Abstract
Fresh-cut processing promotes enzymatic browning of fresh fruits and vegetables, which negatively affects the product appearance and impacts their nutrition. We used RNA-sequencing to analyze the transcriptomic changes occurring during the browning of fresh-cut eggplant fruit samples from both browning-sensitive and browning-resistant cultivars to investigate the molecular mechanisms involved in browning. A total of 8347 differentially expressed genes were identified, of which 62 genes were from six gene families (i.e., PPO, PAL, POD, CAT, APX, and GST) potentially associated with enzymatic browning. Furthermore, using qRT-PCR, we verified 231 differentially regulated transcription factors in fresh-cut eggplant fruits. The enzyme activities of PPO, POD, PAL, and CAT in ‘36’ were significantly higher than those of ‘F’ fresh-cut for 15 min. Both PPO and POD play a major role in the browning of eggplant pulp and might therefore act synergistically in the browning process. Meanwhile, qPCR results of 18 browning related genes randomly screened in 15 eggplant materials with different browning tolerance showed variant-specific expression of genes. Lastly, gene regulatory networks were constructed to identify the browning-related genes. This work provides a basis for future molecular studies of eggplants, and lays a theoretical foundation for the development of browning-resistant fresh-cut fruits and vegetables.
Similar content being viewed by others
Introduction
Solanum melongena L., commonly known as eggplant, is an essential vegetable crop. It is a good source of dietary minerals, vitamins, and anthocyanins, with a high oxygen radical absorbance capacity and low caloric value1. Eggplant has been shown to have important cardiovascular protecting, anticancer, and anti-aging effects2. However, fresh-cut eggplants brown easily which can negatively affect their flavor, odor, nutrients, and commercial value. Browning has become one of the most vital limitations in eggplant processing. Therefore, understanding the mechanism of enzymatic browning and controlling the occurrence of browning have become the focus of research in the fresh-cut fruit and vegetable industry. Furthermore, breeding new eggplant varieties resistant to browning has become an important method to fundamentally solve the browning of eggplant fruit.
Browning, a common post-harvest problem for many fruits and vegetables, can be divided into two categories: enzymatic and non-enzymatic browning3. Non-enzymatic browning occurs as a result of various chemical reactions such as Maillard reactions, caramelization, oxidation of vitamin C, and polyphenol polymerization in daily life4. Enzymatic browning is considered as the main reaction that causes browning in post-harvest storage and processing of fruits and vegetables, and has been the focus of postharvest research5. It refers to the physiological and biochemical processes in which phenolic substances in plant organs or tissues are oxidized to form quinones under the action of polyphenol oxidase under aerobic conditions6. Quinones then polymerize to form brown or black substances, resulting in tissue discoloration. Enzymatic browning occurs only in the presence of phenols, enzymes, and oxygen7. As an important substrate for enzymatic browning of fruit and vegetable tissues, phenols are widely distributed in the roots, stems, leaves, flowers, and fruits of plants—with varying abundance between different types of plants8. Phenolic compounds are important secondary metabolites in plants, which are synthesized mainly through phenylalanine metabolic pathways9. Phenylalanine ammonia-lyase (PAL) is the starting enzyme of the phenylpropane metabolic pathway10. The phenylalanine amino group is deaminated by PAL to form trans-cinnamic acid, which is hydroxylated by cinnamate-4-hydroxylase (C4H) to form trans-4-coumaric acid. Then, under the actions of coumarate 3-hydroxylase (C3H), 4-coumarate-CoA ligase (4CL), hydroxycinnamoyl-CoA: shikimate/quinate hydroxy cinnamoyl transferase (HCT), chalcone synthase (CHS), cinnamoyl-CoA reductase (CCR), and other enzymes, it enters the metabolic pathways of lignin synthesis, chlorogenic acid synthesis, and flavonoid (anthocyanin, procyanidin, rutin) synthesis11. Furthermore, enzymatic browning is mostly a result of transforming phenols into o-quinones—catalyzed by polyphenol oxidase (PPO)12. Other enzymes, such as peroxidase (POD), catalase (CAT), superoxide dismutase (SOD), ascorbate peroxidase (APX), and glutathione S-transferase (GST) also play a role in inducing or inhibiting the enzymatic browning of fresh-cut fruits and vegetables13. Therefore, it is essential to study these genes that encode key enzymes in eggplant enzymatic browning and phenolic compound formation, which may lay the foundation for better understanding of the browning mechanism in fresh-cut fruits and vegetables.
With the development of high-throughput sequencing, technologies such as genomics, transcriptomics, proteomics, and metabolomics have been widely used to study specific biological processes and molecular mechanisms of organisms14. In recent years, RNA-sequencing (RNA-seq) as an efficient and fast transcriptome research method has changed people's understanding of transcriptomics, and has been increasingly used to explore differences in gene expression in plants, such as licorice (Glycyrrhiza glabra L.)15, luffa (Luffa cylindrica (L.) Roem.) 16, potato (Solanum tuberosum L.) 17, strawberry (Fragaria ananassa Duch)18, and pear (Pyrus betulifolia Bunge)19. In a study by Zhu et al. on fresh-cut luffa browning, they found that there were 15 genes with significantly different expressions in enzymatic browning, including PPO, POD, PAL, CAT, and 4 WRKY transcription factors (TFs)20. However, there are few reports on the browning mechanism of fresh-cut eggplant fruit.
We identified two cultivars of eggplant materials, ‘F’ and ‘36’, as either resistant or sensitive to browning, respectively, by fresh-cutting more than 60 eggplant materials and recording their respective browning times. In the current study, we aimed to (1) conduct transcriptomic profiling of the two types of materials, (2) identify the key differentially expressed genes (DEGs) related to enzymatic browning and phenolic compound formation, and (3) provide new insights into the molecular mechanisms of eggplant browning.
Results
Overview of transcriptomic data for cultivars ‘F’ and ‘36’
The browning-sensitive cultivar (‘36’) and browning-resistant cultivar (‘F’) of eggplant showed different degrees of browning with treatment time (Fig. 1). As shown in Table 1, a total of 111.55 Gb clean data were obtained after 18 samples were analyzed and sequenced using a reference transcriptome (ftp://ftp.kazusa.or.jp/pub/eggplant/SME_r2.5.1.fa.gz). The effective data volume of each sample was 5.73–7.04 Gb; 93.61–94.50% of bases had a quality above Q30, and the average GC content was 45.56%. Reads were compared to the reference genome to obtain the genome alignment of each sample, with an alignment rate of 83.50–95.14%. We also analyzed the distribution of gene expression levels in each sample (Fig. 2). We determined the minimum, the first quartile (25%), the median (50%), the third quartile (75%), and the maximum value of gene expression level for each sample. The symmetry and degree of dispersion of the data distribution were observed, with remarkably high gene expression levels. Heatmap of Pearson's correlation coefficients between all pairs of samples is shown in Supplementary Figure S1.
Identification and classification of DEGs
Identification of DEGs between the two eggplant cultivars was one of the aims of the present study. To achieve this goal, we compared the transcriptional profiles of cultivars ‘36’ and ‘F’, which were respectively used as a test and a control for screening DEGs. According to the standard criteria (fold change > 2; p-value < 0.05), we analyzed the difference in the expression of the same gene between treatment pairs. For cultivar ‘36’, we identified 4095 DEGs between fresh-cut for 2 min and CK0 (control before fresh-cut) (1971 up- and 2124 down-regulated genes), 2212 DEGs between 15 min and CK0 (857 up- and 1355 down-regulated genes), and 1957 DEGs between 15 and 2 min (996 up- and 961 down-regulated genes). For cultivar ‘F’, we identified 20 DEGs between fresh-cut for 2 min and CK0 (11 up- and 9 down-regulated genes), 1313 DEGs between 15 min and CK0 (1188 up- and 125 down-regulated genes), and 1087 DEGs between 15 and 2 min (1014 up- and 73 down-regulated genes). Furthermore, there were 2299 DEGs between the CK0 of ‘36’ and ‘F’ (1363 up- and 936 down-regulated genes), 2810 DEGs between fresh-cut for 2 min of ‘36’ and ‘F’ (1633 up- and 1177 down-regulated genes), and 3820 DEGs between fresh-cut for 15 min of ‘36’ and ‘F’ (1336 up- and 2484 down-regulated genes) (Fig. 3, Supplementary Table S1). The DEGs were then subjected to Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses (Supplementary Table S2–S3).
Functional annotation and classification of DEGs
To determine the function of DEGs, GO and KEGG pathway analyses were carried out. For cultivar ‘36’, the treatment pair with the most DEGs was the 2 min/CK0 pair. In this treatment pair, GO functional enrichment analysis was carried out (Fig. 4a). “Metabolic process,” “cell,” and “binding” were the dominant terms in the “biological process,” “cellular component,” and “molecular function” categories, respectively. We also identified a relatively large number of genes associated with “catalytic activity,” “antioxidant activity,” “biological regulation,” “response to stimulus,” “single-organism process,” and “cellular process,” with only a few genes related to “cell killing,” “nucleoid,” and “protein binding TF activity.” As shown in Fig. 4b,c, GO functional enrichment analysis of the treatment pairs ‘F’15 min/‘F’CK0 and ‘36’15 min/‘F’15 min showed similar results as the ‘36’2 min/‘36’CK0 treatment pair.
(a) GO functional enrichment of the ‘36’ 2 min vs ‘36’ CK0 groups. (b) GO functional enrichment of the ‘F’ 15 min vs ‘F’ CK0 groups. (c) GO functional enrichment of the ‘36’ 15 min vs ‘F’ 15 min groups. The figures was drawn with DEseq (v3.5.1). (http://www.bioconductor.org/packages/3.8/bioc/html/DESeq.html).
To further identify important metabolic pathways, we used KEGG pathway analysis to determine the biological functions of DEGs. Pathway entries with a number of corresponding DEGs greater than 2 were screened and sorted by the − log10 p-value of each entry. Phenylpropanoid biosynthesis (ko00940), phenylalanine metabolism (ko00360), tyrosine metabolism (ko00350), glutathione (GSH) metabolism (ko00480), flavonoid biosynthesis (ko00941), linoleic acid metabolism (ko00591), as well as starch and sucrose metabolism (ko00500) were found in the top 20 enrichment biological pathways of these compared. Phenylpropanoid biosynthesis (ko00940), phenylalanine metabolism (ko00360), tyrosine metabolism (ko00350), and GSH metabolism (ko00480) were the more significant pathways for enrichment among the three groups, which had a small p-value (p < 0.05) compared to that of the other pathways (Fig. 5a–c). The top 20 KEGG enrichment analysis of DEGs in all pairwise comparisons are shown in Supplementary Table S4.
(a) KEGG enrichment of the ‘36’ 2 min vs ‘36’ CK0 groups. (b) KEGG enrichment of the ‘F’ 15 min vs ‘F’ CK0 groups. (c) KEGG enrichment of the ‘36’ 15 min vs ‘F’ 15 min groups. The figures was drawn with DEseq (v3.5.1). (http://www.bioconductor.org/packages/3.8/bioc/html/DESeq.html).
Identification of potential genes associated with enzymatic browning
Changes in the transcriptome of the eggplant after fresh-cut treatment were examined by cluster analysis of the gene expression patterns, which were presented in a heat map (Fig. 6). We analyzed the DEGs involved in the tyrosine metabolism pathway. There were 10 genes in total, including 7 PPO genes, 4 of which encoded PPO and 3 encoded catechol oxidase. Two of the four PPO encoding genes (Sme2.5_25992.1_g00001.1, Sme2.5_11776.1_g00001.1) were significantly up-regulated in ‘36’ vs ‘F’, while two genes (Sme2.5_08192.1_g00002.1, Sme2.5_04286.1_g00003.1) had the opposite result (Fig. 6). Furthermore, three genes encoding catechol oxidase (Sme2.5_33651.1_g00001.1, Sme2.5_01441.1_g00006.1, Sme2.5_06843.1_g00001.1) were significantly up-regulated in ‘36’ vs ‘F’. In the phenylpropanoid biosynthesis/phenylalanine metabolism pathway (ko00940/360), 64 DEGs were identified, including 22 POD-encoding genes and 3 PAL-encoding genes (Sme2.5_00864.1_g00009.1, Sme2.5_16832.1_g00001.1, Sme2.5_16832.1_g00002.1) (Fig. 6). This confirmed that the expression pattern of most of these genes were consistent. In total, 28 DEGs were found to be involved in GSH metabolism (ko00480), including 18 GST-encoding genes and 10 genes encoding L-ascorbate oxidase, most of which were down-regulated in ‘36’ vs ‘F’. In addition, 2 CAT-encoding DEGs were identified (Fig. 6). These results showed that the gene expression patterns of ‘36’ and ‘F’ were obviously changed after fresh-cut treatment.
The heat maps for the browning-related genes. The red color in the picture indicates high expression genes, while the green color indicates low expression genes. The heat map was drawn with pheamap (v1.0.12) (Kolde, R. pheatmap: Pretty Heatmaps, r package version 1.0.12 (2019). https://CRAN.R-project.org/package=pheatmap).
Identification of transcription factor genes
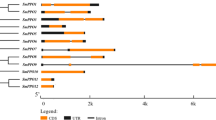
To screen for TFs that may be involved in the enzymatic browning process in fresh-cut eggplant, we analyzed the structures of TFs that were differentially abundant in the ‘F’ and ‘36’ cultivar transcriptomes and detected 231 differential TFs from 40 TF families (Fig. 7a), including genes encoding 27 ERF TFs, 19 MYB TFs, 17 NAC TFs, 16 WRKY TFs, 14 bHLH TFs, 14 C2H2 TFs, 12 HSF TFs, and 10 GRAS TFs, among other types of TFs (Fig. 7a). Subsequently, the TF gene expression levels following the various treatments were analyzed, and the genes were grouped into the following two groups (Fig. 7b): Group I, in which the expression levels of most TFs were higher in ‘F’ than in ‘36’, and Group II, which exhibited the opposite expression pattern. These results imply that the complex browning process of fresh-cut eggplant may be regulated by these candidate TFs.
(a) Number of transcription factors (TFs) involved in fresh-cut eggplant browning. (b) Heat map presenting the expression patterns of different TFs in response to diverse treatments. The heat map was drawn with pheamap (v1.0.12) (Kolde, R. pheatmap: Pretty Heatmaps, r package version 1.0.12 (2019). https://CRAN.R-project.org/package=pheatmap).
Gene regulation network construction
The gene regulation network data for Solanum lycopersicum, Solanum tuberosum, Arabidopsis, and Oryza sativa L. were downloaded from the STRING database. The KEGG pathways ko00350 (tyrosine metabolism) and ko00360 (phenylalanine metabolism) were targeted, respectively. BLAST comparisons were used to construct the eggplant gene regulation network based on the similarities between eggplant and these four types of plants. In the gene regulatory network map constructed based on ko00350 (tyrosine metabolism), a total of 212 were interacting with each other, and 7 core genes (Sme2.5_00661.1_g00006.1, Sme2.5_05449.1_g00003.1, Sme2.5_00484.1_g00015.1, Sme2.5_04286.1_g00003.1, Sme2.5_11776.1_g00001.1, Sme2.5_11841.1_g00001.1, and Sme2.5_00165.1_g00008.1) were differentially expressed in the transcriptome data (Fig. 8). Among them, two genes were annotated as PPO (Sme2.5_04286.1_g00003.1, Sme2.5_11776.1_g00001.1); and Sme2.5_00165.1_g00008.1 was annotated as tyrosine aminotransferase (Fig. 8). Furthermore, a gene regulatory network based on ko00360 (phenylalanine metabolism) was obtained for a total of 370 genes, including 11 core genes, among which 9 genes were differentially expressed in the transcriptome data (Sme2.5_11841.1_g00001.1, Sme2.5_00864.1_g00009.1, Sme2.5_00029.1_g00004.1, Sme2.5_16832.1_g00002.1, Sme2.5_05449.1_g00003.1, Sme2.5_00484.1_g00015.1, Sme2.5_00001.1_g00047.1, Sme2.5_01597.1_g00002.1, and Sme2.5_01597.1_g00004.1) (Fig. 9). Furthermore, Sme2.5_16832.1_g00002.1 was annotated as PAL and Sme2.5_00001.1_g00047.1 was annotated as trans-cinnamate 4-monooxygenase (Fig. 9).
Gene regulatory networks for the KEGG pathway ko00350 (tyrosine metabolism). Cytoscape v3.6.0 (https://cytoscape.org/) was used to create the illustrations.
Gene regulatory networks of the KEGG pathway ko00360 (phenylalanine metabolism). Cytoscape v3.6.0 (https://cytoscape.org/) was used to create the illustrations.
mRNA regulatory network associated with PPO genes
Pearson correlation coefficient was calculated between all the mRNAs and four significant differentially expressed PPO genes (Sme2.5_06843.1_g00001.1, Sme2.5_25992.1_g00001.1, Sme2.5_33651.1_g00001.1, Sme2.5_04286.1_g00003.1). The mRNAs with correlation coefficient greater than 0.9 was considered to be genes that are co-expressed with the PPO genes, of which a total of 212 mRNA correlation coefficients met this criteria (Fig. 10). These related DEGs, such as zinc finger protein WIP4 (Sme2.5_01070.1_g00005.1) and wound-induced proteinase inhibitor 2 (Sme2.5_04984.1_g00003.1), may have a synergistic effect with PPO and participate in the browning process of fresh-cut eggplant, requiring further study.
mRNA regulatory network associated with polyphenol oxidase (PPO) genes. Cytoscape v3.6.0 (https://cytoscape.org/) was used to create the illustrations.
Validation of DEGs with quantitative real-time PCR (qRT-PCR)
The heat map for the browning-related genes indicated that the expression levels varied dependent on both the cultivar and treatment (Fig. 6). To confirm the accuracy of the RNA-seq data, 18 browning-related genes were selected, and their expression levels investigated by qRT-PCR. The list of primer sequences are shown in Supplementary Table S5, whereas the quantitative results are shown in Fig. 11. The expression of DEGs were consistent with the transcriptome. As verified by qRT-PCR, the expression levels of PPO, POD, and PAL encoding genes in ‘36’ were significantly higher than that in ‘F’, whereas the activity of antioxidant enzymes (APX, GST) showed an opposite trend. The results of qRT-PCR were consistent with the analysis of the transcriptome, confirming the reliability of the RNA-seq data.
qRT-PCR of eggplant materials with different browning tolerance
We detected the expression levels of 18 browning-related genes in 15 eggplant materials with different browning tolerance by qRT-PCR. As shown in Fig. 12, the expression patterns of the four PPO-encoding genes were consistent, and the expression levels of all PPO-encoding genes were high in the browning-sensitive materials, while low in the browning-resistant materials. The expression levels of all PPO-encoding genes were the highest in the No. 1 browning-sensitive eggplant material (‘36’), and the lowest in the No. 15 eggplant material (‘F’) (Fig. 12). The genes related to the antioxidant enzymes, such as POD, CAT, APX, and GST, were found to be significantly differentially expressed during browning development. The expression levels of 3 POD DEGs, 1 CAT DEG, 1 APX DEGs, and 3 GST DEGs in browning-sensitive eggplant materials were lower than that in browning-resistant eggplant materials (Fig. 12). Furthermore, the expression pattern of the three WRKY TFs were consistent with the genes related to the antioxidant enzymes, suggesting their involvement in the antioxidant process (Fig. 12). Conversely, the DEGs associated with phenylpropanoid metabolism (2 PAL, 1 CHS) in browning-sensitive eggplant materials were higher than that in browning-resistant eggplant materials (Fig. 12).
Activities of PPO, POD, PAL, SOD, and CAT
As shown in Fig. 13, the enzyme activities of PPO, POD, PAL, and CAT in ‘36’ were significantly higher than those of ‘F’ fresh-cut for 15 min. The increase of PPO activity is often accompanied by browning, and the degree of browning is positively correlated with the activity. Both PPO and POD play a major role in the browning of eggplant pulp, and they may play a synergistic role in the browning process. The increase of PAL activity promotes the accumulation of phenolic substrates and the occurrence of browning. SOD is a key enzyme in the cell membrane protection system, which can effectively remove the accumulation of intracellular reactive oxygen species (ROS) and the occurrence of browning. Therefore, the SOD activity of ‘F’ is significantly higher than that of ‘36’ (Fig. 13). The expression changes of SmePPO, SmePOD, SmePAL, and SmeCAT genes were consistent with the changing trend of the related enzymes (Fig. 6). The expression of related genes can be promoted during fruit browning, which can be translated into the corresponding enzymes to accelerate fruit browning.
Discussion
Fresh-cut fruits and vegetables are ready-to-eat products made from fresh fruits and vegetables, which are cleaned, peeled, cut, dressed, packaged, and kept refrigerated. They are becoming more popular due to their convenience21. However, browning not only affects sensory quality, nutritional quality, and food quality of fruits and vegetables, which reduces commodity value, but also hinders product circulation, shortens the shelf life, and even seriously affects people's visual perception and appetite, making it unacceptable to consumers15. It has become one of the main obstacles restricting the development of fruits and vegetables storage, transportation, and processing. High-throughput sequencing analyses have revealed many genes related to the browning of vegetables and fruits20, but a more comprehensive understanding of gene expression during fresh-cut eggplant browning and strategies for browning control are still needed. The present study investigated the causes of browning and antioxidant capacity in a fresh-cut browning-sensitive cultivar (‘36’) compared to a browning-resistant cultivar (‘F’).
Enzymatic browning is considered the main reason for browning of fresh-cut fruits and vegetables. Enzymes related to phenolic oxidation (i.e., PPO, POD, CAT, and SOD) play a crucial role in the browning of fresh-cut fruits and vegetables. It is widely accepted that the enzymes encoded by the PPO and POD genes function together to induce browning22. Ali et al. found that the enzymatic browning of fresh-cut lotus root can be repressed by applying oxalic acid to decrease PPO and POD activity23. Similarly, an ascorbic acid (AsA) and aloe vera gel combined-treatment suppressed POD and PPO activities, providing indirect evidence that PPO and POD function together to influence the browning of fresh-cut lotus root24. Zheng et al. showed that the 0.1 mM melatonin treatment significantly reduced the expression of genes related to enzymatic browning and phenolic synthesis pathways, decreased POD and PPO activity, and thus reduced the surface browning of fresh-cut pear25. Furthermore, inhibition of PPO-encoding genes by artificial microRNAs in potato has been shown to lead to low PPO protein levels and low browning in potato tubers26. Gonzalez et al. found that specific editing of a single member of the StPPO gene family through the CRISPR/Cas9 system reduced PPO activity by 69% and enzymatic browning by 73% in potato tubers27, whereas evidence from another study indicated that overexpression of PPO in transgenic sugarcane led to higher PPO activity and darker juice color28. In the present study, 7 PPO-encoding genes involved in tyrosine metabolism and 22 POD-encoding genes were differentially regulated in ‘36’ and ‘F’. The expression level of most of the PPO-encoding genes in ‘36’ was significantly higher than that in ‘F’, and the expression level of PPO (Sme2.5_25992.1_g00001.1) was ~ 550-fold higher in ‘36’ than in ‘F’ (Fig. 6). It is generally accepted that the browning mechanism involves the interaction between the polyphenol substrate and PPO in the presence of oxygen, including two reaction processes: (1) hydroxylation of monophenols to diphenols, and (2) oxidation of diphenols to quinones. The Vmax of the hydroxylation reaction is 1.8 mM/min and results in colorless products, while the Vmax of the oxidation reaction is 24.5 mM/min with resultant coloration of quinones. The latter quinone reaction leads to the accumulation of melanin, which leads to browning of plant tissue. The specific structure of the polyphenol substrate determines the specific reaction time of plant tissue browning29. Additionally, the SOD and CAT enzymes play a pivotal role in the browning of fresh-cut fruits and vegetables. As the most important enzyme for scavenging free radicals, CAT can enhance the antioxidant properties of plants by catalyzing the degradation of H2O230. UV-C treatment has been found to inhibit surface browning of fresh-cut apple by improving ROS metabolism and enhancing the activities of SOD and CAT31. Meanwhile, Zhou et al. reported that a methyl jasmonate treatment induced enzymatic browning of fresh-cut potato tubers and enhanced PPO, POD, and CAT activities32. In our study, two CAT-encoding genes were identified. It was significantly up-regulated in the browning-sensitive cultivar (‘36’) (Fig. 6). Therefore, we postulate that CAT enzyme activity increased rapidly when the plant tissue was cut and damaged, thus removing ROS and enhancing the oxidation tolerance.
Phenols are vital substrates of enzymatic browning in fresh-cut fruits and vegetables, with the type and content of phenolic compounds significantly determining browning characteristics33. Therefore, total phenolic content plays a major role in the browning of stored fruits. Phenolic levels have been found to increase in eight cultivars of eggplant during storage, whereas the phenolic concentration varied significantly among fresh-cut samples of eggplant cultivars34. In 2019, a study on the mechanism of the enzymatic browning reaction in potato processing showed that phenolic compounds are closely related to enzymatic browning of potato, and chlorogenic acid is an important substrate for enzymatic browning of potato35. The phenolic compounds identified in banana peels in recent years were mainly dopamine, followed by chlorogenic acid and coumarin, with dopaminerase oxidation being the main factor for banana browning36. Chlorogenic acid and epicatechin are the phenolic compounds with the highest contents in apple fruits37, while in caffeic acid and ferulic acid are the main substrates in mangos38. Mishra et al. also showed that browning of eggplant was related to the increased content of polyphenols34, and Liu et al. found that the variation of chlorogenic acid content in fresh-cut eggplant may be closely related to the browning of fresh-cut eggplant39. Biosynthesis of phenol compounds is involved in the pentose phosphate, shikimate, flavonoid, and phenylpropanoid pathways in plants40, which involve a variety of key enzymes, such as PAL, 4CL, CHS, F3′H, and COMT. PAL is an important enzyme in the phenylpropane metabolic pathway, catalyzing l-phenylalanine ammonia hydrolysis to generate trans-cinnamic acid, and then undergo a series of transformations to form various phenols, lignin, anthocyanins, alkaloids, and other compounds. The produced phenols provide a substrate for the browning reaction41. In a study of the expression of PAL and HSP in fresh-cut banana fruit, it was observed that mechanical injury (fresh-cutting) resulted in the accumulation of PAL mRNA in fruits, which led to a rapid increase in PAL activity. Mechanical injury also increased the synthesis rate of ethylene, which accelerated the decomposition of phenylalanine and the synthesis of a large number of phenolic substances. At this point, the phenolic compounds combined with PPO, activated by mechanical injury, to induce browning. Through linear regression analysis, it was found that the increase in PAL activity in fruit induced by the mechanical injury was highly correlated with the accumulation of phenolic substances42. Moreover, CHS is the key enzyme in anthocyanin biosynthesis43. Melatonin treatment at 0.1 mM increased the expression of PAL and CHS, enhanced their activities, and reduced the browning of fresh-cut pear fruit25. Studies of fresh-cut sweet potatoes treated with ultrasound have found that the induction of PAL was positively correlated with higher total phenolic content, thereby enhancing antioxidant capacity of the fresh-cut sweet potatoes against browning44. An 80% oxygen treatment was found to increase the activities of PAL and POD and the total phenolic content of fresh-cut potato, which effectively enhanced its antioxidant capacity45. Conversely, Gong et al. reported that browning inhibition has no observable correlation with either total phenol or POD activity46. Zhang et al. identified one PAL-encoding gene, one CHS-encoding gene, and two 4CL-encoding genes that were significantly up-regulated in Chinese walnut husks that browned, but another PAL-encoding gene was significantly up-regulated in anti-browning husks47. In the present study, many genes connected with phenylpropanoid biosynthesis/phenylalanine metabolism were differentially regulated in the browning-resistant cultivar compared with the browning-sensitive cultivar. Three PAL-encoding genes and one CHS-encoding gene were up-regulated in the browning-sensitive cultivar (Fig. 6). However, four COMT-encoding gene was down-regulated in the browning-sensitive cultivar (Fig. 6). These results suggest that fresh-cutting treatment induced the expression of the key enzyme genes in phenylalanine metabolism and flavonoid biosynthesis, and promoted the synthesis of polyphenols in plant cell tissues to improve their antioxidant capacity.
AsA and GSH also play important roles in maintaining the stability of proteins, structural integrity of the biofilm system, and defense against membrane lipid peroxidation. Together with antioxidant enzymes such as GST, APX, dehydro-AsA reductase (DAR), monodehydro-AsA reductase (MDAR), and GSH reductase (GR), the AsA-GSH circulatory system can effectively eliminate free radicals48. The coordinated operation of each component in the AsA-GSH cycle enables the accumulation of ROS in plants to be removed, which plays an important role in the enzymatic ROS removal mechanism of fresh-cut fruits and vegetables. Under normal conditions, the production and clearance of ROS in plants are in dynamic balance. When plants are subjected to mechanical stress, the content of ROS increases significantly. Excessive ROS will destroy biological molecules such as proteins and destroy the integrity and function of membranes. In the enzymatic defense system, GST catalyzes the reaction of GSH with membrane lipid peroxides, thus reducing the damage to the membrane structure caused by mechanical stress49. GST participates in the AsA-GSH cycle50, catalyzing H2O2 to generate oxidized GSH (GSSG) and H2O, and decompose organic hydroperoxides into alcohols, H2O, and GSSG, which are then reduced by GR to GSH51. With AsA as the electron donor, APX catalyzes the reduction of H2O2 to MDHA and H2O. MDHA has two reduction pathways: (1) reduced to AsA by MDHAR and (2) reduced to dehydro-AsA (DHA) and then reduced to AsA through DHAR with reduced GSH as an electron donor52. AsA is involved in the oxidation protection system against free radicals; it is easily soluble in water and has sensitive antioxidant properties. Therefore, a change in its content level is considered an important indicator for monitoring fruit and vegetable quality53. l-Ascorbate oxidase plays a catalytic role in physiological changes, which can oxidize AsA to DHA and reduce H2O2, thus eliminating free radicals in cells and delaying senescence54. Browning and PPO activities of fresh-cut artichoke bottoms were significantly inhibited by Cg supplemented with AsA55. Salminen et al. found that 3% AsA and 0.1% green tea extract prevented the browning of fresh-cut apple slices for up to 14 days56. GST is present in almost all plants with up to 90 genes encoding GST in plants. Most of these genes are differentially expressed under stress induction and play an important role in the mechanism of enzymatic ROS scavenging57. We found that the expression levels of 18 GST-encoding genes and 10 genes encoding l-ascorbate oxidase involved in GSH metabolism were significantly higher in the browning-resistant cultivar compared with the browning-sensitive cultivar for all treatments (Fig. 6), indicating that the browning-resistant cultivar had a stronger antioxidant capacity. This could explain why the fruit did not easily brown after cutting.
The browning of fruits and vegetables is closely related to ROS metabolism in plants58. WRKY TFs play an vital role in regulating various physiological processes in plants, including plant growth and development, as well as responses to biological, abiotic, and oxidative stress59. Furthermore, Wang et al. revealed that WRKY TF plays an important induction role in the ROS metabolic pathway60. Overexpression of ATWRKY30 increased the activity of antioxidant enzymes (i.e., CAT, SOD, POX, and APX) in wheat plants61. Zhang et al. ported that one WRKY TF was differentially regulated in the fresh-cut browning and white husks of Chinese walnut47. There were four WRKY TFs significantly differentially expressed in fresh-cut luffa fruit20. These findings provide direct evidence that WRKY TFs play a crucial role in fresh-cut vegetables and fruits. In the current study, 16 WRKY TFs was observed and the expression level of most of the WRKY TFs were significantly up-regulated in the browning-resistant cultivar compared with the browning-sensitive cultivar (Fig. 6). The relationship between WRKY TFs and fresh-cut eggplant browning was revealed in our study. In addition, there are other TFs, such as MYB, ERF, NAC, and bHLH, that show different levels of transcription. The changes of these TFs may be involved in the browning process of fresh-cut eggplant. However, further studies on their structural and functional properties are needed to determine their potential influence on the browning of fresh-cut eggplant fruits.
Gene expression is often affected by other genes, thus forming a complex regulatory network of interaction and influence between genes. Gene regulatory network construction is an important technique in bioinformatics and systems biology that can be used to comprehensively study the mechanisms underlying cellular and metabolic processes by integrating gene expression data, biological information, computational analysis, and mathematical models. Furthermore, analysis of gene regulatory networks can allow researchers to explore the origin and evolution of life62. We set up two gene expression regulatory networks based on KEGG pathways ko00350 (tyrosine metabolism) and ko00360 (phenylalanine metabolism), respectively. We screened some core genes that showed differentially expressed transcriptome results, such as Sme2.5_16832.1_g00002.1, which was annotated as PAL and the Sme2.5_00001.1_g00047.1, which was annotated as trans-cinnamate 4-monooxygenase (Figs. 8 and 9). They play an important regulatory role in phenylalanine metabolism involved in phenolic synthesis. Moreover, a total of 212 mRNA were co-expressed with 4 PPO genes (correlation coefficient greater than 0.9) by Pearson correlation coefficient (Fig. 11), suggesting that they may be involved in the process of fresh-cut eggplant browning. However, further studies on their structural and functional properties are needed to determine their potential influence on the browning of fresh-cut eggplant fruits.
Finally, we detected the expression levels of 18 browning-related genes in 15 eggplant materials with different browning tolerance by qRT-PCR (Fig. 12). The results showed that the transcriptional expression of browning related genes was variety specific. These results suggest that these genes regulate the browning process of fresh-cut eggplant by participating in these biological processes, but further study is still needed for confirmation.
Conclusion
62 genes from six gene families (i.e., PPO, PAL, POD, CAT, APX, and GST) were differentially regulated in fresh-cut eggplant fruit from transcriptoms and the enzyme activities of PPO, POD, PAL, and CAT in ‘36’ were significantly higher than ‘F’ for 15 min, while SOD was down-regulated. Two gene expression regulatory networks was set up based on tyrosine metabolism (ko00350) and phenylpropanoid biosynthesis (ko00940/360) and 16 genes were differentially expressed in transcriptomic data. 18 browning related genes randomly screened in 15 eggplant materials with different browning tolerance showed variety specific.
Material and methods
Plant materials
The 15 eggplant materials were obtained from a breeding line produced by our lab at the Shanghai Key Laboratory of Protected Horticultural Technology of the Shanghai Academy of Agricultural Sciences, Shanghai, China. Mature commercial fruits with the same growth trend, uniform size, no diseases and insect pests, and no obvious mechanical damage were selected for the experiment. All plant studies involving (Solanum melongena L.) were carried out in accordance with relevant institutional, national, and international guidelines and legislation.
Preliminary screening of eggplant materials with different browning tolerance
Fresh-cutting treatment was performed on more than 60 eggplant materials planted in the Zhuanghang Experimental Station of Shanghai Academy of Agricultural Sciences, and the browning occurrence time of each material was recorded. Specifically, 15 eggplant materials were preliminary screened (as shown in Table 2), and subsequent experiments were conducted. In the subsequent experiments, all the eggplant material names were replaced by the numbers in Table 2. Finally, the eggplant cultivars ‘36’ was considered as the most browning-sensitive, while ‘F’ was considered as the most browning-resistant.
Fresh-cut treatments
The eggplant cultivars ‘36’ (browning-sensitive) and ‘F’ (browning-resistant) were obtained from a breeding line produced by our lab at the Shanghai Key Laboratory of Protected Horticultural Technology of the Shanghai Academy of Agricultural Sciences, Shanghai, China. The fruit samples were carefully peeled with a stainless-steel knife. Three treatments were established based on the time elapsed since cutting: control, fresh-cut for 0 min (36 CK0 and F CK0, respectively); treatment 1, fresh-cut for 2 min (36 2 min and F 2 min, respectively); and treatment 2, fresh-cut for 15 min (36 15 min and F 15 min, respectively). After the samples were left for the prescribed amount of time, they were frozen in liquid nitrogen and kept at − 80 °C until RNA extraction. Moreover, after peeling, 15 eggplant materials were immediately sampled and frozen in liquid nitrogen for qPT-PCR.
Total RNA extraction, library construction, and RNA sequencing
Total RNA was isolated using a mirVana RNA Isolation Kit (Ambion-1561; Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s protocol. The concentrations and quality of the RNA samples were assessed using a NanoDrop ND-2000 spectrophotometer (Thermo Fisher Scientific). The A260/280 ratios of individual samples were all above 2. The 28S/18S ratio and RIN values were determined using an Agilent 2100 system (Agilent, Santa Clara, CA, USA). A TruSeq Stranded mRNA LT Sample Prep Kit (Illumina, San Diego, CA, USA) was used to construct the sequencing library, and then samples were sequenced using the HiSeq 2500 system (Illumina) after quality control analysis using an Agilent 2100 Bioanalyzer.
RNA sequence analysis and DEG identification
Raw data (raw reads) were processed using Trimmomatic v0.3663. The reads containing ploy-N and low-quality reads were removed to obtain clean reads. Then, the clean reads were mapped to the reference genome using HISAT264. The reads were reassembled using StringTie v2.1.765. Then, gene structure extension and novel transcript identification were performed by comparing the reference genome and the known annotated genes using Cuffcompare v866.
The gene expression level was indicated by a fragments per kb per million (FPKM) value67. To calculate the different expression levels of genes among the samples, the htseq-count68 was used to acquire the number of reads in each sample. Two functions (estimatSizeFactors and nbinomTest) in the DESeq69 R package (http://www.bioconductor.org/packages/3.8/bioc/html/DESeq.html) were used to normalize the data and calculate the p-value and fold-change. A p-value < 0.05 and fold-change > 2 or < 0.5 were set as thresholds for significantly differential expression.
Functional annotation and classification of DEGs
The sequences of the DEGs were aligned to the KEGG database using BLASTX, retrieving proteins with the highest sequence similarity with the given sequences along with functional annotations for their proteins70. GO annotations of the DEGs were obtained using Blast2GO v6.071. Web Gene Ontology Annotation Plot v2.0 was used to perform GO functional classification with a Pearson’s chi-square test72. The DEGs were mapped to GO terms according to the analyses, and the number of DEGs in each term was calculated73.
Quantitative real-time PCR validation
qRT-PCR was performed to verify the accuracy of the gene expression profile obtained from the RNA-seq data. Total RNA was extracted using the Quick RNA isolation Kit (Beijing Tsingke Biotech, Beijing, China) according to the manufacturer’s protocol. First-strand cDNA was synthesized from 2 μg total RNA using M-MLV reverse transcriptase (Beijing Tsingke Biotech) and Oligo (dT)18 in a 25 μL reaction. Real-time PCR was performed with SYBR Green PCR mix (TaKaRa, Shiga, Japan). SmPKG was used as an endogenous control gene for qRT-PCR analyses. Relative expression levels of the target genes were calculated using the 2−ΔΔCt method. The primers used are listed in Supplementary Table S5.
Gene regulation network construction
The gene regulation network data for Solanum lycopersicum, Solanum tuberosum, Arabidopsis, and Oryza sativa L. were downloaded from the STRING database. BLAST comparisons were used to construct the eggplant gene regulation network based on the similarities between eggplant and these four types of plants. Cytoscape v3.6.0 (https://cytoscape.org/) was used to create the illustrations.
Ethical standards
The conducted experiments comply with the laws of China
Data availability
Data supporting the results and conclusions are included in both the article and additional files. All the transcriptome data have been deposited in the NCBI Sequence Read Archive (SRA) under accession number: PRJNA679701 (http://www.ncbi.nlm.nih.gov/sra).
References
Saini, D. K. & Kaushik, P. Visiting eggplant from a biotechnological perspective: A review. Sci. Hortic. 253, 327–340. https://doi.org/10.1016/j.scienta.2019.04.042 (2019).
Kumar, A., Sharma, V., Jain, B. T. & Kaushik, P. Heterosis breeding in eggplant (Solanum melongena L.): Gains and provocations. Plants https://doi.org/10.3390/plants9030403 (2020).
Manzocco, L., Calligaris, S., Mastrocola, D., Nicoli, M. C. & Lerici, C. R. Review of non-enzymatic browning and antioxidant capacity in processed foods. Trends Food Sci. Technol. 11, 340–346. https://doi.org/10.1016/s0924-2244(01)00014-0 (2001).
Wu, S. Glutathione suppresses the enzymatic and non-enzymatic browning in grape juice. Food Chem. 160, 8–15. https://doi.org/10.1016/j.foodchem.2014.03.088 (2014).
Du, Y., Dou, S. & Wu, S. Efficacy of phytic acid as an inhibitor of enzymatic and non-enzymatic browning in apple juice. Food Chem. 135, 580–582. https://doi.org/10.1016/j.foodchem.2012.04.131 (2012).
Friedman, M. Food browning and its prevention: An overview. J. Agric. Food Chem. 44, 631–653. https://doi.org/10.1021/jf950394r (1996).
Cantos, E., Tudela, J. A., Gil, M. I. & Espín, J. C. Phenolic compounds and related enzymes are not rate-limiting in browning development of fresh-cut potatoes. J. Agric. Food Chem. 50, 3015–3023. https://doi.org/10.1021/jf0116350 (2002).
Randhir, R., Lin, Y. T. & Shetty, K. Stimulation of phenolics, antioxidant and antimicrobial activities in dark germinated mung bean sprouts in response to peptide and phytochemical elicitors. Process Biochem. 39, 637–646. https://doi.org/10.1016/s0032-9592(03)00197-3 (2004).
Min, T. et al. Transcription profiles reveal the regulatory synthesis of phenols during the development of lotus rhizome (Nelumbo nucifera Gaertn). Int. J. Mol. Sci. https://doi.org/10.3390/ijms20112735 (2019).
Ariel Valinas, M., Luciana Lanteri, M., ten Have, A. & Balbina Andreu, A. Chlorogenic acid biosynthesis appears linked with suberin production in potato tuber (Solanum tuberosum). J. Agric. Food Chem. 63, 4902–4913. https://doi.org/10.1021/jf505777p (2015).
Li, J. et al. Multivariate analysis of fruit antioxidant activities of blackberry treated with 1-methylcyclopropene or vacuum precooling. Int. J. Anal. Chem. https://doi.org/10.1155/2018/2416461 (2018).
Gandhi Kelvin, D. et al. Plant polyphenol oxidase: Biochemical properties and browning of fruits and vegetables. Indian J. Agric. Biochem. 31, 1–8. https://doi.org/10.5958/0974-4479.2018.00001.1 (2018).
Lamikanra, O. & Watson, M. A. Effects of ascorbic acid on peroxidase and polyphenoloxidase activities in fresh-cut cantaloupe melon. J. Food Sci. 66, 1283–1286 (2001).
Frishberg, A. et al. Cell composition analysis of bulk genomics using single-cell data. Nat. Methods 16, 327. https://doi.org/10.1038/s41592-019-0355-5 (2019).
Gao, Z. et al. RNA-Seq based transcriptome analysis reveals the molecular mechanism of triterpenoid biosynthesis in Glycyrrhiza glabra. Bioorg. Med. Chem. Lett. 30, 127102. https://doi.org/10.1016/j.bmcl.2020.127102 (2020).
Lou, L., Su, X., Liu, X. & Liu, Z. Transcriptome analysis of Luffa cylindrica (L.) Roem response to infection with Cucumber mosaic virus (CMV). Gene https://doi.org/10.1016/j.gene.2020.144451 (2020).
Chen, L., Lu, Y., Hu, Y. & Xue, X. RNA-Seq reveals that sucrose-free medium improves the growth of potato (Solanum tuberosum L.) plantlets cultured in vitro. Plant Cell Tissue Organ 140, 505–521. https://doi.org/10.1007/s11240-019-01743-y (2020).
Pang, Q. et al. Triacontanol promotes the fruit development and retards fruit senescence in strawberry: A transcriptome analysis. Plants https://doi.org/10.3390/plants9040488 (2020).
Hao, L. et al. A constitutive and drought-responsive mRNA undergoes long-distance transport in pear (Pyrus betulaefolia) phloem. Plant Sci. https://doi.org/10.1016/j.plantsci.2020.110419 (2020).
Zhu, H. et al. De novo sequencing and analysis of the transcriptome during the browning of fresh-cut Luffa cylindrica “Fusi-3” fruits. PLoS One https://doi.org/10.1371/journal.pone.0187117 (2017).
Siddiqui, M. W., Chakraborty, I., Ayala-Zavala, J. F. & Dhua, R. S. Advances in minimal processing of fruits and vegetables: A review. J. Sci. Ind. Res. 70, 823–834 (2011).
Yan, D., Wen-Zhong, H. & Ai-Li, J. Research advances in biological preservatives of fresh-cut fruits and vegetables. J. Food Saf. Food Qual. 6, 2409–2414 (2015).
Ali, S. et al. Effect of postharvest oxalic acid application on enzymatic browning and quality of lotus (Nelumbo nucifera Gaertn.) root slices. Food Chem. https://doi.org/10.1016/j.foodchem.2019.126051 (2020).
Ali, S. et al. Effect of pre-storage ascorbic acid and Aloe vera gel coating application on enzymatic browning and quality of lotus root slices. J. Food Biochem. https://doi.org/10.1111/jfbc.13136 (2020).
Zheng, H., Liu, W., Liu, S., Liu, C. & Zheng, L. Effects of melatonin treatment on the enzymatic browning and nutritional quality of fresh-cut pear fruit. Food Chem. https://doi.org/10.1016/j.foodchem.2019.125116 (2019).
Chi, M. et al. Reduced polyphenol oxidase gene expression and enzymatic browning in potato (Solanum tuberosum L.) with artificial microRNAs. BMC Plant Biol. 14, 62. https://doi.org/10.1186/1471-2229-14-62 (2014).
Gonzalez, M. N. et al. Reduced Enzymatic Browning in Potato Tubers by Specific Editing of a Polyphenol Oxidase Gene via Ribonucleoprotein Complexes Delivery of the CRISPR/Cas9 System. Front. Plant Sci. 10. https://doi.org/10.3389/fpls.2019.01649(2020).
Vickers, J. E. et al. Overexpression of polyphenol oxidase in transgenic sugarcane results in darker juice and raw sugar. Crop Sci.45, 354–362. https://doi.org/10.2135/cropsci2005.0354 (2005).
Toivonen, P. M. A. & Brummell, D. A. Biochemical bases of appearance and texture changes in fresh-cut fruit and vegetables. Postharvest Biol. Technol. 48, 1–14. https://doi.org/10.1016/j.postharvbio.2007.09.004 (2008).
Huang, R., Xia, R., Hu, L., Lu, Y. & Wang, M. Antioxidant activity and oxygen-scavenging system in orange pulp during fruit ripening and maturation. Sci. Hortic. 113, 166–172. https://doi.org/10.1016/j.scienta.2007.03.010 (2007).
Qi, Z. et al. Correlation between enzymatic browning inhibition by UV-C treatment and reactive oxygen species metabolism of fresh-cut apples. Food Sci. 40, 102–109. https://doi.org/10.7506/spkx1002-6630-20171102-023 (2019).
Zhou, F. et al. Effect of methyl jasmonate on wound healing and resistance in fresh-cut potato cubes. Postharvest Biol. Technol. https://doi.org/10.1016/j.postharvbio.2019.110958 (2019).
Liang, Y. D. & Were, L. Cysteine’s effects on chlorogenic acid quinone induced greening and browning: Mechanism and effect on antioxidant reducing capacity. Food Chem. https://doi.org/10.1016/j.foodchem.2019.125697 (2020).
Mishra, B. B., Gautam, S. & Sharma, A. Free phenolics and polyphenol oxidase (PPO): The factors affecting post-cut browning in eggplant (Solanum melongena). Food Chem. 139, 105–114. https://doi.org/10.1016/j.foodchem.2013.01.074 (2013).
Liu, X. et al. Effect of purslane (Portulaca oleracea L.) extract on anti-browning of fresh-cut potato slices during storage. Food Chem. 283, 445–453. https://doi.org/10.1016/j.foodchem.2019.01.058 (2019).
Escalante-Minakata, P. et al. Comparative study of the banana pulp browning process of “Giant Dwarf” and FHIA-23 during fruit ripening based on image analysis and the polyphenol oxidase and peroxidase biochemical properties. 3 Biotech 8, 30. https://doi.org/10.1007/s13205-017-1048-3 (2017).
Park, S. Y., Kang, T. M., Kim, M. J. & Kim, M. J. Enzymatic browning reaction of apple juices prepared using a blender and a low-speed masticating household juicer. Biosci. Biotechnol. Biochem. 82, 2000–2006. https://doi.org/10.1080/09168451.2018.1497943 (2018).
Prasad, K., Sharma, R. R. & Srivastav, M. Postharvest treatment of antioxidant reduces lenticel browning and improves cosmetic appeal of mango (Mangifera indica L.) fruits without impairing quality. J. Food Sci. Technol. 53, 2995–3001. https://doi.org/10.1007/s13197-016-2267-z (2016).
Liu, X. et al. Study on browning mechanism of fresh-cut eggplant (Solanum melongena L.) based on metabolomics, enzymatic assays and gene expression. Sci. Rep. 11, 6937. https://doi.org/10.1038/s41598-021-86311-1 (2021).
Lin, D. et al. An overview of plant phenolic compounds and their importance in human nutrition and management of type 2 diabetes. Molecules 21, 1374 (2016).
Couture, R., Cantwell, M. I., Ke, D. & Saltveit, M. E. Jr. Physiological attributes related to quality attributes and storage life of minimally processed lettuce. HortScience 28, 723–725. https://doi.org/10.21273/hortsci.28.7.723 (1993).
Chen, J.-Y. et al. Expression of PAL and HSPs in fresh-cut banana fruit. Environ. Exp. Bot. 66, 31–37. https://doi.org/10.1016/j.envexpbot.2008.12.019 (2009).
Zhang, S. et al. Transcriptome analysis revealed expression of genes related to anthocyanin biosynthesis in eggplant (Solanum melongena L.) under high-temperature stress. BMC Plant Biol. https://doi.org/10.1186/s12870-019-1960-2 (2019).
Pan, Y. et al. Ultrasound treatment inhibits browning and improves antioxidant capacity of fresh-cut sweet potato during cold storage. RSC Adv. 10, 9193–9202. https://doi.org/10.1039/c9ra06418d (2020).
Liu, X. et al. Effect of high oxygen pretreatment of whole tuber on anti-browning of fresh-cut potato slices during storage. Food Chem. https://doi.org/10.1016/j.foodchem.2019.125287 (2019).
Gong, Y. et al. Short-time water immersion inhibits browning of fresh-cut potato by enhancing antioxidant capability and tyrosine scavenging. J. Food Process. Preserv. https://doi.org/10.1111/jfpp.14168 (2019).
Zhang, S. et al. De novo transcriptome assembly of the fresh-cut white husk of Juglans cathayensis Dode: Insights for enzymatic browning mechanism of fresh-cut husk of walnut. Sci. Hortic. https://doi.org/10.1016/j.scienta.2019.108654 (2019).
Cuddihy, S. L., Parker, A., Harwood, D. T., Vissers, M. C. M. & Winterbourn, C. C. Ascorbate interacts with reduced glutathione to scavenge phenoxyl radicals in HL60 cells. Free Radic. Biol. Med. 44, 1637–1644. https://doi.org/10.1016/j.freeradbiomed.2008.01.021 (2008).
Galant, A., Preuss, M. L., Cameron, J. C. & Jez, J. M. Plant glutathione biosynthesis: Diversity in biochemical regulation and reaction products. Front. Plant Sci. https://doi.org/10.3389/fpls.2011.00045 (2011).
Rahantaniaina, M. S., Tuzet, A., Mhamdi, A. & Noctor, G. Missing links in understanding redox signaling via thiol/disulfide modulation: How is glutathione oxidized in plants?. Front. Plant Sci. 4, 477. https://doi.org/10.3389/fpls.2013.00477 (2013).
Afzal, M., Afzal, A., Jones, A. & Armstrong, D. A rapid method for the quantification of GSH and GSSG in biological samples. Methods Mol. Biol. 186, 117–122. https://doi.org/10.1385/1-59259-173-6:117 (2002).
Hernandez, O., Torres, C. A., Moya-Leon, M. A., Opazo, M. C. & Razmilic, I. Roles of the ascorbate-glutathione cycle, pigments and phenolics in postharvest “sunscald” development on “Granny Smith” apples (Malus domestica Borkh.). Postharvest Biol. Technol. 87, 79–87. https://doi.org/10.1016/j.postharvbio.2013.08.003 (2014).
Ferrante, A., Martinetti, L. & Maggiore, T. Biochemical changes in cut vs. intact lamb’s lettuce (Valerianella olitoria) leaves during storage. Int. J. Food Sci. Technol. 44, 1050–1056. https://doi.org/10.1111/j.1365-2621.2008.01891.x (2009).
Li, X. et al. Effect of cutting styles on quality and antioxidant activity in fresh-cut pitaya fruit. Postharvest Biol. Technol. 124, 1–7. https://doi.org/10.1016/j.postharvbio.2016.09.009 (2017).
El-Mogy, M. M., Parmar, A., Ali, M. R., Abdel-Aziz, M. E. & Abdeldaym, E. A. Improving postharvest storage of fresh artichoke bottoms by an edible coating of Cordia myxa gum. Postharvest Biol. Technol. https://doi.org/10.1016/j.postharvbio.2020.111143 (2020).
Salminen, W. F. & Russotti, G. Synergistic interaction of ascorbic acid and green tea extract in preventing the browning of fresh cut apple slices. J. Food Process. Preserv. https://doi.org/10.1111/jfpp.13192 (2017).
Chi, Y. et al. Expansion mechanisms and functional divergence of the glutathione S-transferase family in sorghum and other higher plants. DNA Res. 18, 1–16. https://doi.org/10.1093/dnares/dsq031 (2011).
Lin, Y. et al. The role of active oxygen metabolism in hydrogen peroxide-induced pericarp browning of harvested longan fruit. Postharvest Biol. Technol. 96, 42–48. https://doi.org/10.1016/j.postharvbio.2014.05.001 (2014).
Phukan, U. J., Jeena, G. S. & Shukla, R. K. WRKY transcription factors: Molecular regulation and stress responses in plants. Front. Plant Sci. 7, 760. https://doi.org/10.3389/fpls.2016.00760 (2016).
Wang, L. et al. Transcriptional and physiological analyses reveal the association of ROS metabolism with cold tolerance in tea plant. Environ. Exp. Bot. 160, 45–58. https://doi.org/10.1016/j.envexpbot.2018.11.011 (2019).
El-Esawi, M. A., Al-Ghamdi, A. A., Ali, H. M. & Ahmad, M. Overexpression of AtWRKY30 transcription factor enhances heat and drought stress tolerance in wheat (Triticum aestivum L.). Genes https://doi.org/10.3390/genes10020163 (2019).
Singh, G. & Singh, V. Construction and analysis of an interologous protein-protein interaction network of Camellia sinensis leaf (TeaLIPIN) from RNA-Seq data sets. Plant Cell Rep. 38, 1249–1262. https://doi.org/10.1007/s00299-019-02440-y (2019).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. https://doi.org/10.1093/bioinformatics/btu170 (2014).
Kim, D., Langmead, B. & Salzberg, S. L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 12, 357-U121. https://doi.org/10.1038/nmeth.3317 (2015).
Pertea, M. et al. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 33, 290. https://doi.org/10.1038/nbt.3122 (2015).
Trapnell, C. et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511-U174. https://doi.org/10.1038/nbt.1621 (2010).
Roberts, A., Trapnell, C., Donaghey, J., Rinn, J. L. & Pachter, L. Improving RNA-Seq expression estimates by correcting for fragment bias. Genome Biol. https://doi.org/10.1186/gb-2011-12-3-r22 (2011).
Anders, S., Pyl, P. T. & Huber, W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169. https://doi.org/10.1093/bioinformatics/btu638 (2015).
Jin, Y. & Hammell, M. Analysis of RNA-Seq data using TEtranscripts. Methods Mol. Biol. 1751, 153–167. https://doi.org/10.1007/978-1-4939-7710-9_11 (2018).
Sudheesh, S. et al. De novo assembly and characterisation of the field pea transcriptome using RNA-Seq. BMC Genomics https://doi.org/10.1186/s12864-015-1815-7 (2015).
Conesa, A. et al. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21, 3674–3676. https://doi.org/10.1093/bioinformatics/bti610 (2005).
Qi, X., Xie, S., Liu, Y., Yi, F. & Yu, J. Genome-wide annotation of genes and noncoding RNAs of foxtail millet in response to simulated drought stress by deep sequencing. Plant Mol. Biol. 83, 459–473. https://doi.org/10.1007/s11103-013-0104-6 (2013).
Garcia-Seco, D., Zhang, Y., Gutierrez-Manoro, F. J., Martin, C. & Ramos-Solano, B. RNA-Seq analysis and transcriptome assembly for blackberry (Rubus sp. Var. Lochness) fruit. BMC Genomics https://doi.org/10.1186/s12864-014-1198-1 (2015).
Acknowledgements
This work was supported by the Shanghai Municipal Agriculture Commission 02-08-00-12-F-1482 (2020), China Agriculture Research System (Grant No. CARS-25), Excellent Team of Shanghai Academy of Agricultural Sciences (Agricultural Science Innovation 2017 [B-06]), and National Key Research and Development Program of China (2017YFD010190412018). These funding bodies did not play a role in the design of the study, the collection, analysis, and interpretation of data, or in writing the manuscript.
Author information
Authors and Affiliations
Contributions
Z.D.S. and W.X.X. conceived the study idea; L.X.H. and S.J. performed the preparation and treatments of test materials. Z.J. and Z.Z.W. designed and performed the experiments. L.X.H. analyzed the data and wrote the manuscript. Z.A.D. revised the article. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Liu, X., Zhang, A., Zhao, J. et al. Transcriptome profiling reveals potential genes involved in browning of fresh-cut eggplant (Solanum melongena L.). Sci Rep 11, 16081 (2021). https://doi.org/10.1038/s41598-021-94831-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-94831-z
- Springer Nature Limited
This article is cited by
-
Breeding for postharvest physiological deterioration in cassava: problems and strategies
CABI Agriculture and Bioscience (2022)
-
Transcriptomic changes induced by applications of a commercial extract of Ascophyllum nodosum on tomato plants
Scientific Reports (2022)
-
Transcriptomic analysis of the effects of melatonin on genes potentially related to the browning of broccoli (Brassica oleracea L. var. italica Planch) hairy roots
Plant Growth Regulation (2022)