Abstract
Weight gain is a frequent and severe adverse reaction in patients taking antipsychotics. The objective was to further investigate in a natural setting influential risk factors associated with clinically significant weight gain. An observational follow-up study was conducted. Patients when initiating treatment with whatever antipsychotic were included; a structured questionnaire was applied at baseline, 3 and 6 months later; a blood sample was obtained. In a nested case–control approach, patients with an increase ≥ 7% of their initial weight were considered as cases, the remaining, as controls. The results showed that, out of 185 patients, 137 completed the 6-month follow-up (cases, 38; controls, 99). Weight gain gradually and significantly increased in cases (baseline, 65.0 kg; 6 months, 74.0 kg) but not in controls (65.6 kg and 65.8 kg, respectively). Age (adjusted OR = 0.97, 95% CI = 0.96–0.99, p = 0.004), olanzapine (adjusted OR = 2.98, 95% CI = 1.13–7.80, p = 0.027) and quetiapine (adjusted OR = 0.25, 95% = 0.07–0.92, p = 0.037) significantly associated with weight gain. An association was also found for the CNR1 (rs1049353) and INSIG2 (rs7566605) polymorphisms. In conclusion, an increased risk of antipsychotics-induced weight gain was observed for younger age and olanzapine, and a relative lower risk for quetiapine. A potential role of CNR1 rs1049353 and INSIG2 rs7566605 polymorphisms is suggested.
Similar content being viewed by others
Despite the severity and large number of their adverse reactions, antipsychotics are widely used in a variety of groups that include children and elderly people1,2,3; moreover, they are largely prescribed for indications other than schizophrenia4. Among the most troublesome adverse reactions are those related to metabolism, in particular weight gain5,6,7,8. Weight gain contributes to medication discontinuation and relapse; it also accounts for the metabolic syndrome and certainly increases morbidity and mortality9. In addition to environmental factors, genetics have been increasingly associated with weight gain when taking antipsychotics10,11,12,13; in particular, a twins study highlighted the significance of genetics at this regard14.
Numerous clinical trials have been published on patients treated with antipsychotics15; however, prospective follow-up studies carried out in real-life settings are scanty. Hence, the objective of this study was to further investigate influential risk factors as associated with significant weight gain in a natural setting.
Material and methods
General
For this purpose, an observational prospective follow-up study was conducted and, for the analysis, a nested case–control approach was adopted. Patients willing to participate, 14 years old and older, when initiating treatment with whatever antipsychotic and with a body mass index (BMI) lower than 35 kg/m2, were included; a washout period of at least 6 months was needed for those previously on antipsychotics. The recruitment period was March 2010 to December 2014; patients were recruited in hospitals, in community services or in nursing homes.
After obtaining a written informed consent, trained monitors directly interviewed patients with a structured questionnaire; the questionnaire was filled at baseline and then prospectively at 3 and 6 months after recruitment. Information upon the following types and specific variables was collected: (i) social and demographic (age, sex, education, working status); (ii) metabolic diseases ([yes/no]); (iii) psychiatric (diagnoses, type of diagnosis [psychotic/non-psychotic disorder], number of psychiatric diagnoses); (iv) substance use (alcohol intake [yes/no]); smoking ([yes/no]); (v) all medications (for antipsychotics: type of drug, number and dose in chlorpromazine equivalence); and (vi) anthropometrics (height [cm], body weight [kg], and BMI [kg/m2]; height and body weight were assessed using a SECA 217 height rod and a SECA 877 scale, respectively; vii) activity factor16. Indirectly, Charlson index was calculated17.
Genotyping
In addition, a venous blood sample (9 ml) was obtained from study participants at the first visit for genetic analysis. Based on results of published studies as well as a search in the PubMed database (http://www.ncbi.nlm.nih.gov/PubMed), relevant candidate genes previously associated with antipsychotics-induced weight gain were selected. In this manner, a total of 38 polymorphisms in 14 candidate genes were selected for the present study: Ankyrin repeat and kinase domain containing 1 (ANKK1): rs1800497, rs7104979, rs17115461; Brain-derived neurotrophic factor (BDNF): rs6265; Cannabinoid receptor 1 (CNR1): rs1049353; Dopamine receptor D2 (DRD2): rs1799978, rs1799732; Fatty acid amide hydrolase (FAAH): rs324420; Fat mass and obesity associated (FTO): rs76804286, rs9926289, rs9939609; Guanine nucleotide binding protein (G protein), beta polypeptide 3 (GNB3): rs5442, rs5443; 5-hydroxytryptamine (serotonin) receptor 2A (HTR2A): rs6313, rs1805055; 5-hydroxytryptamine (serotonin) receptor 2C (HTR2C): rs6318, rs518147, rs1414334, rs5946226, rs3813928, rs3813929; Insulin induced gene 2 (INSIG2): rs7566605, rs10490624, rs11889497, rs17047764, rs17587100; Leptin (LEP): rs791615, rs75550733, rs7798338, rs7799039; Leptin receptor (LEPR): rs1137101; Melanocortin 4 receptor (MC4R): rs489693, rs2229616, rs8087522, rs11872992, rs17066842, rs17782313; Peroxisome proliferator activated receptor gamma (PPARG): rs1801282.
Purified DNAs were quantified by Picogreen. The amplicons were prepared using the Access Array System for Illumina Platform (Fluidigm). Briefly, this method consist of the following phases: (1) quantification by Picogreen and normalization of the DNA samples to 50 ng/ul, (2) combination of diluted DNA samples with primers (5′-AATGATACGGCGACCACCGAGATCTACACTGACGACATGGTTCTACA-3′ and 5′-CAAGCAGAAGACGGCATACGAGAT-[barcode]-TACGGTAGCAGAGACTTGGTCT-3′) of the Access Array Barcode Library for Illumina Sequencers (Fluidigm), (3) preparation of 20X specific target primers solutions, (4) loading of sample inlets and primer inlets of a primed Access Array Integrated Fluidic Circuit (IFC) and running of the PCR AA48 × 48 Standard v1 protocol and, finally, (5) harvest of PCR products from the IFC. For more details consult the protocol: “Access Array System for Illumina Platform User Guide_G1”.
The finally obtained amplicons were validated and quantified by an Agilent 2100 Bioanalyzer using High Sensitivity chips, and an equimolecular pool of these amplicons was purified by agarose gel electrophoresis to eliminate primers/dimers, and titrated by quantitative PCR using the “Kapa-SYBR FAST qPCR kit forLightCycler480” and a reference standard for quantification. The pool of amplicons was denatured prior to be seeded on a flowcell at a density of 10 pM, where clusters were formed and sequenced using a “MiSeq Reagent Kit v3”, in a 2 × 300 pair-end sequencing run on a MiSeq sequencer. Raw sequences were filtered according to quality under standard Illumina, parameters and positive sequences were mapped against human genome. Following mapping, DNA variant detection was performed using the MiSeqReporter software (Illumina).
Statistics analysis
For the case–control approach, patients who, at 180 days, showed an increase ≥ 7% of their initial weight were considered as cases18; the remaining were considered as controls. Values of qualitative variables are presented as absolute frequencies and percentages; quantitative variables are presented as means with Standard Deviation (SD) or median with interquartile ranges [Q1–Q3]. For comparison among qualitative variables, including Hardy–Weinberg equilibrium, the Pearson’s chi-square test, or the Fisher’s exact test if the expected frequency was lower than 5 in any cell, were used. Genotypes distribution in the two groups in comparison was assessed with the Monte Carlo method for multiple testing19; the estimated p values in this form are in Table 3; for comparison among groups for quantitative variables, a Student t test or a Mann–Whitney U were calculated according to the Kolmogorov–Smirnov or Shapiro–Wilk test. To explore possible correlation between some quantitative variables without normal distribution, Spearman´s Rho was calculated.
Based on the literature, covariates initially considered for adjusting in the analysis were: age, sex, BMI at baseline and the different SNPs considered. Information on exposure to the explanatory variables was that consigned at baseline. Other possible confounders were selected among those with a p < 0.1 significance level in a previous univariate analysis; a forward stepwise multivariate logistic regression was performed with those variables. With the significant variables in this first regression, a second regression analysis was performed, minimizing in this way the possible effect of the missing values in variables which finally were non-significant. A Hosmer–Lemeshow goodness-of-fit test was performed. To learn the influence of each of the several factors on weight gain, adjusted odds ratios (adjusted OR) and their 95% confidence intervals (95% CI) were calculated. For all tests, significance level was set at p < 0.05. The Statistical Package for Social Sciences (version 24.0; SPSS, Inc., Chicago, IL) was used for statistical analyses.
Ethics
The protocol of the study was approved by the Ethics and Clinical Research Committee of the Faculty of Medicine, University of Valladolid, Spain (CEIC); it was included in the Spanish and in the European Medicines Agencies registers. The study was conducted following the principles of the Declaration of Helsinki and all applicable local ethical and legal requirements. All participants provided written informed consent before performance of the study procedures.
Results
Out of 185 patients of the ongoing cohort, 137 completed at least a follow-up period of 6 months and were included in the current analysis; reasons for dropouts are presented in a flow-chart (Fig. 1). Of those patients who completed the specified period, 38 showed a clinically significant increase of their initial weight and were accordingly considered as cases; the remaining 99, who did not, were considered as controls.
Flowchart of the patients’ follow-up. Main baseline characteristics of those patients who fulfil inclusion criteria but were lost to follow-up (n = 39): age, 56.2 years (55.2 years in the final cohort); mean BMI, 23.8 kg/m2 (25.5 in the final cohort); females, 44.7% (63.5% in the final cohort). They did not substantially differ from those who were finally included.
The whole sample was of European origin. The distribution of the main sociodemographic and clinical variables in the two groups is presented in Table 1. Mean age in the whole sample was 55.2 years, it ranged from 15 to 100 years; age was higher in controls than in cases (59.7 years vs 43.4 years, p = 0.001). Though there were more female patients in the cohort (63.5%), no significant differences between cases and controls were found (p = 0.654). As an average, patients had 1.1 psychiatric diagnoses and were having 1.2 antipsychotics; risperidone, followed by quetiapine, olanzapine and aripiprazole were the antipsychotics most frequently used. More cases than controls received two or more antipsychotics during some time in the follow-up period (34.2% vs. 20.2%, p = 0.086); cases received higher doses than controls (median chlorpromazine equivalence, 237.74 mg/d vs. 117.24 mg/d, respectively; p = 0.002). More psychotic diagnoses were observed within the cases without statistically significant difference (39.5% vs. 24.2%, p = 0.08). Previous history of metabolic disease was associated with a minor risk of weight gain (p = 0.018); similarly, higher Charlson index scores were associated with less risk (p = 0.008; Table 1). No differences in overall medications were found between cases and controls (p = 0.072).
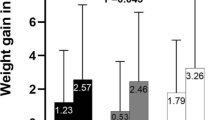
Cases were taller than controls (mean values (SD), 1.64 (0.10) m vs. 1.59 (0.12) m, respectively; p = 0.01); body weight was similar in the two groups at baseline (65.05 (12.71) kg vs. 65.65 (14.49) kg, respectively; p = 0.824), but it differed at the end of the follow-up (73.97 (14.33) kg vs. 65.84 (14.97) kg for cases and controls, respectively; p = 0.003). The average weight increase was 8.92 (4.08) kg for cases and 0.19 (2.99) kg for controls. Weight gradually increased in cases but not in controls (Fig. 2). Regarding BMI, this was different at baseline (cases, 24.23 (4.07) kg/m2; controls, 26.00 (4.24) kg/m2; p = 0.029) but it equalized at the end of follow-up (cases, 27.51 (4.48) kg/m2; controls, 26.07 (4.27) kg/m2; p = 0.082). No differences in physical activity existed between cases and controls, neither at baseline (p = 0.379) nor at the end of the follow-up period (p = 0.608) when measured by the activity factor (Table 1).
In our multivariate model only age, along with exposure to olanzapine or quetiapine, showed a significant association with weight gain (Table 2). Sex was not significant in the model, not even for olanzapine or quetiapine exposure as an interaction term. Dose was not significant either; when taken independently a significant correlation was observed between dose in chlorpromazine equivalents and percentage of weight gain (Rho = 0.31; p = 0.0003). A lower risk of weight gain was observed with increasing patients’ age and for those treated with quetiapine, this similarly occurs when choosing an age range of 18–65; risk of weight gain significantly increased for patients who were treated with olanzapine.
When examined by a 2 × 3 table Pearson’s chi-square test (two degrees of freedom), a trend of significant differences between subjects who gained 7%, or higher, of their baseline body weight, and those who did not, emerged for CNR1 (rs1049353) and INSIG2 (rs7566605) polymorphisms (Tables 3 and 4); CNR1 (rs1049353) was associated with weight change in a dominant model (OR = 0.41, 95% CI = 0.18–0.94, p = 0.034) while INSIG2 (rs7566605) was in a recessive model (OR = 3.03, 95% CI = 1.05–8.78, p = 0.041). After adjusting by age and medication effects, adjusted OR were respectively (OR = 0.50, 95% CI = 0.21–1.21, p = 0.13) and (OR = 2.17, 95% CI = 0.67–6.99, p = 0.195).
Discussion
Our observational study following a nested case–control approach showed that one in three patients on antipsychotics developed an increase of ≥ 7% of their initial weight after a period of 6 months (cases) when compared with those who did not (controls). This significant and severe weight gain associated with antipsychotics has been largely observed elsewhere: at 6 weeks in 45.8% of schizophrenic patients on olanzapine20 and in 20.2% of bipolar patients with the same medication21; at 3 months in 35% of schizophrenic first-episode medication-naive patients22. In a large systematic review and meta-analysis, a significantly higher risk of gaining ≥ 7% of the baseline weight (RR = 2.04, 95% CI = 1.54–2.71, p < 0.001) was also found after 3 to 12 weeks of treatment, and the most severe weight-gain was caused by olanzapine5. Weight gain also has been found in longer follow-up studies at 6, 12 and 36 months23,24,25. Our figures are in this range; follow-up duration, antipsychotics used, psychiatric disease, comorbidities and, particularly, age certainly account for these differences in percentages. Taking as a reference the sample studied by Verma et al.24, with a similar 6-month period of observation, it can be observed that differences in percentages of patients who gained 7% or more than the baseline weight (ours, 27.7%; Verma's, 65%) inversely correlated with average age differences in the two samples (ours, 55.1 years; Verma´s, 29.8 years).
Although percentage of male are generally higher in schizophrenia samples using antipsychotics, female patients are majority in our sample; this is probably due to the more frequent non-psychotic disorders in women, including neurocognitive and affective disorders. The influence of sex in antipsychotic-induced weight gain is still controversial; moreover, sex did not appear to emerge as a critical predictive factor for beneficial effect26. We did not observe differences in weight gain between sexes, what is coincident with previous observations by Zipursky et al.27. Though most of the studies found a higher risk in females1,24,28,29,30 and lower in males in the first months of treatment23, the figures by Zipursky et al.27 remain chiefly reliable because they are based on a survival analysis at 2 years in a study specifically designed to find out predictors of weight gain in people with first-episode psychosis.
As far as age is concerned, younger age seems to be consistently associated with a higher risk of weight gain associated with antipsychotics28,30,31; being children and adolescents even at a greater risk32,33. In fact, younger age, along with non-Caucasian ethnicity, low baseline BMI, along with other variables, are considered one of the 16 items for Weight Gain Risk Factor Assessment Checklist30. In our sample, a higher proportion of patients younger than 35 years showed an increase of weight (43.7%) compared to those of 35–55 years (39.1%), or those older than 55 years (10.2%). According to our estimates, for each year of increasing age, the risk of weight gain diminishes 3%; this estimate remained stable, barely without changes from the crude estimate. Thus, increasing age appears as a protective factor for antipsychotics-associated weight gain in the studied sample. This has been consistently observed in other studies24,29,31,34; it must be underlined the susceptibility observed in children and adolescents to antipsychotics-associated obesity32,35,36. Age is also associated with certain diseases, comorbidities, and the corresponding treatments: i.e., doses of antipsychotics in elderly populations are usually lower than those used in the youngest ones. Moreover, age is related to physiological changes, which could be related to a less propensity for weight gain. None of these factors seem to have a significant modifying effect in our estimate.
Up to eleven different antipsychotics were used by patients in our sample (Table 1); all, but haloperidol, were atypical. Olanzapine was independently and strongly associated with weight gain in our sample (Table 2); this is consistent with previous findings1,21,23,31,37,38,39 and with two meta-analyses, where olanzapine showed the highest risk of weight gain5,40. Although in our study quetiapine increased the initial weight in 0.4 kg as an average, this increase was lower than that observed with the other antipsychotics; this is coincidental with results of a recent meta-analysis5. Most of the patients in the cohort received only one psychiatric diagnosis and had one antipsychotic, though receiving several antipsychotics is a common practice in schizophrenia41,42; this lower use found might be related to the lower proportion of psychotic patients in our sample.
In this study exploring 38 polymorphisms located in 14 candidate genes, two were found to be associated with antipsychotics-induced weight gain in CNR1 and INSIG2 genes (Table 3). The minor T-allele of the frequently studied synonymous variant in the final exon of CNR1 (rs1049353) conferred some protection; it was observed that carriers of the minor allele gained less weight while treated with antipsychotics than the CC homozygotes (OR = 0.41, 95% CI = 0.18–0.94, p = 0.034). To our knowledge, four studies have explored this association so far43,44,45,46. Our results are coincidental with those found by Nurmi et al.43 in a relatively large sample of children with autism treated for the first time with antipsychotics (n = 181); they differ, however, from the results found by the others. In the study by Monteleone et al.44, carried out similarly in a Mediterranean population like ours (MAF, 17% and 22%, respectively), comparable associations would be expected; notwithstanding, subtle differences in design might account for the disparities observed: i.e., patients who had been previously exposed to antipsychotics being already obese could participate in that study (n = 83). Regarding the study by Park et al.45 (n = 78), the differences in allelic distribution of the Asian population they analyzed, with a MAF of 7%, by far lower than ours (22%), make it difficult to detect any association when the exposure in the cases is low, as it was the case (6.7%). The study by Tiwari et al.46 in samples exposed to clozapine and olanzapine with an elevated initial body weight (n = 183) neither found any association in a shorter follow-up period of 14 weeks, though it was found with other polymorphisms in the same gene; all in all, it is concluded that the CNR1 gene may be associated with antipsychotics-induced weight gain in chronic schizophrenia. As for the INSIG2 polymorphism (rs7566605), we found an association with weight gain in a recessive model. A genome-wide study by Skelly et al.47 looked at the same marker in a large sample of patients treated with antipsychotics from the CATIE study1; for the same recessive model, it was observed an analogous trend in magnitude to that first reported by Herbert et al.48 in the general population [in subjects of European ancestry, mean BMI (SD) was 31.9 (0.94) for the CC genotype versus 30.8 (0.43) for combined CG/GG genotypes]. As denoted by these figures, a possible explanation for these inconclusive results might be the elevated baseline BMIs of the patients included in the CATIE study1; otherwise, a study primarily intended to assess a different outcome. Also, the study by Tiwari et al.49 found certain association for the same allele with this variant in a subset of patients with African ancestry who received either clozapine or olanzapine (n = 54). Conversely, two studies exploring the same variant did not find any statistically significant relationship (n = 160 and n = 128, respectively)50,51; patients in both studies, with shorter durations than ours, had been previously exposed to antipsychotics, being stated in one of the studies that it could bias the identified amount of body weight gain51.
The cannabinoid receptor (CNR1 or CB1) is related to food intake and lipogenesis52. Cnr1 knockout mice presents a lean phenotype53,54,55, and, consistently, administration of the inverse agonist rimonabant leads to decreased body weight56,57; on the contrary, endogenous ligands of CNR1 increases dietary food intake58. Insulin induced gene 2 protein (INSIG2) on its part inhibits cholesterol synthesis in the adipose tissue and the liver59; dysregulation of this process may produce obesity and increase insulin resistance. Both CNR1 and INSIG2 are involved in the processing of sterol regulatory element binding proteins (SREBPs): CNR1 receptor activity leads to increased lipogenesis through augmented expression of these SREB proteins60,61 and, on the contrary, INSIG2 blocks these proteins acting as a negative regulator of cholesterol biosynthesis62. Hence, it is conceivable that variants in genes encoding these proteins may alter their normal functioning, accounting for metabolic dysregulation and yielding to weight gain; this is what has been observed in our study: protection with a CNR1 variant and risk with other INSIG2 variant.
Among the study limitations, sample size and heterogeneity are the most remarkable. Heterogeneity is linked to natural settings in which patients are quite different in diagnoses, ages, and treatments; thus, it makes it difficult to identify minor risk factors since larger samples are needed for stratification. Conversely, this setting and the observational nature of our study provides the actual variety of patients on these medications; in addition, a close follow-up has permitted a more detailed collection of the main features of all individuals in the study which includes adherence—dose collected were those that patients were taking. As for genetic association analysis, the stringent inclusion criteria upon use of antipsychotics—most of our patients were drug naive prior to the beginning of the study limiting the influence of one of the most confounding factors in previous studies—and weight along with the prospective character of this study make it suitable to clearly observe the development of the condition studied and assess the genetic risk factors; though, in terms of genetic association studies, the number of subjects is rather small, it still represents a comparably large sample for the specific phenotype. Hidden population always remains as a possible confounder; however, all individuals in our sample were from European origin, they come from the same geographical region, Castile (Spain), without cultural differences. No migrants or adopted children were found in the recruited patients; moreover, the Spanish population is generally like that of Northern and Western Europe origin, but also largely homogeneous within itself63,64.
In summary, our study has identified two major risk factors for weight gain when on antipsychotics, younger age and olanzapine, as well as a third relative protective factor, quetiapine; these influential factors are fully discussed in the light of more information. Additionally, the observations made in the large screening of this study indicate a possible role of certain polymorphisms in CNR1 and INSIG2 genes in antipsychotic-induced weight gain; at this regard, new approaches in the study design considering the likely polygenic character of the genetics influence for weight gain should be adopted. All in all, our results should be interpreted as exploratory and require replication in an independent and larger data set.
References
Lieberman, J. A. et al. Effectiveness of antipsychotic drugs in patients with chronic schizophrenia. N. Engl. J. Med. 353, 1209–1223 (2005).
Pagsberg, A. K. et al. Acute antipsychotic treatment of children and adolescents with schizophrenia-spectrum disorders: A systematic review and network meta-analysis. J. Am. Acad. Child Adolesc. Psychiatry 56, 191–202 (2017).
Gareri, P., De Fazio, P., Manfredi, V. G. L. & De Sarro, G. Use and safety of antipsychotics in behavioral disorders in elderly people with dementia. J. Clin. Psychopharmacol. 34, 109–123 (2014).
Sohn, M., Moga, D., Blumenschein, K. & Talbert, J. National trends in off-label use of atypical antipsychotics in children and adolescents in the United States. Medicine (Baltimore) 95, e3784 (2016).
Barton, B. B., Segger, F., Fischer, K., Obermeier, M. & Musil, R. Update on weight-gain caused by antipsychotics: A systematic review and meta-analysis. Expert Opin. Drug Saf. https://doi.org/10.1080/14740338.2020.1713091 (2020).
Carvajal, A., Martín Arias, L. H. & Jimeno, N. Antipsychotic drugs. in Side Effects of Drugs Annual (ed. Aronson, J.) 33, 89–123 (Elsevier Science BV, 2011).
Galling, B. et al. Type 2 diabetes mellitus in youth exposed to antipsychotics: A systematic review and meta-analysis. JAMA Psychiat. 73, 247–259 (2016).
Brandl, E. J. et al. Genome-wide association study on antipsychotic-induced weight gain in the CATIE sample. Pharmacogenomics J. 16, 352–356 (2016).
De Hert, M., Detraux, J., van Winkel, R., Yu, W. & Correll, C. U. Metabolic and cardiovascular adverse effects associated with antipsychotic drugs. Nat. Rev. Endocrinol. 8, 114–126 (2011).
Li, N. et al. Progress in genetic polymorphisms related to lipid disturbances induced by atypical antipsychotic drugs. Front. Pharmacol. 10, 1669 (2020).
Yoshida, K. & Müller, D. J. Pharmacogenetics of antipsychotic drug treatment: Update and clinical implications. Mol. Neuropsychiatry 5, 1–26 (2019).
Zai, C. C., Tiwari, A. K., Zai, G. C., Maes, M. S. & Kennedy, J. L. New findings in pharmacogenetics of schizophrenia. Curr. Opin. Psychiatry 31, 200–212 (2018).
Zhang, J. P. et al. Pharmacogenetic associations of antipsychotic drug-related weight gain: A systematic review and meta-analysis. Schizophr. Bull. 42, 1418–1437 (2016).
Gebhardt, S. et al. Body weight gain induced by atypical antipsychotics: An extension of the monocygotic twin and sib pair study. J. Clin. Pharm. Ther. 35, 207–211 (2010).
Leucht, S. et al. Sixty years of placebo-controlled antipsychotic drug trials in acute schizophrenia: Systematic review, bayesian meta-analysis, and meta-regression of efficacy predictors. Am. J. Psychiatry 174, 927–942 (2017).
National Research Council, 0. Recommended Dietary Allowances: 10th edition. (National Academies Press, 1989).
Charlson, M. E., Pompei, P., Ales, K. L. & MacKenzie, C. R. A new method of classifying prognostic comorbidity in longitudinal studies: Development and validation. J. Chronic Dis. 40, 373–383 (1987).
Kanders, B., Forse, R. & Blackburn, G. Methods in obesity. in Conn´s Current Therapy (ed. Rakel, R.E.) 524–532 (WB Saunders, 1991).
Westfall, P. & Young, S. Resampling-based multiple testing: Examples and methods for p-value adjustment. (John Wiley & Sons, 1993).
Kemp, D. E. et al. Associations among obesity, acute weight gain, and response to treatment with Olanzapine in adolescent schizophrenia. J. Child Adolesc. Psychopharmacol. 23, 522–530 (2013).
Katagiri, H. et al. Efficacy and safety of olanzapine for treatment of patients with bipolar depression: Japanese subpopulation analysis of a randomized, double-blind, placebo-controlled study. BMC Psychiatry 13, e138 (2013).
Pérez-Iglesias, R. et al. Comparison of metabolic effects of aripiprazole, quetiapine and ziprasidone after 12 weeks of treatment in first treated episode of psychosis. Schizophr. Res. 159, 90–94 (2014).
Pérez-Iglesias, R. et al. Course of weight gain and metabolic abnormalities in first treated episode of psychosis: the first year is a critical period for development of cardiovascular risk factors. Int. J. Neuropsychopharmacol. 17, 41–51 (2014).
Verma, S., Liew, A., Subramaniam, M. & Poon, L. Y. Effect of treatment on weight gain and metabolic abnormalities in patients with first-episode psychosis. Aust. N. Z. J. Psychiatry 43, 812–817 (2009).
Chiliza, B. et al. Changes in body mass and metabolic profiles in patients with first-episode schizophrenia treated for 12 months with a first-generation antipsychotic. Eur. Psychiatry 30, 277–283 (2015).
Raben, A. T. et al. The complex relationship between antipsychotic-induced weight gain and therapeutic benefits: A systematic review and implications for treatment. Frontiers in Neuroscience 11, 741 (Frontiers Media S.A., 2018).
Zipursky, R. B. et al. Course and predictors of weight gain in people with first-episode psychosis treated with olanzapine or haloperidol. Br. J. Psychiatry 187, 537–543 (2005).
Musil, R., Obermeier, M., Russ, P. & Hamerle, M. Weight gain and antipsychotics: A drug safety review. Expert Opin. Drug Saf. 14, 73–96 (2015).
Gebhardt, S. et al. Antipsychotic-induced body weight gain: Predictors and a systematic categorization of the long-term weight course. J. Psychiatr. Res. 43, 620–626 (2009).
Treuer, T. et al. Weight gain risk factor assessment checklist: Overview and recommendation for use. Neuro Endocrinol. Lett. 32, 199–205 (2011).
Lee, S. Y., Park, M. H., Patkar, A. A. & Pae, C. U. A retrospective comparison of BMI changes and the potential risk factors among schizophrenic inpatients treated with aripiprazole, olanzapine, quetiapine or risperidone. Prog. Neuro-Psychopharmacol. Biol. Psychiatry 35, 490–496 (2011).
De Hert, M. et al. Physical illness in patients with severe mental disorders. II. Barriers to care, monitoring and treatment guidelines, plus recommendations at the system and individual level. World Psychiatry 10, 138–151 (2011).
Reekie, J. et al. The effect of antidepressants and antipsychotics on weight gain in children and adolescents. Obes. Rev. 16, 566–580 (2015).
Strassnig, M., Miewald, J., Keshavan, M. & Ganguli, R. Weight gain in newly diagnosed first-episode psychosis patients and healthy comparisons: One-year analysis. Schizophr. Res. 93, 90–98 (2007).
Correll, C. U. & Carlson, H. E. Endocrine and metabolic adverse effects of psychotropic medications in children and adolescents. J. Am. Acad. Child Adolesc. Psychiatry 45, 771–791 (2006).
Vitiello, B. et al. Antipsychotics in children and adolescents: Increasing use, evidence for efficacy and safety concerns. Eur. Neuropsychopharmacol. 19, 629–635 (2009).
Cui, Y., Zheng, Y., Yang, Y., Liu, J. & Li, J. Effectiveness and tolerability of aripiprazole in children and adolescents with Tourette’s disorder: A pilot study in China. J. Child Adolesc. Psychopharmacol. 20, 291–298 (2010).
Perez-Iglesias, R. et al. Weight gain induced by haloperidol, risperidone and olanzapine after 1 year: Findings of a randomized clinical trial in a drug-naïve population. Schizophr. Res. 99, 13–22 (2008).
Zheng, L. et al. Metabolic changes associated with second-generation antipsychotic use in Alzheimer’s disease patients: The CATIE-AD study. Am. J. Psychiatry 166, 583–590 (2009).
Asmal, L. et al. Quetiapine versus other atypical antipsychotics for schizophrenia. Cochrane Database Syst. Rev. 11, CD006625 (2013).
Baandrup, L. Polypharmacy in schizophrenia. Basic Clin. Pharmacol. Toxicol. 126, 183–192 (2020).
Lin, S.-K. Antipsychotic polypharmacy: A dirty little secret or a fashion?. Int. J. Neuropsychopharmacol. 23, 125–131 (2020).
Nurmi, E. L. et al. Moderation of antipsychotic-induced weight gain by energy balance gene variants in the RUPP autism network risperidone studies. Transl. Psychiatry 3, e274 (2013).
Monteleone, P., Milano, W., Petrella, C., Canestrelli, B. & Maj, M. Endocannabinoid Pro129Thr FAAH functional polymorphism but not 1359G/A CNR1 polymorphism is associated with antipsychotic-induced weight gain. J. Clin. Psychopharmacol. 30, 441–445 (2010).
Park, Y. M. et al. Cannabinoid type 1 receptor gene polymorphisms are not associated with olanzapine-induced weight gain. Hum. Psychopharmacol. 26, 332–337 (2011).
Tiwari, A. K. et al. A common polymorphism in the cannabinoid receptor 1 (CNR1) gene is associated with antipsychotic-induced weight gain in schizophrenia. Neuropsychopharmacology 35, 1315–1324 (2010).
Skelly, T., Pinheiro, A. P., Lange, L. A. & Sullivan, P. F. Is rs7566605, a SNP near INSIG2, associated with body mass in a randomized clinical trial of antipsychotics in schizophrenia?. Mol. Psychiatry 12, 321–322 (2007).
Herbert, A. et al. A common genetic variant is associated with adult and childhood obesity. Science 312, 279–283 (2006).
Tiwari, A. K. et al. Association study of polymorphisms in Insulin Induced Gene 2 (INSIG2) with antipsychotic-induced weight gain in European and African-American schizophrenia patients. Hum. Psychopharmacol. Clin Exp 25, 253–259 (2010).
Le Hellard, S. et al. Association between the insulin-induced gene 2 (INSIG2) and weight gain in a German sample of antipsychotic-treated schizophrenic patients: Perturbation of SREBP-controlled lipogenesis in drug-related metabolic adverse effects?. Mol. Psychiatry 14, 308–317 (2009).
Opgen-Rhein, C. et al. Association of HTR2C, but not LEP or INSIG2, genes with antipsychotic-induced weight gain in a German sample. Pharmacogenomics 11, 773–780 (2010).
Silvestri, C. & Di Marzo, V. The endocannabinoid system in energy homeostasis and the etiopathology of metabolic disorders. Cell Metab. 17, 475–490 (2013).
Ravinet Trillou, C., Delgorge, C., Menet, C., Arnone, M. & Soubrié, P. CB1 cannabinoid receptor knockout in mice leads to leanness, resistance to diet-induced obesity and enhanced leptin sensitivity. Int. J. Obes. 28, 640–648 (2004).
Cota, D. et al. The endogenous cennabinoid system affects energy balance via central orexigenic drive and peripheral lipogenesis. J. Clin. Invest. 112, 423–431 (2003).
Di Marzo, V. et al. Leptin-regulated endocannabinoids are involved in maintaining food intake. Nature 410, 822–825 (2001).
Colombo, G. et al. Appetite suppression and weight loss after the cannabinoid antagonist SR 141716. Life Sci. 63, PL113–PL117 (1998).
Rinaldi-Carmona, M. et al. SR141716A, a potent and selective antagonist of the brain cannabinoid receptor. FEBS Lett. 350, 240–244 (1994).
Kirkham, T. C., Williams, C. M., Fezza, F. & Di Marzo, V. Endocannabinoid levels in rat limbic forebrain and hypothalamus in relation to fasting, feeding and satiation: Stimulation of eating by 2-arachidonoyl glycerol. Br. J. Pharmacol. 136, 550–557 (2002).
Dong, X. Y. & Tang, S. Q. Insulin-induced gene: A new regulator in lipid metabolism. Peptides 31, 2145–2150 (2010).
Kim, J., Li, Y. & Watkins, B. A. Endocannabinoid signaling and energy metabolism: A target for dietary intervention. Nutrition 27, 624–632 (2011).
Shi, D. et al. Inhibiting CB1 receptors improves lipogenesis in an in vitro non-alcoholic fatty liver disease model. Lipids Health Dis. 13, 173 (2014).
Yabe, D., Brown, M. S. & Goldstein, J. L. Insig-2, a second endoplasmic reticulum protein that binds SCAP and blocks export of sterol regulatory element-binding proteins. Proc. Natl. Acad. Sci. U. S. A. 99, 12753–12758 (2002).
Bycroft, C. et al. Patterns of genetic differentiation and the footprints of historical migrations in the Iberian Peninsula. Nat. Commun. 10, 551 (2019).
Gayán, J. et al. Genetic structure of the Spanish population. BMC Genom. 11, 326 (2010).
Acknowledgements
Funding for this study was provided by the Ministry of Education of the Regional Government of Castile and Leon [GR129 and VA315U14]; Ministry of Health of the Regional Government of Castile and Leon [BIO/VA20/13] and Spanish Ministry of Health [EC11-395]. The authors acknowledge the cooperation of the researcher network of the Icaro Study.
Author information
Authors and Affiliations
Contributions
N.J.: Study supervision, Data interpretation, Drafting manuscript, Revising manuscript. V.V-G.: Data acquisition, Data analysis, Drafting manuscript, Revising manuscript. I.F.: Data analysis, Data interpretation; Revising manuscript. M.D.: Data acquisition, Revising manuscript. A.C.: Study concept and design; Study supervision; Significant contributions to the manuscript. All the authors approved the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Jimeno, N., Velasco-Gonzalez, V., Fierro, I. et al. Association of CNR1 and INSIG2 polymorphisms with antipsychotics-induced weight gain: a prospective nested case–control study. Sci Rep 11, 15304 (2021). https://doi.org/10.1038/s41598-021-94700-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-94700-9
- Springer Nature Limited