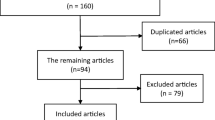
Abstract
Atrial fibrillation (AF) tachycardia causes heart failure and requires more attention. The genetic background of individual heart rate (HR) variations during AF are unclear. We hypothesized that HR-associated single nucleotide polymorphisms (SNPs) reported in Genome-Wide Association Studies (GWAS) are also associated with HR during AF. We enrolled patients with persistent AF (311 for screening and 146 for replication) who underwent AF ablation and were genotyped for the 21 h-associated SNPs reported in GWAS. The patients underwent 24-h Holter monitoring before AF ablation and electrophysiological study after AF ablation during sinus rhythm. Only the GJA1 SNP rs1015451 (T>C) was significantly associated with total HR (TT 110,643 ± 17,542 beats/day, TC 116,350 ± 19,060 beats/day, CC 122,163 ± 25,684 beats/day, P = 8.5 × 10−4). We also confirmed this significant association in the replication set. The intra-atrial conduction was faster in AF patients with the GJA1 minor allele than in those without it. Multivariate analysis revealed the presence of a GJA1 SNP rs1015451 additive model, female gender, lower left ventricular ejection fraction, and higher 1:1 atrioventricular nodal conduction were independently associated with higher HR during AF. The GJA1 SNP might be a new genetic marker for AF tachycardia.
Similar content being viewed by others
Introduction
Atrial fibrillation (AF) is the most common cardiac arrhythmia that causes tachycardia1. There are several treatment strategies for AF. Rate control is one of the most important baseline AF therapies regardless of the stage of AF, and it is used to either prevent the development of heart failure or reduce the symptoms2,3. The Swedish Heart Failure Registry reported that in AF heart failure patients with a reduced ejection fraction, a heart rate (HR) > 100 beats/min was associated with a higher mortality, and β-blocker use was associated with a reduced mortality4.
AF is also the most common cause of tachycardia-induced cardiomyopathy in patients without a history of structural heart disease5. Adequate rate control can reduce the risk of tachycardia-induced cardiomyopathy and worsening heart failure6. However, the HR varies during AF and varies from one individual to another, and the sensitivity to medications used for rate control therapy differs from patient to patient. Some patients are drug-resistant despite using multiple medications. The determinant of the HR during AF has not yet been clarified. It has been reported that the main determinant of the HR during AF is the conduction characteristics of the atrioventricular (AV) node and autonomic tone7,8, but the individual variability of the HR during AF is still not completely elucidated. Genetic differences might explain some of the individual variability in the HR during AF, but there have been a few negative reports about the association of the genetic differences with the HR during AF9,10. However, these studies were investigated under a drug administration with a small sample size.
Previous Genome-Wide Association Studies (GWAS) identified 21 single nucleotide polymorphisms (SNPs) associated with the HR during SR11, and some of those HR-associated SNPs have been reported to be associated with cardiac conduction. We hypothesized that the HR-associated SNPs reported in the GWAS were also associated with the HR during AF.
Results
Relationship between the 21 h-associated SNPs reported in the GWAS and total HR during AF in screening set
Table 1 shows the relationship between the 21 h-related SNPs reported by the GWAS and the total HR during the 24-h Holter monitoring. The GJA1 SNP rs1015451 (T>C) genotypes were significantly associated with the total HR after a Bonferroni correction. The total HR during AF was higher in the persistent AF patients with the GJA1 SNP rs1015451 minor allele than in those without it in the screening set (TT 110,643 ± 17,542 beats/day, TC 116,350 ± 19,060 beats/day, CC 122,163 ± 25,684 beats/day, P = 8.5 × 10−4; TT vs CC: P = 2.5 × 10−3, TT vs TC: P = 1.5 × 10−2, TC vs CC: P = 0.12, Fig. 1). The other SNPs were not significantly associated with the total HR. When the relationship between the HR and SNP was examined using the age, gender, and BMI as covariates, no significant SNP other than GJA1 was observed.
Relationship between the GJA1 single nucleotide polymorphism (SNP) rs1015451 genotypes and total heart rate (HR) in the patients with persistent atrial fibrillation (AF) in the screening set. The GJA1 SNP rs1015451 minor allele (C) was associated with a high total HR in the patients with persistent AF in the screening set (TT 110,643 ± 17,542 beats/day, TC 116,350 ± 19,060 beats/day, CC 122,163 ± 25,684 beats/day, P = 8.5 × 10−4; TT vs CC: P = 2.5 × 10−3, TT vs TC: P = 1.5 × 10−2, TC vs CC: P = 0.12).
Relationship between the clinical characteristics, echocardiographic parameters, and GJA1 SNP rs1015451 genotypes
The age, gender, body mass index, and duration of AF were similar among the 3 GJA1 SNP rs1015451 genotypes. The rates of diabetes, hypertension, strokes, structural heart disease, and heart failure were also similar among the 3 GJA1 SNP genotypes. There were no differences in the left atrial (LA) diameter, LA volume, or left ventricular ejection fraction (LVEF) between the 3 GJA1 SNP genotypes (Table 2).
Relationship between the EPS parameters and the GJA1 SNP rs1015451 genotypes
The relationship between the electrophysiological study (EPS) parameters and GJA1 SNP rs1015451 genotypes is shown in Table 3. The GJA1 SNP rs1015451 genotypes were significantly associated with the intra-atrial conduction time. The intra-atrial conduction times from the high right atrium (HRA) to the His bundle electrogram (HBE) and from the HRA to the distal coronary sinus (CS) were shorter in the patients with the GJA1 SNP rs1015451 minor allele than in those without it (HRA to HBE: TT 39.7 ± 14.0 ms, TC 36.8 ± 15.1 ms, CC 29.6 ± 11.1 ms, P = 6.1 × 10−4, HRA to distal CS: TT 111.6 ± 23.1 ms, TC 108.6 ± 22.2 ms, CC 98.8 ± 18.6 ms, P = 6.4 × 10−3). Furthermore, the AF cycle length was significantly shorter in the persistent AF patients with the GJA1 SNP rs1015451 minor allele than in those without it (TT 155 ± 21 ms, TC 149 ± 18 ms, CC 133 ± 16 ms, P = 2.2 × 10−4). However, the maximum sinus node recovery time (SNRT), corrected SNRT (CSRT), atrial to the His bundle (AH) interval, His bundle to the first ventricular activation (HV) interval, 1:1 AV nodal conduction, and effective refractory period (ERP) of the AV node were similar among the 3 GJA1 SNP rs1015451 genotypes.
Multivariate analysis of the total HR during AF in patients with persistent AF
In the univariate analysis, the GJA1 SNP rs1015451 genotypes, female gender, lower LVEF, higher 1:1 AV nodal conduction, shorter AF cycle length, and shorter atrial conduction time were significantly associated with the higher total HR during AF. A multivariate analysis revealed that the presence of a GJA1 SNP rs1015451 minor allele C, female gender, lower LVEF, and higher 1:1 AV nodal conduction were independently associated with higher HR during AF (Table 4).
Relationship between the GJA1 SNP rs1015451 genotypes and total HR during AF in the replication set
We confirmed the association between the GJA1 SNP rs1015451 and total HR during AF in the replication set (TT 113,139 ± 15,761 beats/day, TC 119,014 ± 18,771 beats/day, CC 128,489 ± 23,424 beats/day, P = 1.2 × 10−3; TT vs CC: P = 1.4 × 10−3, TT vs TC: P = 0.07, TC vs CC: P = 0.05, Fig. 2). The total HR during AF was higher in the persistent AF patients with the GJA1 SNP rs1015451 minor allele than in those without it and also in the replication set. (Fig. 2).
Relationship between the GJA1 single nucleotide polymorphism (SNP) rs1015451 genotypes and total heart rate (HR) in the patients with persistent atrial fibrillation (AF) in the replication set. The GJA1 SNP rs1015451 minor allele (C) was associated with a high total HR in the patients with persistent AF in the replication set (TT 113,139 ± 15,761 beats/day, TC 119,014 ± 18,771 beats/day, CC 128,489 ± 23,424 beats/day, P = 1.2 × 10−3; TT vs CC: P = 1.4 × 10−3, TT vs TC: P = 0.07, TC vs CC: P = 0.05).
Relationship between the GJA1 SNP rs1015451 genotypes and serum MiRNA concentrations
After a quality check, we analyzed 353 miRNAs, in which there were no missing data. To normalize the signals across the different microarrays tested, Norm Finder software was used to assess the stability of the miRNAs. The hsa-miR-149-3p was selected as a normalizer.
We investigated the relationship between the GJA1 SNP rs1015451 genotypes and plasma concentrations of the 353 miRNAs. Although only two miRNAs, miR-4708-3p (P = 4.6 × 10−2) and miR-4448 (P = 4.6 × 10−2), had P values of < 0.05, those were not significantly associated with GJA1 SNP rs1015451 genotypes after a Bonferroni correction. The top 10 miRNAs are shown in the Supplementary Table.
Discussion
In our study, the GJA1 SNP rs1015451 (T>C) minor allele was associated with a higher HR, shorter atrial conduction time, and shorter AF cycle length in the patients with persistent AF.
AF is the most common cardiac arrhythmia that causes tachycardia and heart failure1,2,3. The HR during AF differs from person to person, and many factors are involved in the AF HR. The determinants of the AF HR have not been completely identified. Also, the genetic background of individual HR variations during AF has not been examined. The 21 SNPs associated with the HR during sinus rhythm have been reported by a previous GWAS11. We investigated whether those SNPs were also associated with the HR during AF in patients with persistent AF. We initially clarified that only the GJA1 SNP rs1015451 (T>C) minor allele was associated with a higher HR during AF in patients with persistent AF. As a pertinent issue, the GJA1 SNPs including rs1015451 have recently been reported to be associated with heart failure and a reduced ejection fraction12.
Cardiac conduction is mediated by gap junction channels formed by connexin protein subunits. The GJA1 gene encodes connexin-43 (Cx43), which is one of the main connexin proteins13,14. Three types of connexins, Cx40, 43, and 45, are expressed differently depending on the site of the human cardiomyocytes15,16. Cx43 is the most expressed connexin in the atrial working myocardium, and it is also abundantly expressed in the working ventricular myocardium. Cx40 is abundantly expressed in the intraventricular conduction systems that control fast conduction, such as in the His bundle and Purkinje fibers. Cx45 is mainly expressed in the sinoatrial node and AV node, which have slow conduction properties. On the other hand, the expression level of Cx43 at the sinus node and AV node is very low16,17. In our study, the GJA1 SNP rs1015451 was associated with the conduction time of the atria, but it was not associated with the AV node conduction or SNRT. Those results were consistent with the localization of Cx43.
The GJA1 SNP rs1015451 minor allele (C) was also associated with a short AF cycle. Many studies on the AF cycle length have used an F-wave frequency analysis of the surface electrocardiography (ECG), and studies using an AF cycle length measurement from the intracardiac ECG have been few18,19. These two methods are reported to be correlated20,21, but the intracardiac ECG is more accurate. We analyzed the intracardiac ECG to evaluate the AF cycle length accurately. The HR during AF is influenced by many factors, such as the conduction characteristics of the AV node and autonomic tone. In our study, a shorter AF cycle was also associated with a higher HR during AF. The GJA1 SNP rs1015451 minor allele was associated with a higher HR during AF through a faster atrial conduction and shorter AF cycles, independent of the AV node conduction.
The SNP rs1015451 is called the GJA1 gene SNP11 because the GJA1 gene is the closest gene. However, the GJA1 SNP rs1015451 is located outside of the GJA1 gene, approximately 370 kB away from the GJA1 gene. Variants that are in a strong linkage disequilibrium (LD) with the GJA1 SNP rs1015451 (r2 > 0.8), are located in uncharacterized LOC105377979 and do not spread to the GJA1 gene (Supplementary Fig. 1). Previous studies have suggested that variations in the intergenic regions might regulate transcription factor binding and chromatin modification22. We investigated the expression quantitative trait locus (eQTL) data acquired from 429 human LA appendage samples, 432 human left ventricle samples and 532 human peripheral nerves available from the Genotype-Tissue Expression (GTEx) website (http://gtexportal.org; V7 release) for the cis-eQTL effects of GJA1 SNP rs1015451. We analyzed the genes located within 1 Mb upstream and downstream to GJA1 SNP rs1015451, but we found no genes, including GJA1, for which the expression was significantly associated with the GJA1 SNP rs101545. In addition, there were no LD SNPs around the GJA1 SNP rs101545 associated with the GJA1 expression (Supplementary Fig. 2-1,2). On the other hand, the expression level of the GJA1 gene in the peripheral nerves was significantly higher in patients with the GJA1 SNP rs1015451 minor C allele (Supplementary Fig. 2-3), suggesting that epigenetic regulation could be involved in this process. We investigated the relationship between the GJA1 SNP rs1015451 genotypes and plasma concentrations of 353 miRNAs, and they were not significantly associated with the GJA1 SNP rs1015451 genotypes. More functional studies focused on this intergenic region on chromosome 6q22 will be required to understand its potential effect on GJA1.
There have been some negative reports about the association of the genetic SNPs with AF rate control. Barret et al. reported that no SNP was associated with acute HR control after the administration of diltiazem9. Kolek et al. reported that no SNP was significantly associated with a poor AF HR control10. In our study, the GJA1 gene SNP was associated with the HR during AF in patients with persistent AF in screening and replication sets.
To the best of our knowledge, this is the first report to demonstrate that for the GJA1 gene, encoding the gap junction Cx43, the SNP is associated with the HR during AF. This finding has a clinical implication in that early detection of high-risk AF patients with AF tachycardias who are prone to develop heart failure, and early intervention in these patients are needed to avoid heart failure.
This study had some study limitations. It was a retrospective study conducted at a single center, and the total number of cases was small, and therefore the statistical power of this study was inadequate. Therefore, we must investigate the association between the HR during AF and these SNPs using a larger sample of cases in future research.
Conclusion
The GJA1 gene, encoding the gap junction protein (Cx43), SNP rs1015451 minor allele is associated with a high HR in AF patients. The GJA1 SNP rs1015451 might be a new genetic marker for AF tachycardia.
Methods
Participants
This was a single-center retrospective study. We enrolled 311 patients (243 men and 68 women, mean age 63 ± 10 years) with persistent AF who underwent radiofrequency catheter ablation (RFCA) at the Hiroshima University Hospital from November 2009 to March 2016 for screening. We also enrolled 146 consecutive Japanese persistent AF patients (114 men, 32 women; mean age, 61 ± 10 years) who underwent RFCA at the Hiroshima University Hospital from April 2016 to July 2018 for replication. Persistent AF was defined as AF that lasted longer than 7 days. All procedures involving the human genome use were approved by the Institutional Ethics Committee of the Graduate School of Biomedical Science at the Hiroshima University. Written informed consent was obtained from all participants prior to participation in the study. All methods were performed in accordance with the relevant guidelines and regulations.
Genotyping
We obtained peripheral blood from all participants and extracted the genomic DNA from leukocytes using the QIAamp DNA Blood Mini Kit (QIAGEN, Hilden, Germany) according to the standard protocol. For all subjects, we genotyped the 21 h-associated SNPs reported in the GWAS (MYH6, GJA1, ACHE, CD46, FADS1, SLC35F1, LINC00477, KIAA1755, CCDC141, SYT10, HCN4, GNB4, FLRT2, CHRM2, NKX2-5, GNG11, B3GNT7, FNDC3B, RFX4, CPNE8, TFPI) using a TaqMan assay.
Echocardiography
Transthoracic echocardiography was performed at our institution using a commercially-available system (Vivid E9, GE Healthcare, Milwaukee, WI, USA; or iE33, Philips Medical Systems, Andover, MA, USA) before the RFCA. Experienced echocardiographers who were blinded to the genotyping results conducted all echocardiographic examinations and analyzed the echocardiographic parameters.
Twenty-four-hour Holter monitoring
Twenty-four-hour Holter monitoring was performed one day before the RFCA. Antiarrhythmic drugs, including β-blockers and calcium channel blockers, were stopped at least five half-lives before performing the 24-h Holter monitoring. Only amiodarone was routinely discontinued at least 2 weeks prior to performing the 24-h Holter monitoring. We confirmed that all the subject were AF all the time. The total HR during the AF was recorded in all of the subjects.
Electrophysiological study and RFCA
We also confirmed that all the patients were AF at the beginning of their AF ablation.
The AF cycle length was recorded on the LA posterior wall and four pulmonary veins. An average cycle length of 5 s before the pulmonary vein isolation (PVI) was calculated. We defined the shortest cycle length of them all as the AF cycle length.
A continuous PVI was performed in all subjects using an open-irrigation 3.5-mm tip deflectable catheter (THERMOCOOL SMARTOUCH; Biosense Webster) under the guidance of a 3-dimensional electro-anatomical mapping system (CARTO3, Biosense Webster) with computed tomography integration (CARTOMERGE, Biosense Webster) to achieve electrical isolation of the left- and right-sided pulmonary veins. We confirmed the PVI entrance and exit block and then rechecked these under the infusion of isoproterenol plus adenosine triphosphate23.
After the PVI, an EPS was performed during stable SR. Four 5-French-gauge catheters were used: one catheter was a 10-polar electrode catheter positioned in the CS, and three catheters were quadripolar electrode catheters positioned in the HRA, His bundle, and right ventricle, all with a 5-mm interelectrode distance. The AH and HV intervals were measured during the baseline intracardiac ECG. The SNRT was measured as the recovery interval after a 30-s stimulation from the HRA. The CSRT was defined as the recovery interval in excess of the sinus cycle (i.e., CSRT = maximum SNRT − sinus cycle length)23. The ERP was measured using extrastimulus pacing with varied coupling intervals at a basic cycle length of 600 ms. The ERP was defined as the longest coupling interval that failed to propagate through that tissue. The maximal rate of 1:1 AV nodal conduction was determined using incremental atrial pacing.
miRNA
We investigate the relationships between the GJA1 SNP rs1015451 and serum concentrations of 2555 miRNAs. The total RNA was extracted from individual serum samples using the 3D-Gene RNA Extraction Reagent from a liquid sample kit (Toray Industries, Inc., Kanagawa, Japan). A total of 2555 miRNA sequences were detected using the 3D-Gene miRNA Labeling kit and 3D-Gene Human miRNA Oligo Chip (Toray Industries, Inc). We analyzed the relationships between the GJA1 SNP rs1015451 genotype and serum concentrations of the miRNAs in the AF patients24.
Statistical analysis
Continuous are presented as the mean ± standard deviation. The deviation from the Hardy–Weinberg equilibrium was tested among the cases and controls using an ordinary χ2 test. An additive mode of inheritance was assumed where the SNPs were coded as 0, 1, and 2 in the linear regression model. The association between the GJA1 SNP rs1015451 genotype and serum concentrations of the miRNAs was also analyzed by the linear regression model where the SNPs were coded as 0, 1, and 2. A multivariate analysis was performed by means of multiple regression analysis. The Bonferroni-corrected P-value threshold was P < 0.002 (0.05/21 SNPs) for relationships between the 21 h-associated SNPs and HR in the AF patients. Values of P < 0.05 were considered significant for the other analyses. The statistical analyses were conducted using the R3.3.1 and the JMP statistical package (version 13, SAS Institute, Cary, NC).
Data availability
The data in this study are available from the corresponding author upon reasonable request.
References
Go, A. S. et al. Prevalence of diagnosed atrial fibrillation in adults: National implications for rhythm management and stroke prevention: the AnTicoagulation and risk factors in atrial fibrillation (ATRIA) Study. JAMA 285, 2370–2375. https://doi.org/10.1001/jama.285.18.2370 (2001).
Kirchhof, P. et al. 2016 ESC Guidelines for the management of atrial fibrillation developed in collaboration with EACTS. Eur. Heart J. 37, 2893–2962. https://doi.org/10.1093/eurheartj/ehw210 (2016).
Rienstra, M. et al. Gender-related differences in rhythm control treatment in persistent atrial fibrillation: Data of the Rate Control Versus Electrical Cardioversion (RACE) study. J. Am. Coll. Cardiol. 46, 1298–1306. https://doi.org/10.1016/j.jacc.2005.05.078 (2005).
Li, S. J. et al. Prognostic significance of resting heart rate and use of β-blockers in atrial fibrillation and sinus rhythm in patients with heart failure and reduced ejection fraction: Findings from the Swedish Heart Failure Registry. Circ. Heart Fail. 8, 871–879. https://doi.org/10.1161/CIRCHEARTFAILURE.115.002285 (2015).
Gupta, S. & Figueredo, V. M. Tachycardia mediated cardiomyopathy: Pathophysiology, mechanisms, clinical features and management. Int. J. Cardiol. 172, 40–46. https://doi.org/10.1016/j.ijcard.2013.12.180 (2014).
Van-Gelder, I. C., Rienstra, M., Crijns, H. J. & Olshansky, B. Rate control in atrial fibrillation. Lancet 388, 818–828. https://doi.org/10.1016/S0140-6736(16)31258-2 (2016).
Sandberg, F. et al. Non-invasive assessment of the effect of beta blockers and calcium channel blockers on the AV node during permanent atrial fibrillation. J. Electrocardiol. 48, 861–866. https://doi.org/10.1016/j.jelectrocard.2015.07.019 (2015).
Lauri, T., Alan, K., William, K. & Fred, M. Determinants of the ventricular rate during atrial fibrillation. J. Am. Coll. Cardiol. 16, 1194–1200. https://doi.org/10.1016/0735-1097(90)90552-z (1990).
Barrett, T. W. et al. Association of atrial fibrillation risk alleles and response to acute rate control therapy. Am. J. Emerg. Med. 34, 735–740. https://doi.org/10.1016/j.ajem.2016.01.034 (2016).
Kolek, M. J. et al. A genome-wide association study to identify genomic modulators of rate control therapy in patients with atrial fibrillation. Am. J. Cardiol. 114, 593–600. https://doi.org/10.1016/j.ajem.2016.01.034 (2014).
Marcel, D. H. et al. Identification of heart rate-associated loci and their effects on cardiac conduction and rhythm disorders. Nat. Genet. 45, 621–631. https://doi.org/10.1038/ng.2610 (2013).
Evans, K. L. et al. Genetics of heart rate in heart failure patients (GenHRate). Hum. Genom. https://doi.org/10.1186/s40246-019-0206-6 (2019).
Severs, N. J. Pathophysiology of gap junctions in heart disease. J. Cardiovasc. Electrophysiol. 5, 462–475. https://doi.org/10.1111/j.1540-8167.1994.tb01185.x (1994).
Saffitz, J. E., Laing, J. G. & Yamada, K. A. Connexin expression and turnover: Implications for cardiac excitability. Circ. Res. 86, 723–728. https://doi.org/10.1161/01.RES.86.7.723 (2000).
Severs, N. J. et al. Immunocytochemical analysis of connexin expression in the healthy and diseased cardiovascutar system. Microsc. Res Tech. 52, 301–322. https://doi.org/10.1002/1097-0029(20010201)52:3%3c301::AID-JEMT1015%3e3.0.CO;2-Q (2001).
Davis, L. M., Rodefeld, M. E., Green, K., Beyer, E. C. & Saffitz, J. E. Gap junction protein phenotypes of the human heart and conduction system. J. Cardiovasc. Electrophysiol. 6, 813–822. https://doi.org/10.1111/j.1540-8167.1995.tb00357.x (1995).
Sander, V. & Sven, K. Connexin diversity in the heart: Insights from transgenic mouse models. Front. Pharmacol. 4, 1–14. https://doi.org/10.3389/fphar.2013.00081 (2013).
Platonov, P. G., Corino, V. D., Seifert, M., Holmqvist, F. & Sörnmo, L. Atrial fibrillatory rate in the clinical context: Natural course and prediction of intervention outcome. Europace. 16, 110–119. https://doi.org/10.1093/europace/euu249 (2014).
Haissaguerre, M. et al. Atrial fibrillatory cycle length: Computer simulation and potential clinical importance. Europace. 9, 64–70. https://doi.org/10.1093/europace/eum208 (2007).
Romero, I., Fleck, E. & Kriatselis, C. Frequency analysis of atrial fibrillation surface and intracardiac electrograms during pulmonary vein isolation. Europace. 13, 1340–1345. https://doi.org/10.1093/europace/eur104 (2011).
Bollmann, A. et al. Frequency analysis of human atrial fibrillation using the surface electrocardiogram and its response to ibutilide. Am. J. Cardiol. 81, 1439–1445. https://doi.org/10.1016/s0002-9149(98)00210-0 (1998).
Harismendy, O. et al. 9p21 DNA variants associated with coronary artery disease impair interferon-γ signaling response. Nature 470, 264–268. https://doi.org/10.1038/nature09753 (2011).
Tomomori, S. et al. Maintenance of low inflammation level by the ZFHX3 SNP rs2106261 minor allele contributes to reduced atrial fibrillation recurrence after pulmonary vein isolation. PLoS ONE 13, e0203281. https://doi.org/10.1371/journal.pone.0203281 (2018).
Tomomori, S. et al. Chromosome 4q25 variant rs6817105 bring sinus node dysfunction and left atrial enlargement. Sci. Rep. 8, 14565. https://doi.org/10.1038/s41598-018-32453-8 (2018).
Acknowledgements
We thank the members of the clerical and medical staff of the Hiroshima University Hospital for their assistance and the ENAGO Group (English editing system) for editing the initial draft of this manuscript. Dr. Y. Nakano was supported by JSPS KAKENHI Grant Number 17K09501.
Author information
Authors and Affiliations
Contributions
The conception and design of the study, analysis and interpretation of the data, and drafting of the manuscript were done by S.O., H.O., and Y.N. The critical discussion and revision of the manuscript for its intellectual content was done by S.O., H.O., K.C., Y.K., and Y.N. The analysis and interpretation of the data was performed by S.O., Y.O., H.O., T.T., N.H., Y.O., Y.I., S.M., and Y.N. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Okamura, S., Onohara, Y., Ochi, H. et al. Minor allele of GJA1 gene polymorphism is associated with higher heart rate during atrial fibrillation. Sci Rep 11, 2549 (2021). https://doi.org/10.1038/s41598-021-82117-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-82117-3
- Springer Nature Limited
This article is cited by
-
Association between the APOE gene polymorphism and lipid profile and the risk of atrial fibrillation
Lipids in Health and Disease (2021)