs), the DEGs analysis revealed 10 differentially expressed HaNAC genes in the leaves under PEG stress. HaNAC58, HaNAC84, HaNAC149, HaNAC105, HaNAC57, HaNAC146, HaNAC23, HaNAC121, and HaNAC145 were up-regulated, whereas only HaNAC85 was down-regulated (Table 2). Besides, three DEGs (HaNAC76, HaNAC62, HaNAC24) were up-regulated in the roots (Table 2). Of the differentially expressed HaNAC genes, most were up-regulated, demonstrating that these HaNAC genes might exert a positive regulatory role in drought response.
HaNACs related to abiotic stress and their stress response
Here, we determined which HaNACs potentially played a role in abiotic stress. Of note, 46 scientific papers from 2004 to 2019 have been reported that NACs could enhance plant abiotic stress resistance (mainly salt and drought resistance) (Table S6); Thus, 47 related NAC protein sequences were downloaded and a phylogenetic tree was constructed using HaNACs and these 47 NACs protein sequences. Notably, most NAC proteins enhancing abiotic resistance in plants were classified into the SNAC branch (Fig. 7). These NAC members in this branch were called abiotic stress-related NACs (SNAC). The 35 tolerant NACs were distributed in the SNAC group, while only 12 tolerant NACs were in the larger Non-SNAC group.
A phylogenetic tree generated using 47 reported NACs enhancing plant tolerance to abiotic stress and HaNACs from sunflower. Red dots indicated NACs reported could improve abiotic stress tolerance in plants. Most of these 47 NAC genes were densely distributed in the red subgroup, here called the SNAC group, and the other as Non-SNAC group.
Based on the correlation of function with evolution, it can be assumed that the HaNACs on the SNAC branch are strongly associated with abiotic stress. Therefore, it is necessary to detect the expression of some HaNACs under salt- and PEG-abiotic stress, and these HaNACs were chosen from the SNAC group and the same branch as tolerant NACs in the Non-SNAC group. Four-leaf stage sunflowers were treated with 15% PEG or 150 mM NaCl for 24 h, and the relative expressions of these HaNACs were determined by qRT-PCR. Results showed that the gene expression pattern was similar under PEG and NaCl stress in the same tissues and gene expression varied widely in different tissues. In both the SNAC and Non-SNAC groups, the expressions of some genes were up-regulated after stress. Of note, the numbers and folds of up-regulated genes in the SNAC group were more and higher. In leaves, HaNAC146 and HaNAC105 were most significantly up-regulated, while HaNAC70, HaNAC23, HaNAC84, HaNAC55, and HaNAC35 were most significantly up-regulated in roots (Fig. 8). These up-regulated genes might play an important role in plant response to abiotic stress.
Relative expression of some HaNACs under 15% PEG or 150 mM NaCl stress for 24 h, including those in the SNAC group and those in the same branch as the reported NACs in the Non-SNAC group. (A–D) showed the conditions of gene expression in leaves; (E–G) represented those in roots; (A, C, E, G) showed the results of SNAC gene expression; (B, D, F, H) indicated those of Non-SNAC genes; (A, B, E, F) showed the results of gene expression to PEG stress and (C, D, G, H) as the results to NaCl stress.
Discussion
Sunflower is an economically significant crop with medium salt and drought resistance9. Since a substantial fraction of land in the world is highly saline, improving the ability of sunflower to tolerate abiotic stress would facilitate the utility of this saline-alkali wasteland for agricultural production. The NAC gene family, as a plant-specific transcription factor family, plays important functions in response to abiotic and biotic stresses39,48,49. Limited information has been published on the functional association of the NAC gene family in sunflower. Herein, we reported the first study to systematically analyze the NAC gene family and conduct an expression profiling in sunflower using bioinformatics tools and real-time PCR technology. According to published genomic data, 150 NAC genes have been identified in the sunflower genome (Fig. 1 and Table S1); however, some HaNAC genes are yet to be found because the genome only accounts for about 80% of the entire sunflower genome. A total of 150 HaNACs were divided into 15 subgroups through phylogenetic analysis (Fig. 2), whereby genes performing the same function were clustered into the same group. For instance, the subgroups of AtNAC3, NAP and ATAF contained many classic stress-responsive NAC genes, including ANAC019, ANAC056 (AtNAC2), ANAC055 (AtNAC3), ANAC002 (ATAF1), ANAC081 (ATAF2), and ANAC072 (RD26)49,50, suggesting that HaNACs in these three groups are likely to contribute to the stress response. Conserved motifs and gene structure also indicated that the same group of HaNACs might exert the same biological function (Fig. 3). These gene replication events are generally known to be crucial in the rapid expansion and evolution of gene families. Gene duplication of NAC TFs has been observed in many plant species. In our study, 27 pairs of gene replication events were generated in 150 HaNACs, which may significantly contribute to the expansion of the NAC family in sunflower (Table S3). The analysis of NTLs with transmembrane helixes in HaNACs was found to have similarity with the Arabidopsis thaliana NAC family members51, suggesting that they may function in specific locations by protease degradation via a similar mechanism as NTLs in Arabidopsis52 (Table 1, Figs. S3 and S4). Previous studies revealed that miRNA164s could direct mRNA cleavage of the NAC gene53,54,55. Our study showed that miR164 targeting sites might exist in 12 HaNACs, and miR164 possibly participates in the regulation of HaNAC gene expression.
Cis-elements play an important role in regulating gene expression56,57. In the promoter region of about 2000 bp upstream of HaNAC genes, many cis-acting elements were found, such as light-, methyl jasmonate-, anaerobic-, ABA-, and low temperature-responsive elements, indicating that HaNACs may be associated in sunflower photoperiod, stress and defense pathways. From the stress response perspective, the expression of HaNAC genes under PEG stress as revealed by RNA-seq data showed that some HaNAC genes had obvious expressions to drought stress (15% PEG stress). Through further analysis, we found that most of the stress-responsive NAC genes in the phylogenetic tree were clustered in the same subgroup, which was consistent with related studies in Arabidopsis and rice58,59. This indicated that HaNAC genes in the SNAC group may play a vital role in abiotic stress responses. Moreover, qRT-PCR results also showed that the expression of genes in the SNAC group significantly increased under NaCl and PEG stress, further confirming our previous results (SNAC genes are important in the abiotic stress response pathway). Besides, under the stress, HaNAC70 and HaNAC84 were more highly up-regulated in roots than leaves, whereas HaNAC146 and HaNAC105 were higher up-regulated in leaves. Additionally, HaNAC55 showed a significant up-regulation in the Non-SNAC group, suggesting that the response of HaNACs to abiotic stress is complex or did not get enough research. Altogether, we can infer that sunflower SNAC genes are involved in the response of the plant to abiotic stress, and different SNAC genes may also play a role in different tissues.
Conclusion
In this study, 150 sunflower NAC genes were identified, and their distribution, basic properties, classification, gene structure, evolutionary characteristics, and expression profiles were explored. Among them, eight potential HaNTL genes and 12 HaNAC genes that may be regulated by miR164s were found. Analysis of cis-acting elements and the expression profiles of HaNACs indicated that these genes were involved in response to salt and drought stress. A few important genes that may play an important role in tolerance of sunflower to abiotic stress have been speculated. Notably, this work can provide a solid basis for future functional studies of NAC to improve abiotic stress resistance in sunflower.
Materials and methods
Identification, physicochemical properties and chromosome distribution of sunflower NAC transcription factors
All protein sequences (version 1.0) of sunflower were downloaded from the EnsemblPlants database (http://plants.ensembl.org/). The HMM model file (PF02365) of the NAM domain in the pfam database (http://pfam.xfam.org/) was downloaded, and the software HMMER (version 3.2.1) was used to search for NAC proteins from all protein sequences of sunflower. The obtained proteins were verified in the CCD database (https://www.ncbi.nlm.nih.gov/cdd/). The remaining protein sequences with the complete NAC domain were aligned using the ClustalW method in MEGA X.
The Sequence Manipulation Suite (http://www.detaibio.com/sms2/) was used to analyze the basic physical and chemical properties of HaNACs, such as amino acid number, isoelectric point, and molecular weight.
The position of these genes on the chromosome was determined from the gtf file (version 1.0.42) in the Ensembl Plants database (http://plants.ensembl.org/). MapGene2Chrom web service (http://mg2c.iask.in/mg2c_v2.0/) was used for drawing chromosome distribution, and the pie chart was generated using Excel.
Membrane-bound HaNACs and miR164 target site prediction
The CELLO website (version 2.5) was used to predict the subcellular localization of HaNACs, whereas the transmembrane helix of HaNACs was predicted with TMHMM 2.0 (http://www.cbs.dtu.dk/services/TMHMM-2.0/).
Sunflower miRNA164s and HaNACs mRNA sequences were input to psRNATarget (http://plantgrn.noble.org/psRNATarget/) to predict the action site of miRNA164. The picture was drawn using the gene structure display service (GSDS2.0 http://gsds.cbi.pku.edu.cn/).
Phylogenetic, genetic structure and conserved motifs
The NAC protein sequences of sunflower and Arabidopsis were compared by ClustalW in MEGA X, and a phylogenetic tree was constructed using the Neighbour-Joining method of MEGA X. Then, the reliability of the tree was tested using 1,000 bootstrap tests.
The structures of the HaNACs could be found in the gtf file. The MEME local software (version 5.0.4) of the motif analysis tool was used to find conservative motifs among members of the sunflower NAC family. Except for the maximum number of motifs which is 15, the other parameters were default. TBtools was applied to merge and draw evolution trees, genetic structures, and motifs60.
Syntenic and evolutionary analysis and cis-element analysis
The syntenic relationship of HaNACs was analyzed by the Multiple Collinearity Scan Toolkit (MCScanX). Here, potential gene pairs (E value < 10–5, first five matches) were obtained in the genome of sunflower through BLASTP as input files to analyze segmented and tandem duplications. After that, the relationship map between HaNACs was drawn using TBTools. DnaSP (version 6.12.03) was used to calculate the non-synonymous rate (Ka) and synonymous rate (Ks) of the homologous genes. The type of selection was determined according to the ratio of Ka and Ks, whereas the time of duplication events (T) was determined according to the following formula: T = Ks/2λ, where λ = 1.5 × 10−8 for dicots61.
Further, 2000 bp upstream of the start codon of the HaNAC gene was extracted as an input file for PlantCare (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/), then cis-acting elements were analyzed, and we used TBTools to generate the picture.
RNA-seq data download and expression analysis
Sunflower RNA-seq data for the expression analysis of different tissues and PEG stress were derived from SRA data (Bioproject: PRJNA483306; PRJNA398727). The expression was calculated, and the DEGs were analyzed according to the protocols described by Pertea et al.62 on the software including Hisat2 (version 2.1.0), Stringtie (version 1.3.5) and Ballgown (version 2.18.0). The heat map was generated using TBTools software.
Plant materials and stress treatment
The sunflower used in this research was named as ZADT (Zaoaidatou), the backbone parent for breeding in Xinjiang, China. To understand the expression pattern of HaNAC genes under abiotic stress, seeds were sown, and seedlings were grown in a chamber at the photoperiod of 16 h/day 26 ℃ and 8 h/night at 20 ℃ for 16 days. Then, the seedlings with 4 true leaves were subjected to salt- (150 mM NaCl) and drought- (15% PEG6000) stressed treatments for 24 h, here the control was designed by water treatment. Sunflower roots and leaves were collected and frozen in liquid nitrogen for RNA extraction to detect the expression pattern of HaNAC genes under abiotic stress.
RNA extraction and qRT-PCR
Total RNA was isolated using RNAprep pure Plant Kit (Tiangen, DP432). Reverse transcription was carried out using the PrimeScript™ 1st Strand cDNA Synthesis Kit (Takara, 6110A). The primers (listed in Table S7) were designed using the NCBI Primer-BLAST tool (https://www.ncbi.nlm.nih.gov/tools/primer-blast/), and actin was used as the internal reference gene. Power SYBR™ Green PCR Master Mix (Applied Biosystems, 4367659) and Applied Biosystems QuantStudio 5 Real-Time PCR Systems were used for qRT-PCR. The 2−ΔΔCT method was used to calculate the expression level. Each qRT-PCR procedure was conducted with three biological replicates.
Data availability
Our study complies with relevant institutional, national, and international guidelines and legislation. Permissions were obtained from Xinjiang Academy of Agricultural Sciences for collecting and using sunflower plants for this study. All plant materials used in this study were provided by Xinjiang Academy of Agricultural Sciences. RNA-seq fastq files were deposited in NCBI Sequence Read Archive (SRA) under accession number PRJNA483306 and PRJNA398727. Genome, proteome, and annotation files were available in EnsemblPlants database.
References
Boyer, J. S. Plant productivity and environment. Science 218, 443–448 (1982).
Mwale, S. S., Hamusimbi, C. & Mwansa, K. Germination, emergence and growth of sunflower (Helianthus annuus L.) in response to osmotic seed priming. Seed Sci. Technol. 31, 199–206 (2003).
Kaya, M. D., Ipek, A. & Öztürk, A. Effects of different soil salinity levels on germination and seedling growth of safflower (Carthamus tinctorius L.). Turk. J. Agric. For. 27, 221–227 (2003).
Marschner, H. Mineral nutrition of higher plants. 2nd edn, (CA Academic Press United States US, 1995).
Ashraf, M., McNeilly, T. & Bradshaw, A. D. Selection and heritability of tolerance to sodium chloride in four forage species. Crop Sci. 27, 232–234 (1987).
Bray, E. A. Plant responses to water deficit. Trends Plant Sci. 2, 48–54 (1997).
Dai, A. G. Increasing drought under global warming in observations and models. Nat. Clim. Change. 3, 52–58 (2013).
Radanovic, A., Miladinovic, D., Cvejic, S., Jockovic, M. & Jocic, S. Sunflower genetics from ancestors to modern hybrids: A review. Genes 9, 528 (2018).
Shi, D. C. & Sheng, Y. M. Effect of various salt-alkaline mixed stress conditions on sunflower seedlings and analysis of their stress factors. Environ. Exp. Bot. 54, 8–21 (2005).
Ahmad, S., Ahmad, R., Ashraf, M. Y., Ashraf, M. & Waraich, E. A. Sunflower (Helianthus annuus L.) response to drought stress at germination and seedling growth stages. Pak. J. Bot 41, 647–654 (2009).
Manavella, P. A. & Chan, R. L. Transient transformation of sunflower leaf discs via an Agrobacterium-mediated method: applications for gene expression and silencing studies. Nat. Protoc. 4, 1699 (2009).
Lewi, D. M., Hopp, H. E. & Escandón, A. S. Sunflower (Helianthus annuus L.). Methods Mol. Biol. 343, 291–297 (2006).
Radonic, L. M. et al. Sunflower (Helianthus annuus L.). Methods Mol. Biol. 1224, 47–55 (2015).
Cheng, Q., Lei, Z. H., Huang, Q. X. & Zeng, Y. L. Establishment on tissue culture and plant regeneration system of oil sunflower parent “Zaoaidatou”. Plant Physiol. J. 56, 180–188 (2020).
Saibo, N. J., Lourenco, T. & Oliveira, M. M. Transcription factors and regulation of photosynthetic and related metabolism under environmental stresses. Ann. Bot. 103, 609–623 (2009).
Souer, E., Houwelingen, A. V., Kloos, D., Mol, J. & Koes, R. The No Apical Meristem gene of petunia is required for pattern formation in embryos and flowers and is expressed at meristem and primordia boundaries. Cell 85, 159–170 (1996).
Tran, L. S. P. et al. Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell 16, 2481–2498 (2004).
Lu, M. et al. Overexpression of a stress induced maize NAC transcription factor gene, ZmSNAC1, improved drought and salt tolerance in Arabidopsis. Acta Agron. Sin. 39, 2177–2182 (2013).
Olsen, A. N., Ernst, H. A., Leggio, L. L. & Skriver, K. NAC transcription factors: Structurally distinct, functionally diverse. Trends Plant Sci. 10, 79–87 (2005).
Ooka, H. et al. Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana. DNA Res. 10, 239–247 (2003).
Tak, H., Negi, S. & Ganapathi, T. R. Banana NAC transcription factor MusaNAC042 is positively associated with drought and salinity tolerance. Protoplasma 254, 803–816 (2016).
Zhang, H. et al. Genome-wide identification and expression profile analysis of the NAC transcription factor family during abiotic and biotic stress in woodland strawberry. PLoS One 13, e0197892 (2018).
Moyano, E. et al. Genome-wide analysis of the NAC transcription factor family and their expression during the development and ripening of the Fragaria × ananassa fruits. PLoS ONE 13, e0196953 (2018).
Guan, H. et al. OoNAC72, a NAC-Type Oxytropis ochrocephala transcription factor, conferring enhanced drought and salt stress tolerance in Arabidopsis. Front. Plant Sci. 10, 890 (2019).
Zhang, Y. et al. Genome-wide identification and comprehensive analysis of the NAC transcription factor family in Sesamum indicum. PLoS ONE 13, e0199262 (2018).
Diao, W. et al. Genome-wide analyses of the NAC transcription factor gene family in pepper (Capsicum annuum L.): chromosome location, phylogeny, structure, expression patterns, cis-elements in the promoter, and interaction network. Int. J. Mol. Sci. 19, 1–20 (2018).
Fang, Y., You, J., Xie, K., Xie, W. & Xiong, L. Systematic sequence analysis and identification of tissue-specific or stress-responsive genes of NAC transcription factor family in rice. Mol. Genet. Genomics 280, 547–563 (2008).
Pinheiro, G. L. et al. Complete inventory of soybean NAC transcription factors: Sequence conservation and expression analysis uncover their distinct roles in stress response. Gene 444, 10–23 (2009).
Badouin, H. et al. The sunflower genome provides insights into oil metabolism, flowering and Asterid evolution. Nature 546, 148–152 (2017).
Yuce, M., Taspinar, M. S., Aydin, M. & Agar, G. Response of NAC transcription factor genes against chromium stress in sunflower (Helianthus annuus L.). Plant Cell Tiss. Org. 136, 479–487 (2019).
Moschen, S. et al. Identification of candidate genes associated with leaf senescence in cultivated sunflower (Helianthus annuus L.). PLoS ONE 9, e104379 (2014).
Giordani, T. et al. An analysis of sequence variability in eight genes putatively involved in drought response in sunflower (Helianthus annuus L.). Theor. Appl. Genet. 122, 1039–1049 (2011).
Su, H. Y., Zhang, S. Z., Yin, Y. L., Zhu, D. Z. & Han, L. Y. Genome-wide analysis of NAM-ATAF1,2-CUC2 transcription factor family in Solanum lycopersicum. J. Plant Biochem. Biotechnol. 24, 176–183 (2015).
Gong, W. et al. Genome-wide ORFeome cloning and analysis of Arabidopsis transcription factor genes. Plant Physiol. 135, 773–782 (2004).
Le, D. T. et al. Genome-wide survey and expression analysis of the plant-specific NAC transcription factor family in soybean during development and dehydration stress. DNA Res. 18, 263–276 (2011).
Kadier, Y. et al. Genome-wide identification, classification and expression analysis of NAC family of genes in sorghum [Sorghum bicolor (L.) Moench]. Plant Growth Regul. 83, 301–312 (2017).
Nuruzzaman, M. et al. Genome-wide analysis of NAC transcription factor family in rice. Gene 465, 30–44 (2010).
Kim, S. G. et al. Genome-scale screening and molecular characterization of membrane-bound transcription factors in Arabidopsis and rice. Genomics 95, 56–65 (2010).
Kim, Y. S. et al. A membrane-bound NAC transcription factor regulates cell division in Arabidopsis. Plant Cell 18, 3132–3144 (2006).
Bartel, D. P. MicroRNAs: Target recognition and regulatory functions. Cell 136, 215–233 (2009).
Larue, C. T., Wen, J. & Walker, J. C. Genetic interactions between the miRNA164-CUC2 regulatory module and BREVIPEDICELLUS in Arabidopsis developmental patterning. Plant Signal. Behav. 4, 666–668 (2009).
Li, J. et al. miRNA164-directed cleavage of ZmNAC1 confers lateral root development in maize (Zea mays L.). BMC Plant Biol. 12, 220 (2012).
Larue, C. T., Wen, J. & Walker, J. C. A microRNA-transcription factor module regulates lateral organ size and patterning in Arabidopsis. Plant J. 58, 450–463 (2009).
Dai, X. B., Zhuang, Z. H. & Zhao, P. X. psRNATarget: A plant small RNA target analysis server (2017 release). Nucleic Acids Res. 46, W49–W54 (2018).
Fujii, H. et al. In vitro reconstitution of an abscisic acid signalling pathway. Nature 462, 660–664 (2009).
Wilkinson, S. & Davies, W. J. Drought, ozone, ABA and ethylene: New insights from cell to plant to community. Plant Cell Environ. 33, 510–525 (2010).
Cheong, J. J. & Choi, Y. D. Methyl jasmonate as a vital substance in plants. Trends Genet. 19, 409–413 (2003).
Xu, Z. Y. et al. The Arabidopsis NAC transcription factor ANAC096 cooperates with bZIP-type transcription factors in dehydration and osmotic stress responses. Plant Cell 25, 4708–4724 (2013).
Fujita, M. et al. A dehydration-induced NAC protein, RD26, is involved in a novel ABA-dependent stress-signaling pathway. Plant J. 39, 863–876 (2004).
Wu, Y. R. et al. Dual function of Arabidopsis ATAF1 in abiotic and biotic stress responses. Cell Res. 19, 1279–1290 (2009).
Liang, M. et al. Subcellular distribution of NTL transcription factors in Arabidopsis thaliana. Traffic 16, 1062–1074 (2015).
Inge, D. C. et al. The membrane-bound NAC transcription factor ANAC013 functions in mitochondrial retrograde regulation of the oxidative stress response in Arabidopsis. Plant Cell 25, 3472–3490 (2013).
Guo, H. S., Xie, Q., Fei, J. F. & Chua, N. H. MicroRNA directs mRNA cleavage of the transcription factor NAC1 to downregulate auxin signals for Arabidopsis lateral root development. Plant Cell 17, 1376–1386 (2005).
Kim, J. H. et al. Trifurcate feed-forward regulation of age-dependent cell death involving miR164 in Arabidopsis. Science 323, 1053–1057 (2009).
Nikovics, K. et al. The balance between the MIR164A and CUC2 genes controls leaf margin serration in Arabidopsis. Plant Cell 18, 2929–2945 (2006).
Hapgood, J. P., Riedemann, J. & Scherer, S. D. Regulation of gene expression by GC-rich DNA cis-elements. Cell Biol. Int. 25, 17–31 (2001).
Sheshadri, S. A., Nishanth, M. J. & Simon, B. Stress-mediated cis-element transcription factor interactions interconnecting primary and specialized metabolism in planta. Front. Plant Sci. 7, 1725 (2016).
Nuruzzaman, M., Sharoni, A. M. & Kikuchi, S. Roles of NAC transcription factors in the regulation of biotic and abiotic stress responses in plants. Front. Microbiol. 4, 248 (2013).
Nakashima, K., Takasaki, H., Mizoi, J., Shinozaki, K. & Yamaguchi-Shinozaki, K. NAC transcription factors in plant abiotic stress responses. Biochim. Biophys. Acta 1819, 97–103 (2012).
Chen, C. et al. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202 (2020).
Blanc, G. & Wolfe, K. H. Widespread paleopolyploidy in model plant species inferred from age distributions of duplicate genes. Plant Cell 16, 1667–1678 (2004).
Pertea, M., Kim, D., Pertea, G. M., Leek, J. T. & Salzberg, S. L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgowm. Nat. Protoc. 11, 1650–1667 (2016).
Acknowledgements
This work was financially supported by the Natural Science Foundation of Xinjiang Uygur Autonomous Region(2020D01C020), the Key R & D Projects in Xinjiang Uygur Autonomous Region (2018B01006-2) and “the Belt and Road” Biotechnology Collaborative Innovation Project (BRI-ASTIP-MISACC-02-2017).
Author information
Authors and Affiliations
Contributions
W.L., Y.Z. designed the experiments and analyzed the data. W.L., Y.Z. wrote the manuscript text. W.L., R.W., X.M. and F.Y. performed the experiments. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Li, W., Zeng, Y., Yin, F. et al. Genome-wide identification and comprehensive analysis of the NAC transcription factor family in sunflower during salt and drought stress. Sci Rep 11, 19865 (2021). https://doi.org/10.1038/s41598-021-98107-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-98107-4
- Springer Nature Limited