Abstract
Well-defined large-volume polysomnographic (PSG) data can identify subgroups and predict outcomes of obstructive sleep apnea (OSA). However, current PSG data are scattered across numerous sleep laboratories and have different formats in the electronic health record (EHR). Hence, this study aimed to convert EHR PSG into a standardized data format—the Observational Medical Outcome Partnership (OMOP) common data model (CDM). We extracted the PSG data of a university hospital for the period from 2004 to 2019. We designed and implemented an extract–transform–load (ETL) process to transform PSG data into the OMOP CDM format and verified the data quality through expert evaluation. We converted the data of 11,797 sleep studies into CDM and added 632,841 measurements and 9,535 observations to the existing CDM database. Among 86 PSG parameters, 20 were mapped to CDM standard vocabulary and 66 could not be mapped; thus, new custom standard concepts were created. We validated the conversion and usefulness of PSG data through patient-level prediction analyses for the CDM data. We believe that this study represents the first CDM conversion of PSG. In the future, CDM transformation will enable network research in sleep medicine and will contribute to presenting more relevant clinical evidence.
Similar content being viewed by others
Introduction
Obstructive sleep apnea (OSA) is an significant risk factor in several major health conditions, such as cardiovascular1,2,3,4, neurovascular5,6, and metabolic diseases7,8. OSA is diagnosed on the basis of certain crucial parameters, including the apnea–hypopnea index (AHI) of polysomnography (PSG). PSG is a standard diagnostic sleep test for OSA, and its results hold significant clinical implications for various major diseases. For example, severe OSA with AHI > 30 is known to be correlated with the development of strokes and incident hypertension5,9. Although large-scale prospective cohort studies can be used to empirically prove such important clinical observations, they suffer from the limitations of long follow-up periods and high costs. On the other hand, retrospective studies can only establish statistical associations between the risks of major conditions and PSG results rather than a definitive causal relationship. Furthermore, PSG is a whole-night test, and the capacity for PSG tests per sleep center is thus limited. However, multi-center collaborative studies can be used to conduct more PSGs, and well-defined large-volume PSG databases have the potential to corroborate the validity of conjectured correlations. The analysis of a wide range of electronic health record (EHR) data, including medical conditions, drug exposures, procedures, and measurements, in conjunction with PSG data, and their rapid verification across multiple institutions may enable the procurement of crucial pieces of robust scientific evidence through enhancements in analytic power.
However, as the primary goal of EHR is medical application, rather than research, the reuse of EHR data for academic purposes necessitates the mapping of clinical observations to standard vocabularies10. To this end, Observational Health Data Sciences and Informatics (OHDSI), an international collaborative initiative, has created and applied an open-source standard data format and analytic solutions to diverse health and medical databases across the world11. The Observational Medical Outcome Partnership's (OMOP) common data model (CDM), which is utilized by OHDSI as a standard data format, serves as a guide for the standardization of heterogeneous representations of healthcare data obtained from disparate sources. Conversion of health and medical databases into the CDM format is expected to enable interdisciplinary collaborative large-scale analyses. Such large-scale analyses using open-source analytic tools based on standardized datasets are, in turn, expected to improve the speed and efficiency of population-level estimation and patient-level prediction, thereby enhancing the reliability of clinical decision-making11,12.
To the best of our knowledge, measurements obtained via PSG are yet to be transformed into the CDM format. Linking the diverse data obtained from PSG with the extensive EHR database in a structured CDM format is expected to facilitate multi-center studies and strengthen general analytic power. In this study, we aimed to convert EHR PSG data into the standardized OMOP CDM data format and conduct a pilot feasibility test. Through a pilot feasibility study, we attempted to confirm the possibility of developing a predictive model using existing CDM data and additional PSG data, and to verify the usefulness of the integrated data.
Methods
Study population for CDM conversion
This study included patients who visited the Sleep Center at Seoul National University Bundang Hospital (SNUBH), located in the metropolitan area of Seoul in South Korea, and had undergone PSG between February 2004 and June 2019.
Data source
OMOP CDM data obtained from SNUBH were used in this study. In particular, the data comprised de-identified EHR data based on OMOP CDM version 5.3.1 and accumulated over a period of 16 years—from the opening of SNUBH with the full EHR system in May 2003, till June 2019. The EHR data of more than 2 million patients, including patient demographics, diagnosis, chief complaints, drug exposures, test orders/results, vital signs, surgeries, family histories, and past medical histories, were converted to CDM.
This study was performed in accordance with the relevant guidelines and regulations of the SNUBH Institutional Review Board (IRB) and was approved by the SNUBH IRB. As it is an observational study and the data source was de-identified, this study was approved based on waivers of informed consent or exemptions by the SNUBH IRB (IRB No: X-2002–592-904).
Polysomnographic parameters
We considered all PSGs performed at the Sleep Center of SNUBH as target data to be converted into OMOP CDM, including full-night PSGs, split-night PSGs, PSGs for continuous positive airway pressure (CPAP) titration, and multiple sleep latency tests (MSLTs). In the case of split-night PSGs, the values of the parameters represented only the diagnostic portions in this study. No home sleep apnea tests were included because they are not popular in South Korea. The PSG parameters to be transformed into OMOP CDM included information related to sleep architecture, respiratory activity, positions during sleep, blood oxygen saturation, and limb movement.
We conducted PSGs using an Embla N 7000 (Embla, Reykjavik, Iceland) recording system equipped with standard electrodes and sensors, in the presence of a sleep technician. The entire PSG retinue consisted of electroencephalography, electrooculography, echocardiography, submental and limb electromyography, chest and abdominal plethysmography, nasal pressure manometry, oronasal thermistor, pulse oximetry, and a snoring sensor. Apnea was defined as a pause in the respiratory airflow lasting at least 10 s, and hypopnea was defined as a reduction in the airflow by 50% or more lasting at least 10 s, or the accompaniment of airflow reduction by arousal or an oxygen desaturation by 4% or more13. The PSG data were reviewed and scored by sleep experts using the Embla RemLogic PSG Software (Embla, ON, Canada). The study report from the Embla RemLogic PSG Software has the following parameter (variable) categories: patient information; sleep summary; summary graph; sleep information; arousal statistics; autonomic arousal (plethysmogram) statistics; apnea/hypopnea statistics; apnea-desaturation relation; Cheyne Stokes breathing statistics; breath statistics; snoring statistics; flattening statistics; respiratory mechanic instability statistics; SpO2 statistics; desaturation statistics; heart rate statistics; cardiac events; bruxism; rapid eye movement sleep behavior disorder information; rhythmic movement disorder information; periodic limb movement statistics; and position statistics. Among them, the sleep experts at our sleep center selected the PSG parameters that are commonly employed in the literature to make available in the PSG summary report of our EHR. The selected parameters were automatically exported and imported into our EHR in a structured format.
Strategy to convert PSG data into OMOP CDM
We designed and implemented the following extract–transform–load (ETL) process to transform the PSG data into the OMOP CDM format.
Despite being reported in a structured form, the EHR PSG results considered in this study had been revised approximately 11 times. Hence, we extracted the data corresponding to each revised form and integrated them within the CDM format via standardization. The procedural information for PSG order itself had already been converted into the CDM format. Thus, in this study, we linked the extracted PSG results and the corresponding existing orders in the CDM to connect the PSG procedures with their corresponding results.
The PSG parameters were manually mapped by sleep domain experts (J.-W. Kim and S.-W- Cho) to standard concepts within the Logical Observation Identifiers Names and Codes (LOINC) or Systematized Nomenclature of Medicine–Clinical Terms (SNOMED CT) vocabularies corresponding to the measurement and observation domains. Non-mapped parameters were added to the concept, concept_ancestor, and concept_relationship tables to be used as new custom standard concepts (please see Supplementary Table S1 for the concept mapping information in the case of PSG and Supplementary Table S2 for the concept definitions). More than 2 billion digits were assigned to the concept_id of the new custom concepts. In the concept_ancestor table, the newly added concepts served as their own ancestors and descendants. In the concept_relationship table, the mapping information between source and standard concepts was added. Additionally, we described the bidirectional relationship between PSG and its parameters in the table using the concepts of ‘Panel contains’ and ‘Contained in panel (LOINC)’ relationships.
The extracted PSG data were transformed and loaded into measurement and observation tables with standard concepts. Observation data were linked to the corresponding PSG procedures via the observation_event_id and obs_event_field_concept_id fields. In order to link measurements with corresponding procedures, we used the new modifier_of_event_id and modifier_of_field_concept_id fields that have been proposed by the OHDSI Oncology Working Group14. The procedure_occurrence, measurement, and observation tables were linked to the person and visit_occurrence tables based on their foreign keys. The CDM tables associated with the PSG data are depicted in Fig. 1.
After completing the ETL, we assessed the PSG data quality via exploratory data analysis and developed data quality check rules for data cleaning (please see Supplementary Table S3 for the detailed cleaning rules and the number of records filtered by the rules). Finally, the cleaned PSG data integrated into the existing CDM were utilized for a feasibility test.
Pilot feasibility test using open-source OHDSI analytic tools
We conducted a pilot feasibility test using only full-night PSG tests of patients 18 years or older. The feasibility test was designed to develop and validate a model to predict cardio-neuro-metabolic disease within a target population between a period of 1 day and 1095 days from the target cohort start date of the PSG test. A cardio-neuro-metabolic disease was defined as any condition involving International Classification of Disease, Tenth Revision (ICD-10) codes corresponding to the comorbidities listed in Supplementary Table S4. We included any occurrence of the defied ICD-10 codes without constraints on the frequency.
In the population setting for the patient-level prediction, varying minimum lookback periods of 30 days, 90 days, and 180 days were utilized for the prior observation periods of patients from the target population. Subjects without time-at-risk of 1094 days were also removed. Patients who had experienced prior outcomes were also not considered in this study.
Among the preexisting CDM data, we utilized multiple covariates, such as gender, 5-year age group, Anatomical Therapeutic Chemical (ATC) drug group, SNOMED CT condition group, procedure, measurement value, observation, visit concept count, the CHA2DS2-VASc (congestive heart failure, arterial hypertension, age > 75 years, diabetes mellitus, stroke/transient ischemic attack, vascular disease, age 65–74 years, sex category) score, diabetes complications severity index (DCSI), and the Charlson comorbidity score. Two different covariate settings were tested to determine which PSG parameters could be selected during the cardio-neuro-metabolic disease prediction. One setting (PSG-only covariates) used only gender, age group, and PSG parameters, and the other (all covariates) used all CDM covariates, including the PSG parameters described above as covariates. The observation time windows of the covariates for short, medium, and long terms were set as prior 7 days, 30 days, and 180 days before the cohort start date, respectively.
Three different machine learning models—Lasso Logistic Regression (Lasso), Gradient Boosting Machine (GBM), and Random Forest (RF)—were developed using 25% of the total data for training and 75% for testing. Hyper-parameter training was performed using five-fold cross-validation on the training set. PatientLevelPrediction R package15 version 4.0.5 was used for this purpose.
To evaluate the models, model discrimination was assessed using the area under the receiver operating characteristic curve (AUC).
Results
Conversion results of PSG parameters into OMOP CDM concepts
We converted data from a total of 11,392 tests corresponding to 11,797 sleep studies into the OMOP CDM format. These included 7,191 full-night PSGs, 2,725 split-night PSGs, 1,474 CPAP titration PSGs, and 407 MSLTs. Among the PSG test results stored in EHR, the conversion target parameters converted into CDM are presented in Table 1. These included 7 pertaining to body measurements, 7 to sleep summaries, 6 to sleep stages, 16 to respiratory events, 4 to apnea or hypopnea duration, 8 to sleep position, 5 to arousals, 2 to limb movement, 5 to snoring, 8 to oxygen statistics, 1 to continuous positive airway pressure, 2 to questionnaires, 11 to MSLT, 1 to apnea level manometry test, and 3 to Friedman staging. A total of 85 PSG parameter concepts were converted to the measurement domain and one to the observation domain (Waist/hip ratio). Moreover, 20 (23.3%) PSG codes were mapped to the standard OHDSI vocabulary including LOINC and SNOMED CT, but the remaining 66 (76.7%) could not be mapped and were added as new custom standard concepts.
Characteristics of PSG data
The overall characteristics of the total sleep studies that were converted into OMOP CDM are presented in Table 2. Out of an aggregate of 11,392 sleep tests, 8363 (73.4%) tests were conducted on male patients and 3029 (26.6%) on female patients. There was an average of 1.2 tests per person. Tests of patients aged 40–49 years, 50–59 years, and 60–69 years accounted for approximately 65% of the total number of tests. The number of sleep studies conducted each year exhibited a progressive increment. The prevalence of AHI < 5, mild OSA (5 ≤ AHI < 15), moderate OSA (15 ≤ AHI < 30) and severe OSA (30 ≤ AHI) was 28.5%, 23.8%, 19.3% and 28.4%, respectively. The basic statistics of the associated PSG parameters are provided in Supplementary Table S5.
Performance of the prediction models
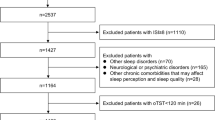
Corresponding to the best performance setting of each prediction models, the number of people eligible for inclusion into the target population, the outcome count, and the number of people lost due to each inclusion step are illustrated in Fig. 2. The target population comprising 5581 full-night PSG tests of patients 18 years or older was reduced to a population comprising 2555 tests of 2542 patients. The outcome rate of cardio-neuro-metabolic disease was observed to be 11.1%.
The AUC performance of the prediction feasibility test based on CDM data achieved an 0.751(0.693–0.810) for the RF model with all covariates. The performance results corresponding to each set of configurations are listed in Table 3. All three models—RF, GBM, and Lasso—performed better when all parameters, such as condition, drug, measurement, and comorbidity score, were utilized as CDM data along with PSG, rather than only the PSG parameters.
The top 20 covariates selected from the RF are presented in Table 4. Among them, 11 were PSG parameters, for example, AHI during right lateral (/h), central apnea index (/h), waking oxygen saturation (%), and snoring time (min). The top 20 covariates selected from the other models are included in Supplementary Table S6.
Discussion
To the best of our knowledge, this study represents the first attempt to convert EHR PSG data into ODHSI OMOP CDM, a standard format for health and medical data. Through this study, we successfully converted more than 11,000 PSGs stored in a tertiary hospital EHR into the OMOP CDM version 5.3.1 format. However, we were able to map only approximately 23% of the 86 parameters present within the PSG data to the existing OMOP CDM standard vocabulary, and new custom standard concept names had to be created for the remaining 77% of the parameters. The method used to create the new custom standard concept can be employed when other sites add non-mapping PSG parameters that are not reported in this study.
The most significant advantage of the standardization of EHR data into the CDM format is the speed and efficiency of large-scale analysis afforded to researchers and clinicians using the open-source analysis tools provided by ODHSI10,12. Furthermore, due to the inapplicability of OMOP CDM to PSG parameters till date, CDM studies using PSG and MSLT test results, which are the most important tests in sleep medicine, are yet to be conducted. In this context, conversion of PSG results into the CDM format also enables utilization of OHDSI's open-source analytical solutions in clinical studies involving PSG results. In addition, the OMOP CDM format has already been used to standardize a comprehensive collection of EHR data, including diagnostic information, specimen test results, imaging test information, procedure and intervention information, drug exposures, past medical histories, and family histories. Therefore, the standardization procedure attempted in this study enables researchers to conduct robust and scalable analyses involving PSG results in conjunction with pre-CDM-converted large-scale EHR data. Collaborative research across a growing number of sites participating in the standardized CDM network is expected to lead to higher performance in population-level estimation and patient-level prediction models that leverage sleep study parameters.
In this study, the performance of the pilot feasibility test in terms of patient-level prediction for cardio-neuro-metabolic disease exhibited a significant improvement when the entire EHR data along with PSG was used, rather than solely the PSG data. This suggests the feasibility of utilizing all EHR data in the OMOP CDM format via CDM conversion of PSG data.
OSA is a broad-spectrum disease with several different subgroups or phenotypes, and each OSA phenotype is likely to be manifested with different levels of severity, both clinically and objectively16. Previous one-size-fits-all approaches based on apnea–hypopnea index suffered from insufficient consideration of these diverse phenotypic subtypes of OSA due to the imperfection of the apnea–hypopnea index as a diagnostic metric with respect to OSA-related symptoms and outcomes17. Several studies have demonstrated that each OSA phenotype exhibits different characteristics and varying risks of disease outcomes16,18. The most important data included in these studies were various metrics of PSG, including all the PSG results, which enabled the classification of OSA into various phenotypes via the phenotyping technique. One study that attempted a structured, data-driven approach based on multiple PSG features of approximately 2,000 OSA patients was able to identify seven subgroups (phenotypes). The aforementioned study also revealed that a unique phenotype that may have been missed during conventional OSA severity classification based on a single metric—apnea hypopnea index—could account for the risk of cardiovascular outcome more effectively19. In our previous study, we also identified four clusters based on various PSG features and there was a significant difference in disease outcome among the clusters, and such a difference could not be found in the standard classification of OSA based only on AHI severity20. Moreover, these characteristic phenotypes may exhibit different patterns depending on race, country, or individual. Therefore, to improve the ability to predict adverse OSA outcomes for a population or an individual, simply having a large number of PSGs is not sufficient—it is necessary to acquire PSGs across various data sources. Therefore, it is advantageous to use standardized data such as OMOP CDM to increase the reproducibility and statistical significance of the analyses. The conversion of data into the OMOP CDM format enables ATLAS, OHDSI's open-source analytic solution, to generate queries that can set the aforementioned OSA phenotypes as target cohorts and queries that can set OSA complications to be predicted as the outcome cohort. This enables verification of the reproducibility of outcome predictions of OSA phenotyping through analysis of the dataset including PSG with the same queries in multiple sleep centers where PSG-CDM standardization has been completed. In addition to the analysis of large-scale PSG data, the clinical relevance of the OSA phenotypes across various populations by region and race will be able to be also verified.
With the increase in CDM conversion of EHR data across medical institutions, research based on CDM-format datasets is expected to be pursued in various fields. However, unlike the CDM conversion of data such as clinical diagnosis results, laboratory sample test results, and drug exposure data, the CDM conversion of medical data based on patient-generated signals, including PSG, is still insufficient. Therefore, till date, CDM-based research has been actively conducted in fields where conversion to the pre-existing standard vocabulary is feasible. Domains where CDM research is most active include pharmacovigilance21,22,23 and pharmacoepidemiology24. For example, a study assessing anti-seizure drug-related adverse reactions in 1344 target epilepsy cohorts determined that the detection rate of the adverse drug reaction based on CDM-format data was comparable to previously published results obtained using traditional data analysis techniques21. In addition, it is possible to implement various designs of research by constructing a target cohort corresponding to a study entry population and an outcome cohort corresponding to a disease outcome population25,26. Examples include a prognostic model validation study predicting hemorrhagic transformation of acute ischemic stroke within a CDM dataset of more than 600,000 patients via the OHDSI international network25, and a survival analysis study using 115 variables in 346 patients diagnosed with intrahepatic cholangiocarcinoma26.
Despite the significant implications, the present study has certain limitations. First, the rate of correspondence between ODHSI's standard OMOP CDM concepts and PSG parameters was as low as approximately 20%. This can be attributed to the fact that the pre-existing OMOP CDM standard vocabulary does not reflect all of the approximately 80 PSG variables considered in this study. The custom standard vocabulary developed to address this limitation in this study is expected to contribute to future studies that utilize PSG parameters in CDM-based EHR studies. When creating the custom concepts, we made it easy to find all PSG parameters by defining the relationship to the PSG order. For concepts that may have varying definitions, the definition of the concept is provided as metadata. For concepts (e.g., %Time of saturation < 60%, %Time of saturation < 70%) in which multiple criteria can exist, a concept was created in a way that has individual concept_ids. Since the MEASUREMENT table does not have a modifier attribute, it would be the best practice to create individual concepts for them. By doing this, the meaning of new concepts can be clarified. As the basic PSG parameters of the PSG recording systems of the various vendors are similar, we think other institutions will also be able to apply the new concept proposed in this study. In addition, we look forward to adding the new concepts to OHDSI's standard vocabulary. Second, in South Korea, insurance for CPAP began in July 2018; before then, it had been recorded in a different form of EHR rather than an order. Thus, in this study, only CPAP orders after July 2018 were converted to CDM and can be used as predictors for the pilot prediction models. There could be an issue where information on orders for CPAP, which may be an important variable in predicting cardio-neuro-metabolic disease, is not complete. However, as the purpose of this study was only to demonstrate the pilot feasibility of the prediction model using CDM including PSG data, predictors should be considered more elaborately when developing a prediction model in the future. Third, different sleep centers represent PSG databases in EHRs in different ways. Many centers store PSG results in EHR as an image file, or simply record OSA severity in a report format. Therefore, significant implementation effort and time is required to extract, transform, and load the PSG results into the CDM format. Furthermore, different levels of digitization of PSG data in different hospitals may cause concerns regarding the different levels of CDM conversion from PSG parameters. However, with the increase in CDM studies including PSG parameters, the electronic representation of PSG data in the EHR system is expected to be facilitated across hospitals. Finally, conversion of data into the CDM format is time-consuming, requiring a substantial amount of resources, in addition to the fundamental requirement of collecting native source data. The need to code subsets of data manually may limit conversion efforts. However, once the native data are converted to the CDM format, EHR systems in the network will be able to use the same queries to identify cohorts. Thus, conversion to CDM is expected to minimize the effort required to develop cohorts and analyze results across multiple sites.
The harmonization across different sites requires collaborative efforts from multidisciplinary experts, including clinical domain experts, terminology experts, and engineers from various sites. When other sites try to map their own PSG data, efforts should be made to use and propose the same vocabulary and the same concept as much as possible by using the mapping result proposed in this study or by participating in the OHDSI community. As the standard terminology for PSG data has not yet been established internationally, if a specific ontology for sleep study can be proposed as OHDSI vocabulary by reviewing previous efforts, such as the Sleep Domain Ontology and the National Sleep Research Resource, it is expected to be helpful in the conversion and expansion of CDM by other sites.
Conclusions
The Observational Medical Outcomes Partnership (OMOP) Common Data Model (CDM) is a standard data format and has been applied to various EHR databases. However, its application to PSG data has not been attempted till date. To the best of our knowledge, this study represents the first attempt to transform PSG data into the OMOP CDM format. Well-defined large-volume OMOP CDM databases of PSG data can potentially enable the identification of clinically relevant OSA phenotypes, estimation of disease outcomes at the population level and prediction of outcomes at the patient-level. We expect the CDM mapping and CDM custom vocabulary of the PSG proposed in this study to contribute to the CDM conversion of PSG databases and future studies leveraging such databases.
Data availability
CDM data are designed to support a distributed research network. Thus, access to the data is restricted on internal private networks. Therefore, the data are not publicly available.
References
Tietjens, J. R. et al. Obstructive sleep Apnea in cardiovascular disease: a review of the literature and proposed multidisciplinary clinical management strategy. J. Am. Heart Assoc. 8, e010440. https://doi.org/10.1161/jaha.118.010440 (2019).
Strausz, S. et al. Obstructive sleep apnoea and the risk for coronary heart disease and type 2 diabetes: a longitudinal population-based study in Finland. BMJ Open 8, e022752. https://doi.org/10.1136/bmjopen-2018-022752 (2018).
Cadby, G. et al. Severity of OSA is an independent predictor of incident atrial fibrillation hospitalization in a large sleep-clinic cohort. Chest 148, 945–952. https://doi.org/10.1378/chest.15-0229 (2015).
Joyeux-Faure, M. et al. Contribution of obstructive sleep apnoea to arterial stiffness: a meta-analysis using individual patient data. Thorax 73, 1146–1151. https://doi.org/10.1136/thoraxjnl-2018-211513 (2018).
Catalan-Serra, P. et al. Increased incidence of stroke, but not coronary heart disease, in elderly patients with sleep Apnea. Stroke 50, 491–494. https://doi.org/10.1161/strokeaha.118.023353 (2019).
Castello-Branco, R. C. et al. Association between risk of obstructive sleep Apnea and cerebrovascular reactivity in stroke patients. J. Am. Heart Assoc. 9, e015313. https://doi.org/10.1161/jaha.119.015313 (2020).
Subramanian, A. et al. Risk of incident obstructive sleep Apnea among patients with type 2 diabetes. Diabetes Care 42, 954–963. https://doi.org/10.2337/dc18-2004 (2019).
Anothaisintawee, T., Reutrakul, S., Van Cauter, E. & Thakkinstian, A. Sleep disturbances compared to traditional risk factors for diabetes development: Systematic review and meta-analysis. Sleep Med. Rev. 30, 11–24. https://doi.org/10.1016/j.smrv.2015.10.002 (2016).
Guillot, M. et al. Association between severe obstructive sleep apnea and incident arterial hypertension in the older people population. Sleep Med. 14, 838–842. https://doi.org/10.1016/j.sleep.2013.05.002 (2013).
Mendonca, E. & Tachinardi, U. Personalized and Precision Medicine Informatics 199–213 (Springer, Berlin, 2020).
Hripcsak, G. et al. Observational health data sciences and informatics (OHDSI): opportunities for observational researchers. Stud. Health Technol. Inf. 216, 574–578 (2015).
FitzHenry, F. et al. Creating a common data model for comparative effectiveness with the observational medical outcomes partnership. Appl. Clin. Inf. 6, 536–547. https://doi.org/10.4338/aci-2014-12-cr-0121 (2015).
The Report of an American Academy of Sleep Medicine Task Force. Sleep-related breathing disorders in adults: recommendations for syndrome definition and measurement techniques in clinical research. Sleep 22, 667–689 (1999).
Group, O. O. W. OHDSI Oncology Working Group, https://www.ohdsi.org/web/wiki/doku.php?id=projects:workgroups:oncology-sg&s%5B%5D=oncology
JM, R., MJ, S., MA, S., PB, R. & PR, R. OHDSI Patient Level Prediction Package, https://github.com/OHDSI/PatientLevelPrediction
Zinchuk, A. & Yaggi, H. K. Phenotypic subtypes of OSA: a challenge and opportunity for precision medicine. Chest 157, 403–420. https://doi.org/10.1016/j.chest.2019.09.002 (2020).
Malhotra, A., Orr, J. E. & Owens, R. L. On the cutting edge of obstructive sleep apnoea: where next?. Lancet Respir. Med. 3, 397–403. https://doi.org/10.1016/s2213-2600(15)00051-x (2015).
Park, S. et al. Polysomnographic phenotype as a risk factor for cardiovascular diseases in patients with obstructive sleep apnea syndrome: a retrospective cohort study. J. Thorac. Dis. 12, 907–915. https://doi.org/10.21037/jtd.2019.12.66 (2020).
Zinchuk, A. V. et al. Polysomnographic phenotypes and their cardiovascular implications in obstructive sleep apnoea. Thorax 73, 472–480. https://doi.org/10.1136/thoraxjnl-2017-210431 (2018).
Kim, J.-W. et al. Polysomnographic phenotyping of obstructive sleep apnea and its implications in mortality in Korea. Sci. Rep. 10, 1–12 (2020).
Choi, S. A. et al. Analysis of antiseizure drug-related adverse reactions from the electronic health record using the common data model. Epilepsia 61, 610–616 (2020).
Seo, S. I. et al. Comparative risk of Clostridium difficile infection between proton pump inhibitors and histamine-2 receptor antagonists: a 15-year hospital cohort study using a common data model. J. Gastroenterol. Hepatol. https://doi.org/10.1111/jgh.14983 (2020).
Choi, Y. I. et al. Effect of age on the initiation of biologic agent therapy in patients with inflammatory bowel disease: Korean common data model cohort study. JMIR Med. Inform. 8, e15124. https://doi.org/10.2196/15124 (2020).
Lai, E. C. et al. Applying a common data model to Asian databases for multinational pharmacoepidemiologic studies: opportunities and challenges. Clin. Epidemiol. 10, 875–885. https://doi.org/10.2147/CLEP.S149961 (2018).
Wang, Q. et al. Development and validation of a prognostic model predicting symptomatic hemorrhagic transformation in acute ischemic stroke at scale in the OHDSI network. PLoS ONE 15, e0226718. https://doi.org/10.1371/journal.pone.0226718 (2020).
Hong, N. et al. Preliminary exploration of survival analysis using the OHDSI common data model: a case study of intrahepatic cholangiocarcinoma. BMC Med. Inform. Decis. Mak. 18, 116. https://doi.org/10.1186/s12911-018-0686-7 (2018).
Acknowledgements
This research was supported by the Bio and Medical Technology Development Program of the National Research Foundation (NRF) funded by the Korean Government, Ministry of Science and ICT, Sejong (NRF-2015M3A9D7066973) and by Seoul National University Bundang Hospital (SNUBH grant #18-2018-013), Seongnam, South Korea. This work was also supported by the Technology Innovation Program (20003883, Advancing and expanding CDM based distributed biohealth data platform) funded by the Ministry of Trade, Industry & Energy (MOTIE, Korea). The authors thank to Mui Van Zandt and Dmytry Dymshyts at IQVIA for helping with the standard concept mapping.
Author information
Authors and Affiliations
Contributions
K.J.W. and Y.S. designed this study. K.J.W. wrote the main manuscript text. K.S. and Y.S. designed and implemented the CDM conversion process and Y.S., R.B., and S.W. performed the data analyses. K.J.W., Y.S., and L.H. inspected the manuscript critically and took part in the revision of manuscript. All authors have read and approved the final manuscript data.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Kim, JW., Kim, S., Ryu, B. et al. Transforming electronic health record polysomnographic data into the Observational Medical Outcome Partnership's Common Data Model: a pilot feasibility study. Sci Rep 11, 7013 (2021). https://doi.org/10.1038/s41598-021-86564-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-86564-w
- Springer Nature Limited
This article is cited by
-
Conceptual design of a generic data harmonization process for OMOP common data model
BMC Medical Informatics and Decision Making (2024)