Abstract
The functional significance of the zinc-finger of the cerebellum (ZIC) gene family in gliomas remains to be elucidated. Clinical data from patients with gliomas, containing expression levels of ZIC genes, were extracted from CCLE, GEPIA2 and The Human Protein Atlas (HPA). Univariate survival analysis adjusted by Cox regression via OncoLnc was used to determine the prognostic significance of ZIC expression. We used cBioPortal to explore the correlation between gene mutations and overall survival (OS). ZIC expression was found to be related to immune cell infiltration in gliomas via TIMER analysis. GO term and KEGG pathway enrichment analyzes were performed with Metascape. PPI networks were constructed using STRING. The expression levels of ZIC1/3/4/5 in gliomas were significantly different from those in normal samples. High expression levels of ZIC1/5 were associated with poor OS in brain low-grade glioma (LGG) patients, while low ZIC3 expression combined was related to favorable OS in glioblastoma multiforme (GBM). ZIC alterations were associated with poor prognosis in LGG patients and related to favorable prognosis in GBM patients. We observed that the expression of ZICs was related to immune cell infiltration in glioma patients. ZICs were enriched in several pathways and biological processes involving Neuroactive ligand-receptor interaction (hsa04080). The PPI network revealed that some proteins coexpressed with ZICs played a role in the pathogenesis of gliomas. Differences in the expression levels of ZIC genes could provide a significant marker for predicting prognosis in gliomas.
Similar content being viewed by others
Introduction
Gliomas are the most common type of primary tumour arising from glial cells, representing 75% of malignant brain tumours1. The World Health Organization (WHO) divides gliomas, including low-grade gliomas (LGG) and high-grade gliomas (HGG), into 4 stages2. WHO stage IV glioma, also known as glioblastoma multiforme (GBM), has a remarkably poor prognosis and accounts for 2.9% of cancer-related deaths but only 1.4% of cancers1,3. Over the years, although great progress has been made in surgery and chemoradiation, the overall median survival of patients is approximately 12.6 months and has not improved significantly4,5. Existing studies have suggested that glioma progression is associated with the regulatory networks of genes6,7. Therefore, investigating biomarkers underlying glioma tumorigenesis might be helpful for improving therapeutic strategies.
Some studies have confirmed that aberrant expression of mRNAs is involved in the carcinogenesis process8. Moreover, glioblastoma cell migration and invasion were suppressed by the expression of some mRNAs9. Human zinc-finger of the cerebellum (ZIC) family genes are indispensable during development. A previous study showed that ZIC5, a member of the ZIC family, drives melanoma aggressiveness by activating FAK and STAT310. In addition, ZIC1 could reprogramme glioma cells into neuron-like cells, which suggested that ZIC family members were associated with glioma11. Whether ZICs have prognostic value in glioma patients remains unclear.
To determine the role of ZICs as prognostic biomarkers in LGG and GBM, we performed a comprehensive bioinformatics analysis using multiple datasets and further explored these data to identify the role of ZICs in glioma.
Methods
Broad Institute Cancer Cell Line Encyclopedia
Broad Institute Cancer Cell Line Encyclopedia (CCLE) (https://portals.broadinstitue.org/ccle), an open-access database containing data on 1457 cell lines and 84,434 genes, is a resource for that uses model cancer cell lines to accelerate cancer research12. In this study, scatterplots for ZIC expression were obtained from CCLE.
GEPIA2
GEPIA2 (http://www.gepia2.cancer-pku.cn/#index) is a comprehensive online platform that provides fast and customized delivery of functionalities from the genotype-tissue expression (GTEx) and TCGA (The Cancer Genome Atlas) databases13. In our study, we used GEPIA2 to analyze the transcriptional expression of ZIC family members among different cancer tissues and their corresponding contiguous normal control samples. The prognostic values of genes were validated using a Kaplan–Meier curve.
The human protein atlas
The Human Protein Atlas (http://www.proteinatlas.org) includes immunohistochemistry-based expression data for almost 20 extremely common cancers, and every tumour type includes 12 individual tumour samples14. In our study, the gene expression of ZICs was explored using immunofluorescence data from brain-related cell lines. Next, the protein expression of ZIC1/3 in human normal tissues was determined by immunohistochemistry and compared with that in glioma tissues.
OncoLnc
OncoLnc (https://www.oncolnc.org) contains survival data from 8647 patients from The Cancer Genome Atlas (TCGA) and includes data from 21 cancer studies. Users can download the survival data or create Kaplan–Meier plots15. In this study, the prognostic value of ZIC expression in LGG and GBM patients was analyzed using this tool. We downloaded the survival data of cancer patients who had been divided into low and high expression groups by median to further analyze the utility of ZICs in predicting survival.
cBioPortal
cBioPortal (http://www.cbioportal.org) is a comprehensive web resource for the exploration and analysis of multidimensional cancer genomics data16. In our study, we used Glioblastoma Multiforme(TCGA, PanCancer Atlas) dataset to analyze the genomic data of ZICs, which included mRNA expression z-scores relative to diploid samples (RNASeq V2RSEM) with a z-score threshold of ± 1.8, mutations and putative copy number alterations with the GISTIC tool. Their relationship of ZIC genetic mutations with overall survival was analyzed with Kaplan–Meier plots, and the log-rank test was used to determine the significance of differences between the survival curves. A P value < 0.05 was considered statistically significant.
TIMER
TIMER (http://cistrome.shinyapps.io/timer/), a user-friendly interactive web resource, identified the proportions of different immune cells based on expression data and the corresponding clinical impact of these cells for systematic evaluation17,18. In this study, we used the “gene module” to analyze the relationship between the infiltration of immune cells and ZIC expression levels. The “survival module” was used to further explore the association between the infiltration of immune cells, ZIC expression and clinical outcome.
Metascape
Metascape (http://metascape.org) is a publicly available tool for gene set enrichment analysis and gene annotation19. In this study, we found more than 100 genes similar to ZICs via the “similar genes detection module” of GEPIA213. Next, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway20,21,22 enrichment analysis and Gene Ontology enrichment analysis of ZICs were performed, providing enriched cellular component (CC), biological process (BP) and molecular function (MF) terms23,24,25,26. The minimum enrichment was set as 3, and a P value cutoff of 0.05 was considered statistically significant.
STRING
STRING (http://string-db.org/) is a powerful web resource covering 5090 organisms that supports functional exploration of genome-wide experimental datasets. Users are allowed to perform gene set enrichment analysis and visualize subsets of enriched proteins as interaction networks27. To construct a protein–protein interaction (PPI) network of differentially expressed ZIC family members, we explored additional proteins related to ZICs with STRING. We used a minimum required interaction score cutoff of ≥ 0.7 (high confidence) to obtain the significant PPIs.
Statistical methods
The association of ZIC mRNA expression with patient survival was analyzed by Cox regression using SPSS software version 20.0. We used this software to evaluate the effect of expression of ZIC family members on prognosis.
Results
Transcriptional levels of ZICs in gliomas tissues
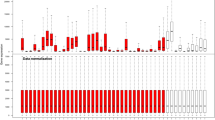
As shown in Fig. 1, the mRNA expression levels of ZIC family members were measured and compared to those in corresponding normal tissues via the GEPIA2 database. Significant upregulation of ZIC1/3/4 was found in brain low-grade glioma patients, while ZIC5 was downregulated. The mRNA expression of ZIC1 were highly expressed in glioblastoma patients, and ZIC3 and ZIC4 were downregulated expression. The results regarding ZICs gene expression between the CCLE database and GEPIA2 were discrepant partly, which could have been caused by the different classifications of brain and central nervous system (CNS) cancers. Brain and CNS cancers are divided into brain low-grade gliomas and glioblastoma multiforme in GEPIA2. The expression of ZIC2 between glioma tissues and corresponding normal samples had no obvious difference (Fig. 1a–j).
Furthermore, the gene expression of ZICs was also evaluated using immunofluorescence data from brain-related cell lines via The Human Protein Atlas (Fig. 2a–e). RNA expression above 1 was considered statistically significant. In our study, ZIC1 and ZIC3 expression were enriched in brain-related cell lines, which means in these cell lines ZIC levels are at least four times higher than any other cell type. After analyzing the mRNA expression patterns of ZICs in glioma, we further explored the protein expression of ZICs in glioma (Fig. 2f,g). According to information provided by the HPA database, ZIC1 was highly expressed in normal brain tissues, and higher expression of ZIC was observed in malignant low-grade glioma than in normal tissues. In addition, lower protein expression of ZIC3 was identified in malignant high-grade glioma tissues compared with normal samples. The protein expression data for ZIC2/4/5 in the HPA database were incomplete. Taken together, our results showed that the gene expression of ZIC1 and ZIC3 were obviously different between glioma and normal samples, and the evidence of gene protein expression was not sufficient which needed further experimental verification..
Representative immunohistochemistry images of ZICs staining in glioma tissues and normal brain tissues (HPA). Specific antibody staining is shown in green, while microtubule staining is shown in red. Brown staining indicates where an antibody has bound to its corresponding antigen in the IHC. Compared with other cell lines, expression above 1 was considered at least elevated (A–E). High ZIC1 protein expression was observed in LGG tissues (F), while low ZIC3 protein expression was observed in GBM tissues (G).
Prognostic significance of ZIC mRNA expression in glioma patients
To explore the utility of ZICs in predicting survival, we analyzed the mRNA expression of ZICs in both LGG and GBM patients via OncoLnc. As shown in Fig. 3, the mRNA expression of ZICs was significantly associated with glioma patient prognosis. Highly expression levels of ZIC1/5 were associated with short overall survival (OS) in LGG patients. (Fig. 3a,d). Downregulated expression of ZIC3 was significantly related to unfavorable OS (Fig. 3f). Next, the patient data were downloaded to further validate these results by univariate and multiple Cox regression analysis. The outcome data are shown in Tables 1 and 2 (p < 0.05 was considered statistically significant). These results indicated that the mRNA expression of ZIC1/5 was a significant independent prognostic predictor for OS in LGG patients, and the mRNA expression of ZIC3 was related to GBM patient prognosis, which suggested that ZIC1/5 and ZIC3 are independent prognostic factors in LGG and GBM patients, individually.
Prognostic value of ZIC family member mRNA expression in LGG and GBM patients (OncoLnc). The framed images contain parameters that were found to have prognostic relevance. High mRNA expression of ZIC1/5 individual was associated with poor OS in LGG patients (a,d). Low mRNA expression of ZIC3 was significantly associated with poor OS in GBM patients (f), while high expression of ZIC4 was significantly related to unfavorable OS in GBM patients (g). ZIC1/5 was not related to the OS in GBM patients (e,h), and ZIC3/4 was not related to the OS in LGG patients (b,c).
Genetic mutation in ZICs and its relationship with the OS of glioma patients
Next, we analyzed genetic alterations in ZICs and their correlation with the overall survival of LGG and GBM patients. In the 514 sequenced LGG patient samples, gene alterations were observed in 59 samples, and the mutation rates for ZIC1/5 were 0.39%, (Fig. 4a). Meanwhile, in 155 GBM patients, gene alterations were found in 8 patients with no mutated cases, which implied that mutation or DNA copy-number alteration of ZICs occurs at a low rate in LGG and GBM (Fig. 4b). Next, the results from the log-rank test and Kaplan–Meier plots showed that genetic alteration in ZICs was associated with poor OS and PFS (Progression-free survival) in LGG patients (Fig. 4c) and favorable OS in GBM patients (Fig. 4d). These results suggested that genetic alteration of ZICs could significantly influence LGG and GBM patient prognosis (all p < 0.05) (Supplementary Figure 1).
Genetic mutations in ZICs and their relationship with the OS of gliomas patients (cBioPortal). The mutation rates of ZIC1and ZIC5 were 0.39% in LGG (a), while they were no mutations in GBM (b). Genetic alterations in ZICs were associated with poor OS and PFS in LGG patients (c) and favorable OS and PFS in GBM patients (d).
Relationship between immune cell infiltration and ZIC expression in patients with gliomas
ZICs are involved in immune cell infiltration and inflammatory responses, and hence, the clinical outcome of glioma patients related to these factors should be considered. We explored the association between immune cell infiltration and ZIC expression via the TIMER database. When summarizing the outcome data, we also took the tumour purity into account28,29. In LGG patients, ZIC5 expression was positively associated with infiltration of B cells, CD8+ T cells, CD4+ T cells, macrophages, neutrophils, and dendritic cells (Fig. 5a, Tables 3 and 4) p < 0.05 was considered statistically significant.
In TCGA database, we determined the correlation between ZIC3 and CCL2, (Fig. 5b) which participate in recruitment of macrophages. CCL2 mRNA levels could be enhanced in low-expressed ZIC3 patients. The EGFR, which constitutively activates and enhances tumorigenicity through RAS-SHC-GRB2 pathway, was correlative with ZIC5 as shown in Fig. 5c.
Functional enrichment analysis of ZICs
After exploring the immune cell infiltration of ZICs in glioma patients, functional enrichment analysis were construct using the genes that were coexpressed with ZIC1/5 and ZIC3 in LGG and GBM, individually. As shown in Fig. 6a, biological processes such as embryonic skeletal system development (GO: 0048706), response to radiation (GO: 0009314), and sensory organ morphogenesis (GO: 0090596) were significantly regulated by ZIC1 alterations in glioma. Cellular component terms, including chromosomal region (GO: 0098687) and nuclear chromosome part (GO: 0044454), were associated with ZIC alterations. In KEGG analysis, Neuroactive ligand-receptor interaction (hsa04080) was related to the functions of ZIC5 alternations in glioma (Fig. 6a,b) (Supplementary Figure 2).
Construction of the PPI network
PPIs of ZIC family members were determined based on their correlation coefficients by STRING. As shown in Fig. 6c, PAX3, which has been implicated in glioma tumorigenesis is co-expressed with ZIC1. Transcriptional regulation was mutually affected by ZIC1 and SHH. ZIC3 acts as a transcriptional activator or is activated in PPIs involved in neurogenesis. We used a minimum required interaction score cutoff of ≥ 0.7 (high confidence) to obtain the significant PPIs, and the topological parameters of the PPI networks are shown in Table 5.
Discussion
Gliomas are the most prevalent malignant brain tumours. There has been significant progress in understanding the biology and molecular pathogenesis of glioblastoma, but translation of these findings into improved outcomes needs more time. ZICs participate in the progression of various cancers, including bladder cancer, prostate cancer, gastric cancer and melanoma10,30,31,32. Although some ZICs have been verified to play important roles in glioma, the distinct roles of ZIC family members in glioma remain to be elucidated33.
In the present study, we used multiple bioinformatics analysis tools to demonstrate that ZIC family members are related to the prognosis of glioma patients. We observed a significant difference in the expression of ZICs between glioma tissues and normal samples. Upregulated expression of ZIC1/5 was confirmed to be a significant predictor of poor OS in LGG patients. Low expression of ZIC3 was a significant prognostic factor in GBM patients. Furthermore, mutation of ZICs was identified in glioma patients, and genetic alteration in ZICs was associated with short OS in LGG patients and favorable OS in GBM patients. These differences between LGG and GBM related to ZIC3 expression confirm that the expression of ZIC3 is negatively correlated with the prognosis of GBM.
Some studies have reported that dendritic cells, which have the ability to induce antitumour immunity, possess immediate tumoricidal activity34. Macrophages are also the most common cell type related to tumour-infiltrating immune cells35,36. In this study, we explored the association between the expression of ZIC family members and the infiltration of immune cells, including B cells, CD4+ T cells, CD8+ T cells, neutrophils, macrophages and dendritic cells. The results revealed that ZIC5 had weak, but statistically significant correlations with the infiltration of immune cells, which indicated that ZIC5 reflect immune status and deserve further research.
GO enrichment and KEGG pathway analyzes of ZIC1/3/5 and their 100 similar genes were performed, and our results showed that the embryonic skeletal system development, response to radiation, sensory organ morphogenesis, chromosomal region and nuclear chromosome part were significantly associated with ZIC alterations. Pathways such as Neuroactive ligand-receptor interaction were found to be regulated by ZIC3 in glioma patients.
Finally, a PPI network was generated to analyze the correlation between the expression of ZICs and their related proteins. NANOG, SOX2 and POU5F1, three stemness-associated genes that are part of the FA-BSA-NP delivery system, could facilitate autophagy inhibition and chemotherapy efficacy in glioma therapy37. PASD1, a coexpression gene of ZIC3, promotes glioma cell proliferation by inhibiting apoptosis in vitro12. ZIC proteins are critical in cerebellar development, and the ZIC-related autoimmune response impacts the pathogenesis of neurologic disorders38. These results suggested that ZIC proteins may play an indispensable role in the progression of gliomas.
Unfortunately, there were some limitations in the present study. Although the mRNA expression levels of different ZIC family members were associated with the OS of glioma patients, all the data were obtained from online databases in our analysis, and thus, further studies consisting of more samples are required to confirm our findings; in addition, the clinical utility of ZIC members in glioma treatment needs to be explored. Moreover, our transcriptional level analysis of immune cells reflected some aspects of the immune status, but the global changes should be considered in future studies. Further studies are needed to investigate the relationships between specific ZICs and glioma (Supplementary Information).
Conclusion
In summary, a bioinformatics analysis was performed to confirm the prognostic value of ZIC family members and explore their mutations and relationship with immune cell infiltration. Via the CCLE, GEPIA2, HPA, cBioPortal and TIMER databases, ZIC1, and ZIC5 were identified as prognostic factors in LGG patients, and ZIC3 was identified as prognostic factors in GBM patients, and these results were replicated by univariate and multivariate Cox regression. PPI networks were constructed to analyze the correlations and shared functions between the ZICs and related proteins. Our results identified ZIC1/3/5 as novel biomarkers and potential therapeutic targets for glioma.
Data availability
The datasets generated and analyzed during this study are available.
Abbreviations
- GBM:
-
Glioblastoma multiforme
- LGG:
-
Brain low-grade glioma
- CNS:
-
Central nervous system
- OS:
-
Overall survival
- PPI:
-
Protein–protein network
- GO:
-
Gene ontology
- KEGG:
-
Kyoto encyclopedia of genes and genomes
- ZIC:
-
Zinc-finger of the cerebellum
References
Lapointe, S., Perry, A. & Butowski, N. A. Primary brain tumours in adults. Lancet 392, 432–446. https://doi.org/10.1016/S0140-6736(18)30990-5 (2018).
Long, S., Li, M., Liu, J., Yang, Y. & Li, G. Identification of immunologic subtype and prognosis of GBM based on TNFSF14 and immune checkpoint gene expression profiling. Aging (Albany NY) 12, 7112–7128. https://doi.org/10.18632/aging.103065 (2020).
Batash, R., Asna, N., Schaffer, P., Francis, N. & Schaffer, M. Glioblastoma multiforme, diagnosis and treatment; recent literature review. Curr. Med. Chem. 24, 3002–3009. https://doi.org/10.2174/0929867324666170516123206 (2017).
Carlsson, S. K., Brothers, S. P. & Wahlestedt, C. Emerging treatment strategies for glioblastoma multiforme. EMBO Mol. Med. 6, 1359–1370. https://doi.org/10.15252/emmm.201302627 (2014).
Razavi, S. M. et al. Immune evasion strategies of glioblastoma. Front. Surg. 3, 11. https://doi.org/10.3389/fsurg.2016.00011 (2016).
Chen, H. et al. SOX9-activated PXN-AS1 promotes the tumorigenesis of glioblastoma by EZH2-mediated methylation of DKK1. J. Cell Mol. Med. https://doi.org/10.1111/jcmm.15189 (2020).
Gao, X. et al. lncTCF7 is a negative prognostic factor, and knockdown of lncTCF7 inhibits migration, proliferation and tumorigenicity in glioma. Sci. Rep. 7, 17456. https://doi.org/10.1038/s41598-017-17340-y (2017).
Galardi, S., Michienzi, A. & Ciafre, S. A. Insights into the regulatory role of m(6)A epitranscriptome in glioblastoma. Int. J. Mol. Sci. 21, E2816. https://doi.org/10.3390/ijms21082816 (2020).
Jiang, K. et al. MOB2 suppresses GBM cell migration and invasion via regulation of FAK/Akt and cAMP/PKA signaling. Cell Death Dis. 11, 230. https://doi.org/10.1038/s41419-020-2381-8 (2020).
Satow, R. et al. ZIC5 drives melanoma aggressiveness by PDGFD-mediated activation of FAK and STAT3. Cancer Res. 77, 366–377. https://doi.org/10.1158/0008-5472.CAN-16-0991 (2017).
Yokota, N. et al. Predominant expression of human zic in cerebellar granule cell lineage and medulloblastoma. Cancer Res. 56, 377–383 (1996).
Ghandi, M. et al. Next-generation characterization of the cancer cell line encyclopedia. Nature 569, 503–508. https://doi.org/10.1038/s41586-019-1186-3 (2019).
Tang, Z., Kang, B., Li, C., Chen, T. & Zhang, Z. GEPIA2: An enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 47, W556-560. https://doi.org/10.1093/nar/gkz430 (2019).
Asplund, A., Edqvist, P. H., Schwenk, J. M. & Ponten, F. Antibodies for profiling the human proteome-the human protein atlas as a resource for cancer research. Proteomics 12, 2067–2077. https://doi.org/10.1002/pmic.201100504 (2012).
Anaya, J. OncoLnc: Linking TCGA survival data to mRNAs, miRNAs, and lncRNAs. PeerJ Comput. Sci. 2, e67. https://doi.org/10.7717/peerj-cs.67 (2016).
Gao, J. et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal 6, pl1. https://doi.org/10.1126/scisignal.2004088 (2013).
Li, T. et al. TIMER: A web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res. 77, e108-110. https://doi.org/10.1158/0008-5472.CAN-17-0307 (2017).
Li, B. et al. Comprehensive analyzes of tumor immunity: Implications for cancer immunotherapy. Genome Biol. 17, 174. https://doi.org/10.1186/s13059-016-1028-7 (2016).
Zhou, Y. et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 10, 1523. https://doi.org/10.1038/s41467-019-09234-6 (2019).
Kanehisa, M. Toward understanding the origin and evolution of cellular organisms. Protein Sci. Publ. Protein Soc. 28, 1947–1951. https://doi.org/10.1002/pro.3715 (2019).
Kanehisa, M., Furumichi, M., Sato, Y., Ishiguro-Watanabe, M. & Tanabe, M. KEGG: Integrating viruses and cellular organisms. Nucleic Acids Res. 49, D545–D551. https://doi.org/10.1093/nar/gkaa970 (2020).
Kanehisa, M. & Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30. https://doi.org/10.1093/nar/28.1.27 (2000).
Zar, J. H. Biostatistical Analysis. 4th edn, (Prentice Hall, 1999).
Hochberg, Y. & Benjamini, Y. More powerful procedures for multiple significance testing. Stat. Med. 9, 811–818. https://doi.org/10.1002/sim.4780090710 (1990).
Cohen, J. A coefficient of agreement for nominal scales. Educ. Psychol. Meas. 20, 37–46 (1960).
Shannon, P. et al. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504. https://doi.org/10.1101/gr.1239303 (2003).
Szklarczyk, D. et al. STRING v11: Protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 47, D607-613. https://doi.org/10.1093/nar/gky1131 (2019).
Aran, D., Sirota, M. & Butte, A. J. Systematic pan-cancer analysis of tumour purity. Nat. Commun. 6, 8971. https://doi.org/10.1038/ncomms9971 (2015).
Aran, D., Sirota, M. & Butte, A. J. Corrigendum: Systematic pan-cancer analysis of tumour purity. Nat. Commun. 7, 10707. https://doi.org/10.1038/ncomms10707 (2016).
Ge, Q. et al. Zic1 suppresses gastric cancer metastasis by regulating Wnt/beta-catenin signaling and epithelial–mesenchymal transition. FASEB J. 34, 2161–2172. https://doi.org/10.1096/fj.201901372RR (2020).
Kandimalla, R. et al. Genome-wide analysis of CpG island methylation in bladder cancer identified TBX2, TBX3, GATA2, and ZIC4 as pTa-specific prognostic markers. Eur. Urol. 61, 1245–1256. https://doi.org/10.1016/j.eururo.2012.01.011 (2012).
Wang, Z. Y., Duan, Y. & Wang, P. SP1-mediated upregulation of lncRNA SNHG4 functions as a ceRNA for miR-377 to facilitate prostate cancer progression through regulation of ZIC5. J. Cell Physiol. 235, 3916–3927. https://doi.org/10.1002/jcp.29285 (2020).
Fu, J. Q. et al. A single factor induces neuronal differentiation to suppress glioma cell growth. CNS Neurosci. Ther. 25, 486–495. https://doi.org/10.1111/cns.13066 (2019).
Tyrinova, T. et al. Defective regulation of membrane TNFalpha expression in dendritic cells of glioblastoma patients leads to the impairment of cytotoxic activity against autologous tumor cells. Int. J. Mol. Sci. 21, E2898. https://doi.org/10.3390/ijms21082898 (2020).
Ostuni, R., Kratochvill, F., Murray, P. J. & Natoli, G. Macrophages and cancer: From mechanisms to therapeutic implications. Trends Immunol. 36, 229–239. https://doi.org/10.1016/j.it.2015.02.004 (2015).
Shan, X. et al. Prognostic value of a nine-gene signature in glioma patients based on tumor-associated macrophages expression profiling. Clin. Immunol. 216, 108430. https://doi.org/10.1016/j.clim.2020.108430 (2020).
Lu, L. et al. The nanoparticle-facilitated autophagy inhibition of cancer stem cells for improved chemotherapeutic effects on glioblastomas. J. Mater. Chem. B 7, 2054–2062. https://doi.org/10.1039/c8tb03165g (2019).
Bataller, L., Wade, D. F., Fuller, G. N., Rosenfeld, M. R. & Dalmau, J. Cerebellar degeneration and autoimmunity to zinc-finger proteins of the cerebellum. Neurology 59, 1985–1987. https://doi.org/10.1212/01.wnl.0000038352.01415.ce (2002).
Zhaocheng Han, J. J., Lv, Y., Wang, R., Cao, K. Transcriptional expression of ZICs as an independent indicator of survival in gliomas. https://doi.org/10.21203/rs.3.rs-28764/v1 (2020).
Acknowledgements
This manuscript has been released as a pre-print at Research.Square (Zhao cheng Han et al.)39. We thank “Nature Research Editing Service” for its language editing of the manuscript. We truly appreciate all the medical workers fighting against the COVID-19 pandemic. Zhaocheng Han wants to thank the support, care and encouragement from Jingnan Jia, and he wants to tell her, “I love you three thousand”.
Funding
Funding from National Major Scientific and Technological Special Project for “Significant New Drugs Development” (No. 2017ZX09304019).
Author information
Authors and Affiliations
Contributions
Z.H. and J.J. designed and performed the analysis, and these authors contributed equally to this work. Z.H. drafted the manuscript. Y.L. and R.W. checked the data and analysis. K.C. reviewed and confirmed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Han, Z., Jia, J., Lv, Y. et al. Transcriptional expression of ZICs as an independent indicator of survival in gliomas. Sci Rep 11, 17532 (2021). https://doi.org/10.1038/s41598-021-93877-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-93877-3
- Springer Nature Limited
This article is cited by
-
AR-regulated ZIC5 contributes to the aggressiveness of prostate cancer
Cell Death Discovery (2022)