Abstract
Although sex determination and differentiation are key developmental processes in animals, the involvement of non-coding RNA in the regulation of this process is still not clarified. The tiger pufferfish (Takifugu rubripes) is one of the most economically important marine cultured species in Asia, but analyses of miRNA and long non-coding RNA (lncRNA) at early sex differentiation stages have not been conducted yet. In our study, high-throughput sequencing technology was used to sequence transcriptome libraries from undifferentiated gonads of T. rubripes. In total, 231 (107 conserved, and 124 novel) miRNAs were obtained, while 2774 (523 conserved, and 2251 novel) lncRNAs were identified. Of these, several miRNAs and lncRNAs were predicted to be the regulators of the expression of sex-related genes (including fru-miR-15b/foxl2, novel-167, novel-318, and novel-538/dmrt1, novel-548/amh, lnc_000338, lnc_000690, lnc_000370, XLOC_021951, and XR_965485.1/gsdf). Analysis of differentially expressed miRNAs and lncRNAs showed that three mature miRNAs up-regulated and five mature miRNAs were down-regulated in male gonads compared to female gonads, while 79 lncRNAs were up-regulated and 51 were down-regulated. These findings could highlight a group of interesting miRNAs and lncRNAs for future studies and may reveal new insights into the function of miRNAs and lncRNAs in sex determination and differentiation.
Similar content being viewed by others
Introduction
Non-coding RNAs (ncRNAs) represent a class of RNA molecules that do not transcribe into protein. It has been demonstrated that as versatile regulators of genome stability, defense against foreign genetic elements, and gene expression, ncRNAs have multiple functions in diverse cellular processes1. Several kinds of ncRNAs exist, and among the ncRNAs, microRNAs (miRNAs) and long ncRNAs (lncRNAs) have been widely studied. MiRNAs, usually 18–26 nucleotides (nt) in length, are a highly conserved, single-stranded class of short small ncRNAs present in plants, animals, and some eukaryotic viruses. Since the discovery of the first miRNAs two decades ago, there are > 250 other species with miRNAs and, to date over 2500 miRNAs have been discovered in humans2. MiRNAs have been indicated to have pervasive roles in the regulation of gene expression and > 60% of human and > 37% of Drosophila protein-coding transcripts are conserved miRNA binding sites, through some degree of selective pressure3,4. MiRNAs function in the form of an effector complex named RNA-induced silencing complex (RISC), along with Argonaute proteins. The interaction between the miRNA-RISC and target mRNAs can mediate gene silencing by multiple inhibitory mechanisms, such as translational effects, mRNA deadenylation, and mRNA degradation. MiRNA-target recognitions usually occurr through seed-pairing, and miRNA recognition elements or miRNA binding sites are positioned mostly at the 3ʹ untranslated region (3ʹ-UTR) of mRNAs. However, it was also suggested that it occurs at 5ʹ-UTRs and/or in coding regions. The miRNA regulation strength can be affected by the perfect base pairing between the target site and miRNA seed region, and the factors that influence the consequence of hybridization, such as the number of miRNA recognition elements of the same mRNA and their relative position5.
LncRNAs are the most heterogeneous class of ncRNAs ranging from 200 to 100,000 nt in length. Abundant evidence showed that lncRNAs could involved in regulating gene expression by a variety of mechanisms such as inhibiting RNA polymerase activity, binding to the promoters of target genes, and degrading target mRNAs6. LncRNAs can also act as targets of miRNAs, repressing the interaction between coding genes and miRNAs, or encoding certain miRNAs as precursors7. Moreover, it has also been found to compete with miRNAs through interactions with protein coding genes, and miRNAs can reduce the stability of lncRNAs8,9,10.
Sex dimorphism is one of the most pervasive and diverse features in the animal kingdom11. MiRNAs and lncRNAs have been demonstrated to play significant roles in sex differentiation, gametogenesis, and sexual reproduction in vertebrates. For example, at the critical time of sex determination of mice, it has been revealed that miR-124 is able to prevent Sox9 expression in ovarian cells, which is the first evidence that an miRNA is involved in the process of sex determination and gonad development12. In chickens, male-biased miR-107 has been shown to mediate the post-transcriptional regulation of estrogen signaling by directly decreasing the expression of nuclear receptor subfamily 5 group A member 1, and its downstream genes, Cyp19a1a13. In teleosts, such as in Atlantic halibut (Hippoglossus hippoglossus)14, rainbow trout (Oncorhynchus mykiss)15,16, yellow catfish (Pylodictis fulvidraco)17, Nile tilapia (Oreochromis niloticus)18,19,20,21, common carp (Cyprinus carpio)22, olive flounder (Paralichthys olivaceus)23, and medaka (Oryzias melastigma)24,25, sexual dimorphic miRNA expression was detected in gonads during sex differentiation and development. Moreover, some miRNAs were predicted to be target genes related to sex determination and differentiation. In the protogynous hermaphroditic fish Epinephelus coioides, cyp19a1a expression, and E2 levels are necessary for sex reversal, furthermore, a positive feedback loop estradiol-17β/miRNA-26a/cyp19a1a involved in the control of E2 production26. In mammals, lncRNA X-inactive specific transcript is required for silencing of one X-chromosome during development in females27. In mice, the lncRNA Dmrt1-related gene has been shown to form a trans-splicing RNA isoform with dmrt1, which disrupts the coding region and replaces the 3′-UTR of dmrt1, resulting in decreased dmrt1 protein level28. In the Chinese soft-shell turtle (Pelodiscus sinensis), numerous lncRNAs were shown to regulate the expression of sox9, dmrt1, sox3, sox8 and cyp19a29.
The tiger pufferfish (Takifugu rubripes) is one of the most valuable commercial fish cultured in Asia. On the market, male fish are more highly valued than females as the mature testes of tiger pufferfish are regarded as a delicacy, and all-male stocks are preferred in aquaculture. Therefore, elucidating the mechanism of sex determination and differentiation is necessary in this species. In 2012, a missense single-nucleotide polymorphism (SNP) in the amhr2 gene has been identified as a likely candidate for the master sex determination polymorphism30. However, sex determination is a complex process, involving a large network of interactions among genes as well as between environment and genes, and the factors involved in gonadal sex differentiation of such an important fish species have yet to be fully elucidated. Screening the sex-related mRNAs and ncRNAs in the undifferentiated gonads at the critical stage of molecular sex determination might provide new information on the role of ncRNAs in gonadal function and help to clarify the regulatory network during early sex differentiation. In 2018, we reported the dimorphic expression patterns of sex-related mRNAs in the undifferentiated gonad of T. rubripes through transcriptome analysis31. However, screening the sexual dimorphic expression of miRNAs and lncRNAs at early sex differentiation stages have not been conducted yet. Thus, the present study aimed to identify sex-biased miRNAs and lncRNAs in the undifferentiated gonads, which might provide a basis for a better understanding of the functions of miRNAs and lncRNAs in regulating the sex differentiation process in T. rubripes.
Results
Characterization of gonadal small RNA of T. rubripes
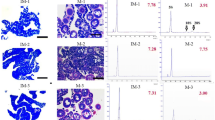
Gonadal sections of 40 days after hatching (dah) T. rubripes larvae confirmed that the characteristic of morphological sex differentiation was not observed in any of the specimens (Fig. 1). Based on the construction and sequencing of small RNA libraries, a total number of 16,653,352 raw reads was generated from female gonads and 19,117,245 from male gonads. After removing low-quality reads, adaptors, and contaminants, 13,151,941 (78.97%) clean reads from female gonads and 16,775,235 (87.75%) from male gonads were obtained. In total, 5,769,544 (57.69%) XX sequences and 9,720,240 (66.55%) XY sequences of the clean reads matched to the T. rubripes genome. Of these matched sequences, 2,193,958 (in XX) and 4,245,186 (in XY) matched miRNAs were found in the Rfam database (see Supplementary File S2). Size distribution analysis of small RNAs showed that 22 nt was the abundant size in both XX and XY gonads followed by 23 and 21 nt (Fig. 2).
A total of 107 known miRNAs and 97 novel miRNAs were expressed in XX fish, while 106 known miRNAs and 112 novel miRNAs were expressed in XY fish. In total, 107 conserved miRNAs and 124 novel miRNAs were identified in the gonads. The most abundant conserved miRNA was fru-miR-100, with 453,080 reads in female gonads and 906, 233 reads in male gonads. In addition, fru-miR-30d, fru-miR-181a-5p, fru-miR-21, fru-miR-22a, fru-miR-92, fru-miR-199, fru-miR-125a, fru-let-7d, fru-miR-25, and fru-miR-200b were found more than 100,000 times in male gonads, and fru-miR-92, fru-miR-30d, fru-miR-22a, fru-miR-21, and fru-miR-181a-5p were found more than 100,000 times in female gonads. Moreover, the expression of novel miRNAs was lower in T. rubripes gonads; novel_1 was the most abundant (52,989 reads in male gonads and 34,118 reads in female gonads (see Supplementary File S3).
Target prediction and function annotation of miRNAs
A total of 12,517 target genes (12,464 were annotated) were predicted (see Supplementary File S4). Among them, fru-miR-15b was predicted to regulate the expression of foxl2, and novel-167, novel-318, and novel 538 were predicted to target to dmrt1. Some miRNAs target key genes related to TGF-β signaling; for example, amh was predicted to be the target of novel-548, gata2 was predicted to be the target of novel-240, and smad4 was predicted to be the target of novel-277, novel-407, and novel-512. Other miRNAs were predicted to target key genes in pathways involving sex steroid synthesis; for example, novel-358 and fru-miR-132 were predicted to target star, novel-451 was predicted to target cyp17a12, and fru-miR-15a, novel-496, fru-miR-122, and novel-538 were predicted to target cyp11b. Novel-128, fru-miR-187, and novel-542 were predicted to regulate the expression of cyp11a1.
Five mature miRNAs were down-regulated and three mature miRNAs were up-regulated in male gonads compared with female gonads (Fig. 3A). In total, 1,076 potential target genes of the differentially expressed miRNAs were identified. Enrichment analysis of 631 target genes indicated that they were enriched in the biological process gene ontology (GO) terms regulation of response to stimulus (19), regulation of cell communication (16) and regulation of signal transduction (16), and regulation of signaling (16). At the molecular function level, the most enriched GO term was ATPase activity, coupled (12) (Fig. 3B). According to KEGG analysis, most of the target genes were enriched in MAPK signaling (15), tight junction (11), RNA transport (10), and focal adhesion (12) (Fig. 4).
(A) Expression of miRNAs in the ovaries and testes of T. rubripes. The x-axis shows the fold change of expression levels between the XX and XY gonads. The y-axis shows the statistical significance of the change in miRNA expression. Blue dots indicate miRNAs that were equally expressed, red dots indicate upregulated miRNAs in XX gonads, and green dots indicate downregulated miRNAs in XX gonads. (B) Gene ontology (GO) analysis of the predicted target genes of the differentially expressed miRNAs. The GO enrichment of the predicted target genes in molecular functions, cellular components, and biological processes is shown.
Overview of lncRNA sequencing
In total, 950,603,34 raw reads were produced and 922,149,64 clean reads were generated by removing reads mapped to rRNA and low-quality reads in female gonads. While in the male gonads, 857,388,18 raw reads were produced and 830,504,86 clean reads were generated. Of the clean reads, 75.66% and 66.67% were efficiently mapped against the T. rubripes reference genome in female and male libraries, respectively (see Supplementary File S5).
Analysis of differentially expressed lncRNAs and their putative target genes
Illumina RNA-seq analysis identified 31,362 mRNAs, including 30,187 known transcripts and 1175 putative transcripts. Of the remaining 2774 lncRNA transcripts, only 523 (18.85%) were previously identified, and 2251 (81.15%) were novel (see Supplementary File S6). The average mRNA length was 2732 bp (open reading frame, ORF, 573 bp) and the average length of lncRNAs was 2104 bp (ORF, 225 bp) in T. rubripes gonads (Fig. 5A,B). Moreover, both the annotated and novel mRNAs (12) contained, on average more exons than lncRNAs (5) (Fig. 5C).
A total of 130 lncRNA transcripts displayed sex-biased expression in T. rubripes gonads. Of which, 51 were down-regulated and 79 were up-regulated in male gonads compared with female gonads (Fig. 6A and Supplementary File S7). Overall, 2243 lncRNAs were predicted to target 13,093 genes (see Supplementary File S8). Interestingly, the predicted target genes of several sex-biased lncRNAs were sex-differentially expressed genes; these included lnc_000338, lnc_000370, lnc_000690, XLOC_021951, and XR_965485.1, which target gsdf, thyroid hormone receptor-associated protein 3, thyroid hormone receptor beta, methyltransferase like protein, and zp3, respectively. Enrichment analysis of 1122 target genes revealed that there were more target genes involved in cellular component organization (81) at the biological process level and structural molecule activity (47) and endopeptidase activity (39) at the molecular function level (Fig. 6B). KEGG analysis showed that the target genes were mainly enriched in neuroactive ligand-receptor interaction (24), calcium signaling (20), cGMP-PKG signaling (18), and Oxytocin signaling (17) (see Supplementary File S9).
(A) Expression of lncRNAs in the ovaries and testes of T. rubripes. The x-axis shows the fold change of expression levels between the XX and XY gonads. The y-axis shows the statistical significance of the change in lncRNA expression levels. Blue dots indicate lncRNAs that were equally expressed, red dots indicate upregulated lncRNAs in XX gonads, and green dots indicate downregulated lncRNAs in XX gonads. (B) Gene ontology (GO) analysis of the predicted target genes of the differentially expressed lncRNAs. The GO enrichment of the predicted target genes in molecular functions, cellular components, and biological processes is shown.
Validation of miRNAs and lncRNAs by qPCR
The relative expression levels of five miRNAs (Fig. 7A) and four lncRNAs (Fig. 7B) from qPCR are shown in Fig. 7A. Fru-miR-212, fru-142, novel_128 and novel_167 were expressed at higher levels in XX than in XY, while fru-mir-1 was expressed at higher levels in XY than in XX at 40 dah (p < 0.05). The expression levels of Lnc_000569, Lnc_001034 and Lnc_000338 were significantly higher in XY, while the level of Lnc_000370 was significantly higher in XX (p < 0.05). The results are consistent with the sequencing data, demonstrating that the process used to identify putative miRNAs and lncRNAs was sufficiently stringent.
Discussion
Recently, several studies have screened the sex-biased expressed miRNAs between female and male gonads, however, few studies have focused on the larvae at the early sex differentiation stages in teleosts19,23. Additionally, lncRNAs have been widely shown to play an essential role in the process of sex determination and differentiation in mammals, but their potential role in other vertebrates has only been reported in the Chinese soft-shell turtle29. In this work, the dominant size of small RNAs in both male and female gonads was 22 nt, followed by 23 and 21 nt, within the typical size range for Dicer-derived products. This phenomenon is similar to that in O. niloticus19and C. carpio23. Additionally, in another study of adult T. rubripes, the predominant size of small RNAs were 26–28 nt in the ovaries and testes, with most having a length of 27 nt32. In the gonads of O. niloticus and D. rerio, a difference in length distribution of small RNAs between adults and larvae has also been found18,33. This may be due to the higher expression of piRNAs in the gonads of those adult fish. PiRNAs are a class of 26–32 nt ncRNAs, which are expressed mainly in the germline and play a significant role in germline development.
In the present study, fru-miR-100, fru-miR-30d, fru-miR-181a-5p, fru-miR-21, fru-miR-22a, fru-miR-92, fru-miR-199, fru-miR-125a, fru-let-7d, fru-miR-25, and fru-miR-200b were the conserved miRNAs that were highly expressed in the gonads of T. rubripes. The results implied that these miRNAs probably play a critical and basal physiological role in the process of gonadal development in fugu. miR-100 is also abundant in the undifferentiated gonads of O. niloticus19, and in the mature ovaries and testes of Pelteobagrus fulvidraco, and H. hippoglossus14,17. In T. ovatus, the miR-30 family is highly expressed in the testes34. In O. niloticus18 and D. rerio35, the miR-181a family is also abundantly expressed. In P. fulvidraco, compared with XY, it was observed that miR-21-5p and miR-21-3p levels increased more than four fold in YY testes and XX ovaries17. Taken together, this indicated that the miR-100, miR-30, miR-181a, and miR-21 families play an essential role in the gonadal development of teleosts. The miR-17/92 cluster is a typical highly conserved and well-studied miRNA cluster, and has pivotal roles in the regulation of the cell cycle, proliferation, and apoptosis36. MiR-92 is abundant in the gonads of T. rubripes, similar to the results found in zebrafish (D. rerio) and yellow catfish (P. fulvidraco)17,35,37. A recent study has demonstrated that during the early stages of embryogenesis of D. rerio, maternal miR-92a-3p can suppress the cyclin dependent kinase 1 inhibitor (Wee 1 homolog 2), which regulates cell cycle progression. A significantly higher rate of embryonic developmental arrest at the one-cell embryo stage was observed due to the inhibition of maternal miR-92a-3p expression38. let-7 is one of the miRNAs family initially identified in Caenorhabditis elegans as a key developmental switch and plays a crucial role in controlling the transition from larva to adult39. Eight members of the let-7 family (let-7a/b/d/e/g/h/i/j) were identified in T. rubripes gonads in the present study. As a critical regulator of gene expression, let-7 miRNAs are abundant in gonadal tissues of mammals and are involved in multiple physiological processes, such as the regulation of sexual maturity, sperm formation and oocyte maturation40,41,42. In teleosts, the let-7 family was also highly expressed in the gonads of Trachinotus ovatus34, C. carpio22, P. olivaceus23, Megalobrama amblycephala43, Danio rerio35, and Oryzias latipes25, consistent with the expression in T. rubripes, indicating functional conservation across vertebrates and suggesting that the let-7 family might be crucial for reproductive physiology. In P. olivaceus, female-biased expression of miR-200b was found, and the expression level in the testes was sevenfold lower than in the ovaries23. In mice, it was shown that Dicer, a ribonuclease essential for miRNA biogenesis, is crucial for Sertoli cell maturation, survival, and ultimately sustenance of germ cell development44. Ablation of Dicer in Sertoli cells caused the loss of several miRNAs, including miR-125a-3p, and subsequent up-regulation of SOD-1, a protein linked to apoptosis45.
In the present study, five mature miRNAs were down-regulated and three mature miRNAs were up-regulated in undifferentiated XY gonads compared with XX gonads. We therefore selected five miRNAs for qPCR analysis, and Fru-miR-212, fru-142, novel_128 and novel_167 were expressed at higher levels in XX than in XY, while fru-mir-1 was expressed at higher levels in XY than in XX at 40 dah (p < 0.05). In another study of adult T. rubripes, some sexually dimorphic miRNAs were also found in gonads. For example, fru-miR-214, fru-miR-143-3p, fru-miR-202-5p, fru-miR-24-3p and fru-miR-145b-5p exhibited higher expression levels in the ovaries than in the testes, while fru-miR-2478-3p and fru-miR-2898-3p exhibited higher expression levels in the testes than in the ovaries32. The differentially expressed miRNAs in gonads are totally different in larval and adult fugu, which suggested that different miRNAs may be involved in the different gonadal developmental stages. Similar phenomena were observed in tilapia, for example, by transcriptomic analysis of gonads collected at five different time points (30, 50, 75, 100, and 165 days after fertilization). Xiao et al. found stage-specific expression patterns of miRNAs18. Tao et al. quantified the expression of miRNAs in the undifferentiated gonads at the critical stage of molecular sex determination (5 dah), and obtained many different sex-biased expression miRNAs which different from tnat obtained in Xiao et al.18,19. Since limited reports focus on the different stage-specific expression levels, especially in teleosts, more work should be performed to elucidate the function miRNAs during sex differentiation and gonadal development, and the underlying mechanisms. Target prediction suggested that some miRNAs target key genes that participate in the process of sex differentiation of T. rubripes. Notably, novel-167 is enriched in the gonads of XX T. rubripes, and dmrt1 was its candidate direct target gene. In vertebrates, several DM domain genes (dmrt genes) have been shown to be required for gametogenesis and gonadal differentiation. Dmrt1 seems to have a more significant role and likely regulates testicular differentiation in all vertebrates46. Our previous study reported that XY T. rubripes have higher expression levels of dmrt1 in undifferentiated gonads compared with XX individuals31. During the early life stages of T. rubripes, 17-beta estradiol treatment was able to induce decrease of dmrt1 levels and feminization in XY individuals47,48. Therefore, dmrt1 may play a pivotal role during T. rubripes testicular differentiation, as reported in other species, and novel-167 is possibly involved in the regulation of dmrt1 transcription in T. rubripes.
Similar to previous studies in other vertebrates, the identified T. rubripes lncRNAs had shorter transcripts, lower expressions level, and fewer exons than the identified mRNAs49. T. rubripes lncRNAs (average, 2104 nt) were shorter than those in chicken (2941 nt)50, but longer than those in mouse (550 nt), human (1000 nt), zebrafish (1113 nt), goat (1180 nt), and Chinese soft-shell turtle (1717 nt)29,51,52,53. Additionally, on average, lncRNA transcripts in T. rubripes contained more exons than those in mouse (3.7), human (2.9), zebrafish (2.8), goat (2.2), and Chinese soft-shell turtle (2.0)29,51,52,53.
This study aimed to identify lnRANs involved in the onset of sexual differentiation in T. rubripes. We found that 130 out of 2774 lncRNAs were differentially expressed in male and female gonads. Among them, four lncRNAs were randomly selected for qPCR, and the qPCR results were consistent with the sequencing data. Moreover, some sex-differentially expressed genes are predicted to be the target of sex-biased lncRNAs, indicating these lncRNA-gene pairs may play an important role in fugu sexual differentiation. Remarkably, a new member of the TGF-β superfamily, gsdf was found to be a potential target of lncRNA000338. gsdf restricts expression in gonadal somatic cells in teleosts and gain- and loss-of-functional analysis demonstrated it is essential for teleost sex determination and differentiation54,55. In the male undifferentiated gonads of T. rubripes, higher expression levels of gsdf were observed, suggesting that it may have a similar role as described in other telosts31. Further study should be focused on validating the functional analysis of gsdf and lncRNAs, and the relationships and regulatory mechanisms between them.
Materials and methods
Samples collection, RNA extraction, and histological observation of gonads
Eighty larvae with 1.92 ± 0.19 cm average full length at 40 dah were obtained from a fishery in Dalian, China in April 2018. Then, 60 T. rubripes were anesthetized, and the gonads were dissected, flash-frozen in liquid nitrogen and stored at − 80 °C. Since they are tiny, the gonads of each individual were placed into a tube one by one. For sex verification of each larva, a piece of tissue sample was stored in a 1.5 ml tube containing 100% alcohol in a freezer at − 20 °C. After sex verification, gonads with the same gender (20 gonads/sex) were pooled, and RNA was extracted from male and female fish as described in our previous study31. Briefly, DNA was extracted from the tissues according to the manufacturer’s protocol for the TIANamp Marine Animals DNA kit (Tiangen, Beijing, China), and the genetic sex of each fish was identified using SNP markers (a region containing exon 9 of the amhr2 gene). As shown in Supplementary File S1, the genotype of males was C/G (XY) and that of females was C/C (XX). Previous studies have demonstrated there is a perfect concordance between the SNP genotype and phenotypic sex30,31,56. Histological observation of gonads was also conducted as described in our previous study31. All experiments with fugu were approved by the Animal Study Ethical Committee of Fishery Resources Enhancement Laboratory at Dalian Ocean University (Dalian, China). Experiments were performed in accordance with the lab animal protection regulations and guidelines of the People’s Republic of China (Order of the State Council of the People’s Republic of China No. 676), Liaoning Province (Order No. 143 of the people’s government of Liaoning province).
Library preparation and sequencing
RNA purity and concentration were examined as described in our previous study31. The small RNA libraries were prepared using an RNA-Seq library preparation kit (NEBNext Multiplex Small RNA Library Prep Set for Illumina, NEB, USA.) following the manufacturer’s recommended instructions. After quality certification and the cluster generation, two transcriptome libraries were sequenced on an Illumina Hiseq 2500 platform.
For lncRNA library construction, rRNA was firstly removed (Epicentre Ribo-zero rRNA Removal Kit, Epicentre, USA). After the rRNA-depleted RNA fragmentation, two sequencing libraries were generated according to the manufacturer’s protocol using the NEBNext Ultra Directional RNA Library Prep Kit for Illumina (NEB, USA). Subsequently, the library fragments were purified using an AMPure XP system (Beckman Coulter, Beverly, USA) to preferentially select cDNA fragments of 150–200 bp, and then the libraries sequencing was carried out on an Illumina Hiseq 4000 platform (paired-end 150 bp reads).
Small RNA sequence analysis
Data processing was carried out according to previously described procedures34. Clean reads were obtained by removing adapter sequences, low-quality sequences, sequences containing poly-N, and tags originating from protein coding genes, repeat sequences, rRNA, tRNA, snRNA, and snoRNA. Then, reads between 18 and 32 nt in length were selected for mapping to the T. rubripes genome using Bowtie, and the mapped small RNA tags were used to look for known miRNAs. The MiRBase20.0 (ftp://mirbase.org/pub/mirbase/20/) was used as reference, and modified software programs mirdeep2 and sRNA-tools-cli were used to obtain the potential miRNAs and draw the secondary structures. Two software programs, miREvo and mirdeep2, were integrated to predict novel miRNAs. miFam.dat (http://www.mirbase.org/ftp.shtml) was used to look for families for conserved miRNAs, while Rfam (http://rfam.sanger.ac.uk/search/) was used to look for Rfam families for novel miRNA.
Transcriptome (mRNA) sequence analysis and identification of candidate lncRNAs
Clean data were obtained by removing low-quality reads, reads containing adapters, and reads containing poly-N from raw data. Bowtie2 v2.2.8 was used to build the index of the reference genome, and HISAT2 v2.0.4 was used to align the clean reads to the reference genome (ftp://ftp.ncbi.nlm.nih.gov/genomes/all/GCF_000180615.1_FUGU5). The lncRNA transcriptome was assembled using StringTie (v1.3.1) in a reference-based approach57.
Three types of coding potential analysis softwares, coding-Non-Coding-Index, coding Potential Calculator, and Pfam Scan (v1.3) were combined to screen the non-protein coding RNA candidates from putative mRNAs. The phylogenetic codon substitution frequency (PhyloCSF) metric (v20121028) was used to build the multi-species genome sequence alignments with default parameters. After the above analysis, transcripts without coding potential predicted by either/all of the four tools were candidate lncRNAs.
Prediction and annotation of miRNA and lncRNA targets
Predictions of target genes of miRNAs was analyzed by miRanda. Since lncRNA can cis target their neighboring genes, the coding genes form 10 kb downstream and upstream of all identified lncRNAs were searched and their functions were analyzed. Pearson’s correlation coefficients between expression levels of lncRNAs and mRNAs were calculated with custom scripts58.
Differential expression of identified miRNAs and lncRNA, and enrichment analysis of their target genes
In order to compare the differential expression of miRNA in female and male gonads, the expression levels of miRNAs were calculated by the transcript per million (TPM) approach firstly34. Then, based on TPM normalized counts, the analysis was carried out using the DEGseq (2010) R package. The criteria used for screening the differentially expressed miRNAs were q-value < 0.05 and |log2(fold change)| > 1.
Cuffdiff (v2.1.1) was used to calculate fragments per kilobase of transcript per million mapped reads values for both lncRNAs and mRNAs in each sample59. The Ballgown suite includes functions for interactive exploration of the transcriptome assembly, visualization of transcript structures and feature-specific abundances for each locus, and post-hoc annotation of assembled features to annotated features60. Transcripts with an adjusted P-value < 0.05 and |log2(fold change)| > 1 were assigned as differentially expressed.
GOseq-based Wallenius non-central hyper-geometric distribution and KOBAS software were used for GO and KEGG pathway enrichment analysis of the putative target genes61,62. GO terms or KEGG pathway with corrected P < 0.05 were considered significantly enriched in differentially expressed genes.
Validation of sex-biased miRNAs and lncRNAs
Five miRNAs and four lncRNAs were randomly chosen for qPCR validation according to previously described methods31. The expression of miRNAs and lncRNAs were determined with a LightCycler480II Real-Time PCR System, in which U6 and β-actin were used as reference genes, respectively. The sequences of primers, including miRNA-specific stem-loop RT primers are listed in Table 1. All Reactions were carried out in triplicate, and the relative expression of the selected miRNAs and lncRNAs was calculated by the 2−ΔΔCT method. The Student t-test was conducted to evaluate the statistical significance between female and male gonads. Statistical analysis was carried out using SPSS version 22.0 software (IBM, Chicago, IL, USA). A p-value of < 0.05 was deemed to be significant.
Data availability
The Illumina sequences data of this study have been submitted to the Short Read Archive of the National Center for Biotechnology Information (NCBI) (Accession Numbers: PRJNA646544, PRJNA646995 and PRJNA647059).
References
Holoch, D. & Moazed, D. RNA-mediated epigenetic regulation of gene expression. Nat. Rev. Gene 16, 71–84 (2015).
Kozomara, A., Birgaoanu, M. & Griffiths-Jones, S. miRBase: from microRNA sequences to function. Nucl. Acids Res. 47, D155-162 (2019).
Friedman, R. C. et al. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 19, 92–105 (2009).
Agarwal, V. et al. Predicting microRNA targeting efficacy in Drosophila. Genome Biol. 19, 152. https://doi.org/10.1186/s13059-018-1504-3 (2018).
Pinhal, D. et al. Genome-wide microRNA screening in Nile tilapia reveals pervasive isomiRs’ transcription, sex-biased arm switching and increasing complexity of expression throughout development. Sci. Rep. 8, 8248. https://doi.org/10.1038/s41598-018-26607-x (2018).
Marchese, F. P., Raimondi, I. & Huarte, M. The multidimensional mechanisms of long noncoding RNA function. Genome Biol. https://doi.org/10.1186/s13059-017-1348-2 (2016).
Jeggari, A., Marks, D. S. & Larsson, E. miRcode: A map of putative microRNA target sites in the long non-coding transcriptome. Bioinformatics 28, 2062–2063 (2012).
Liu, Q. et al. LncRNA loc285194 is a p53-regulated tumor suppressor. Nucl. Acids Res. 41, 4976–4987 (2013).
Faghihi, M. A. et al. Evidence for natural antisense transcript-mediated inhibition of microRNA function. Genome Biol. 11, R56. https://doi.org/10.1186/gb-2010-11-5-r56 (2010).
Franklin, J. L. et al. Malignant transformation of colonic epithelial cells by a colon-derived long noncoding RNA. Biochem. Biophys. Res. Commun. 440, 99–104 (2013).
Herpin, A. & Schartl, M. Plasticity of gene-regulatory networks controlling sex determination: Of masters, slaves, usual suspects, newcomers, and usurpators. EMBO Rep. 16, 1260–1274 (2015).
Real, F. M. et al. A microRNA (mmu-miR-124) prevents Sox9 expression in developing mouse ovarian cells. Biol. Reprod. 89, 78–78 (2013).
Miao, N. et al. Identification of male-biased microRNA-107 as a direct regulator for nuclear receptor subfamily 5 group A member 1 based on sexually dimorphic microRNA expression profiling from chicken embryonic gonads. Mol. Cell Endocrinol. 429, 29–40 (2016).
Bizuayehu, T. T. et al. Differential expression patterns of conserved miRNAs and isomiRs during Atlantic halibut development. BMC Genomics. 13, 1–14 (2012).
Juanchich, A. et al. Characterization of an extensive rainbow trout miRNA transcriptome by next generation sequencing. BMC Genom. 17, 164. https://doi.org/10.1186/s12864-016-2505-9 (2016).
Juanchich, A. et al. Identification of differentially expressed miRNAs and their potential targets during fish ovarian development. Biol. Reprod. 88, 121–111 (2013).
Jing, J. et al. Sex-biased miRNAs in gonad and their potential roles for testis development in yellow catfish. PLoS ONE 9, e107946. https://doi.org/10.1371/journal.pone.0107946 (2014).
Xiao, J. et al. Identification and characterization of microRNAs in ovary and testis of Nile tilapia (Oreochromis niloticus) by using solexa sequencing technology. PLoS ONE 9, e86821. https://doi.org/10.1371/journal.pone.0086821 (2014).
Tao, W. et al. Integrated analysis of miRNA and mRNA expression profiles in tilapia gonads at an early stage of sex differentiation. BMC Genom. 17, 1–13 (2016).
Wang, W. et al. Coordinated microRNA and messenger RNA expression profiles for understanding sexual dimorphism of gonads and the potential roles of microRNA in the steroidogenesis pathway in Nile tilapia (Oreochromis niloticus). Theriogenology 85, 970–978 (2016).
Danillo, P. et al. Genome-wide microRNA screening in Nile tilapia reveals pervasive isomiRs’ transcription, sex-biased arm switching and increasing complexity of expression throughout development. Sci. Rep. 8, 1–18 (2018).
Wang, F. et al. Identification and profiling of Cyprinus carpiomicroRNAs during ovary differentiation by deep sequencing. BMC Genom. 18, 333. https://doi.org/10.1186/s12864-017-3701-y (2017).
Gu, Y., Zhang, L. & Chen, X. Differential expression analysis of Paralichthys olivaceus microRNAs in adult ovary and testis by deep sequencing. Gen. Comp. Endocrinol. 204, 181–184 (2014).
Lau, K. et al. Identification and expression profiling of microRNAs in the brain, liver and gonads of marine medaka (Oryzias melastigma) and in response to hypoxia. PLoS ONE 9, e110698. https://doi.org/10.1371/journal.pone.0110698 (2014).
Qiu, W. et al. Identification and expression analysis of microRNAs in medaka gonads. Gene 646, 210–216 (2018).
Yu, Q. et al. An estradiol-17β/miRNA-26a/cyp19a1a regulatory feedback loop in the protogynous hermaphroditic fish, Epinephelus coioides. Mol. Cell. Endocrinol. 504, 116089. https://doi.org/10.1016/j.mce.2019.110689 (2020).
McHugh, C. A. et al. Te Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature 521, 232–236 (2015).
Zhang, L. et al. A novel ncRNA gene from mouse chromosome 5 trans-splices with dmrt1 on chromosome 19. Biochem. Biophys. Res. 400, 696–700 (2010).
Zhang, J. et al. Screening and characterization of sex differentiation-related long non-coding RNAs in Chinese soft-shell turtle (Pelodiscus sinensis). Sci. Rep. 8, 8630. https://doi.org/10.1038/s41598-018-26841-3 (2018).
Kamiya, T. et al. A transspecies missense SNP in Amhr2 is associated with sex determination in the tiger pufferfish, Takifugu rubripes (fugu). PLoS Genet. 8, e1002798 (2012).
Yan, H. W. et al. Identification of genes involved in gonadal sex differentiation and the dimorphic expression pattern in Takifugu rubripes gonad at the early stage of sex differentiation. Fish Physiol. Biochem. 44, 1275–1290 (2018).
Wongwarangkana, C. et al. Deep sequencing, profiling and detailed annotation of microRNAs in Takifugu rubripes. BMC Genom. 16, 457. https://doi.org/10.1186/s12864-015-1622-1 (2015).
Houwing, S. et al. A role for Piwi and piRNAs in germ cell maintenance and transposon silencing in zebrafish. Cell 129, 69–82 (2007).
He, P. P. et al. Identification and characterization of microRNAs in the gonad of Trachinotus ovatus using Solexa sequencing. Comp. Biochem. Physiol. D 30, 312–320 (2019).
Presslauer, C. et al. Dynamics of miRNA transcriptome during gonadal development of zebrafish. Sci. Rep. 7, 43850. https://doi.org/10.1038/srep43850 (2017).
Mogilyansky, E. & Rigoutsos, I. The miR-17/92 cluster: A comprehensive update on its genomics, genetics, functions and increasingly important and numerous roles in health and disease. Cell Death Differ. 20, 1603–1614 (2013).
Vaz, C. et al. Deep sequencing of small RNA facilitates tissue and sex associated microRNA discovery in zebrafish. BMC Genom. 16, 1–16 (2015).
Presslauer, C. E. et al. miR-92a-3p controls cell cycle progression in zebrafish. bioRxiv 16, 201 (2019).
Reinhart, B. J. et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 403, 901–906 (2000).
Luo, M. M. et al. MicroRNA profiles and potential regulatory pattern during the early stage of spermatogenesis in mice. Sci. China Life Sci. 58, 442–450 (2015).
Gilchrist, G. C. et al. MicroRNA expression during bovine oocyte maturation and fertilization. Int. J. Mol. Sci. 17, 396. https://doi.org/10.3390/ijms17030396 (2016).
Shen, G. et al. Upstream and downstream mechanisms for the promoting effects of IGF-1 on differentiation of spermatogonia to primary spermatocytes. Life Sci. 101, 49–55 (2014).
Lan, T. et al. Comparative expression analysis of let-7 microRNAs during ovary development in Megalobrama amblycephala. Fish Physiol. Biochem. 45, 1101–1115 (2019).
Papaioannou, M. D. et al. Sertoli cell Dicer is essential for spermatogenesis in mice. Dev. Biol. 326, 250–259 (2009).
Papaioannou, M. D. et al. Loss of dicer in Sertoli cells has a major impact on the testicular proteome of mice. Mol. Cell. Proteomics. 10, M900587. https://doi.org/10.1074/mcp.M900587-MCP200 (2011).
Zarkower, D. DMRT genes in vertebrate gametogenesis. Curr. Top. Dev. Biol. 102, 327–356 (2013).
Lee, K. H. et al. Germ cell degeneration in high-temperature treated pufferfish, Takifugu rubripes. Sex Dev. 3, 225–232 (2009).
Hu, P. et al. Expression profiles of sex-related genes in gonads of genetic male Takifugu rubripes after 17β-estradiol immersion. J. Oceanol. Limnol. 37, 1113–1124 (2019).
Ponting, C. P., Oliver, P. L. & Reik, W. Evolution and functions of long noncoding RNAs. Cell 136, 629–641 (2009).
Li, Z. H. et al. Integrated analysis of long non-coding RNAs (lncRNAs) and mRNA expression profiles reveals the potential role of lncRNAs in skeletal muscle development of the chicken. Front. Physiol. 7, 687–687 (2016).
Cabili, M. N. et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Gene Dev. 25, 1915–1927 (2011).
Pauli, A. et al. Systematic identification of long noncoding RNAs expressed during zebrafish embryogenesis. Genome Res. 22, 577–591 (2012).
Gao, X. et al. Screening and evaluating of long noncoding RNAs in the puberty of goats. BMC Genom. 18, 164. https://doi.org/10.1186/s12864-017-3578-9 (2017).
Jiang, D. N. et al. Gsdf is a downstream gene of dmrt1 that functions in the male sex determination pathway of the Nile tilapia. Mol. Reprod. Dev. 83, 497–508 (2016).
Zhang, X. et al. Autosomal gsdf acts as a male sex initiator in the fish medaka. Sci Rep. 6, 19738 (2016).
Matsunaga, T. et al. An efficient molecular technique for sexing tiger pufferfish (fugu) and the occurrence of sex reversal in a hatchery population. Fish Sci. 80, 933–942 (2014).
Mihaela, P., Daehwan, K., Geo, M. P., Jeffrey, T. L. & Steven, L. S. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 11(9), 1650–1667 (2016).
Langfelder, P. & Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 9, 559 (2008).
Trapnell, C. et al. Transcript assembly and quantification by RNA-seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511–515 (2010).
Alyssa, C. et al. Ballgown bridges the gap between transcriptome assembly and expression analysis. Nat. Biotechnol. 33, 243–246 (2015).
Young, M.D., Wakefield, M.J., Smyth, G.K. & Oshlack, A. goseq: Gene Ontology testing for RNA-seq datasets. R Bioconductor. (2010).
Mao, X., Cai, T., Olyarchuk, J. G. & Wei, L. Automated genome annotation and pathway identification using the KEGG orthology (KO) as a controlled vocabulary. Bioinformatics 21, 3787–3793 (2005).
Acknowledgements
This research was supported by the Youth Program of National Natural Science Foundation of China (31902347),the National Key R&D Program of China (2017YFB0404000), and the General Project of Education Department of Liaoning Province (JL201904). We are also deeply grateful to Mingzheng Luo, Yu Liu, Wanrong Lu, Yihan Li, and other students from our research team, who were involved in the rearing process and sampling of specimens.
Author information
Authors and Affiliations
Contributions
H.Y. and Q.L. conceived and designed the experiments; J.J. and X.S. analyzed the data; L.Z., Z.Y. and Y.W. performed the experiments; H.Y. wrote of the the original draft paper; Q.L. and Y.L. critically revised the manuscript; and H.Y. had the primary responsibility for final content. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Yan, H., Liu, Q., Jiang, J. et al. Identification of sex differentiation-related microRNA and long non-coding RNA in Takifugu rubripes gonads. Sci Rep 11, 7459 (2021). https://doi.org/10.1038/s41598-021-83891-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-83891-w
- Springer Nature Limited