Abstract
Type I interferon (IFN) plays a crucial role in innate and adaptive immunity, and aberrant IFN responses are involved in systemic autoimmune diseases, such as systemic lupus erythematosus (SLE) and dermatomyositis (DM). Type I IFNs can be induced by transcribed retrotransposons. The regulation of retrotransposons and type I IFN and the downstream IFN pathways in SLE, DM, and autoimmune blistering disease (AIBD) were investigated. The gene expression levels of retrotransposons, including LINE-1, type I-III IFNs, and IFN-stimulated genes (ISGs) in peripheral blood cells from patients with DM (n = 24), SLE (n = 19), AIBD (n = 14) and healthy controls (HCs, n = 10) were assessed by quantitative polymerase chain reaction. Upregulation of retrotransposons and IFNs was detected in DM patient samples, as is characteristic, compared to HCs; however, ISGs were not uniformly upregulated. In contrast, retrotransposons and IFNs, except for type II IFN, such as IFN-γ, were not upregulated in SLE. In AIBD, only some retrotransposons and type I interferons were upregulated. The DM, SLE, and AIBD samples showed coordinated expression of retrotransposons and type I IFNs and distinct spectra of IFN signaling. A positive correlation between LINE-1 and IFN-β1 was also detected in human cell lines. These factors may participate in the development of these autoimmune diseases.
Similar content being viewed by others
Introduction
Autoimmune diseases can affect any organ and are classified into two categories: organ-specific diseases (e.g., autoimmune blistering disease (AIBD), type I diabetes, etc.) and systemic diseases (e.g., systemic lupus erythematosus (SLE), dermatomyositis (DM), etc.) and are associated with autoantibody production1. SLE usually manifests as constitutional symptoms and mucocutaneous initial symptoms and affects various organs, including the kidney, brain, heart, and lung, with specific autoantibodies, such as anti-double stranded DNA (dsDNA), nucleosome, Sm, histone, C1q, ribosomal P, Ro/SSa, La/SSb, and ribonucleoprotein (RNP) antibodies (Abs)2,3. Skin lesions occur in as many as 75–80% of SLE patients4. The lesions are categorized as acute (malar rash), subacute (papulosquamous lesions, lupus tumidus and chilblain lupus), chronic (discoid lupus), and bullous LE. Although acute cutaneous lupus is nearly always associated with systemic lupus, discoid lupus is infrequently (3–5%) associated with systemic disease3. DM is an idiopathic inflammatory myopathy with interstitial pneumonia and characteristic cutaneous manifestations, accompanied by specific autoantibodies such as anti-TIF1-γ, NXP2, MDA5, SAE, Mi2, and ARS Abs2,5. Pemphigus vulgaris (PV) and bullous pemphigoid (BP) represent two major AIBD disorders and affect only the skin and mucosa6. Autoantibodies, anti-desmoglein (Dsg)1 and Dsg3 Abs are seen in PV, whereas type XVII collagen at the dermo-epidermal junction and anti-BP180 and BP230 Abs are detected in BP7.
Type I interferon (IFN) is reportedly upregulated in autoimmune diseases, such as SLE and DM8,9. Type I IFN comprises 13 IFN-α subtypes, 2 IFN-β subtypes, and less-studied subtypes, such as IFN-ε, IFN-κ, and IFN-ω10,11. The in vitro antiviral and antiproliferative potencies vary among these subtypes, which may be due to their distinct affinities for the subunits of IFN-α/β receptor (IFNAR) 1 and 212,13. Although plasmacytoid dendritic cells (pDCs) produce the majority of type I IFNs upon viral infection, all cell types can secrete type I IFNs14. Type II IFN, such as IFN-γ, is secreted from T lymphocytes and natural killer (NK) cells15. Type III IFNs, IFN-λ1, IFN-λ2, IFN-λ3, and IFN-λ4, are antiviral cytokines. Their functions are similar to those of type I IFNs. The binding of either a type I IFN to the IFNAR1/2 complex or a type III IFN to the IL-10R2/IFNLR1 complex triggers the activation of Janus kinase 1 (JAK1) and tyrosine kinase 2 (TYK2)16. This leads to the recruitment and phosphorylation of signal transducers and activators of transcription 1 and 2 (STAT1 and STAT2). STAT1 and 2 form a heterodimer that recruits IFN-regulatory factor 9 (IRF9) to form IFN-stimulated gene factor 3 (ISGF3)17,18. The binding of type II IFN dimers to the IFN-γ receptor 1 and 2 (IFNGR1/2) complex leads to the activation of JAK1 and JAK2, which subsequently phosphorylate STAT1. Phosphorylated STAT1 homodimers form the IFN-γ activation factor (GAF)19. Both ISGF3 and GAF induce the expression of genes regulated by IFN-stimulated response elements (ISREs) and gamma-activated sequence (GAS) promoter elements, respectively, resulting in the expression of IFN-stimulated genes (ISGs)20. Dysregulation of various IFNs may contribute to autoimmune pathogenesis21. In SLE, dysregulated IFN-γ is associated with autoantibody positivity, and upregulation of IFN-α/β activity can be observed in a pre-disease state9,22. Since dysregulation of various IFNs is seen in autoimmune diseases, the IFN and related gene expression profiles in peripheral blood can be useful in understanding the pathogenesis.
Retrotransposons are transposable elements involved in the pathogenesis of autoimmune diseases23,24. They are divided into two groups: those with long terminal repeats (LTRs) and non-LTRs25. LTR retrotransposons are recognized as endogenous retroviruses (ERVs) derived from exogenous retroviruses. In particular, the HERV-K virus is the youngest human endogenous retrovirus and is integrated almost solely as complete proviral copies in the human genome. Among non-LTR retrotransposons, the long interspersed nuclear element-1 (LINE-1) autonomous family of retroelements is active in mammalian genomes. It comprises approximately 17% of the human genome, yielding one-half million copies, and the transcription of LINE-1 is normally suppressed by the methylation of the respective promoter. Transcribed LINE-1 induces the production of type I IFNs26. Alu sequences are short interspersed elements (SINEs) composed of two monomers linked by an A-rich linker sequence (A5TACA6). SINE-VNTR-Alu (SVA) is a composite element comprising an Alu-like CCCTCT hexamer region, a region of variable numbers of tandem repeats (VNTRs), and an HERV-K-derived region known as SINE-R. While all non-LTR retrotransposons share a poly-A tail, they are otherwise structurally different25.
We have investigated the roles of retrotransposons in the pathogenesis of autoimmune diseases, especially in SLE and DM. Furthermore, we examined whether type I IFN production enhances the JAK-STAT signaling pathway and induces ISGs using patient clinical samples and established human cell lines.
Results
Upregulation of retrotransposons with reduced LINE-1 promoter methylation in AIBD and DM
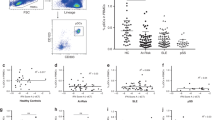
To assess the involvement of retrotransposons in autoimmune diseases, we initially performed qPCR analyses of LINE-126, HERVK14C, and SVA using peripheral blood cells from healthy controls (HCs) and patients with AIBD, SLE, or DM (Fig. 1A). Characteristically, the expression of all retrotransposon mRNAs was significantly upregulated in DM (LINE-1, HC vs. DM, P < 0.001; HERVK14C, HC vs. DM, P < 0.0001; and SVA, HC vs. DM, P < 0.001), and the expression of LINE-1 was increased in AIBD (LINE-1: HC vs. AIBD, P < 0.05). In contrast, no statistically significant difference was detected between HC and SLE. Since methylation of the LINE-1 promoter region plays a major role in LINE-1 transcriptional regulation27, we analyzed the methylation levels of three promoter sites of LINE-1 by pyrosequencing (Fig. 1B). The methylation level of the LINE-1 promoter was significantly reduced in AIBD and DM patients but not in SLE patients, in comparison to that in HCs (HC vs. AIBD, P < 0.05; HC vs. DM, P < 0.0001). DNA methylation is generally catalyzed by DNA methyltransferase (DNMT) 1, DNMT3A, DNMT3B, lymphoid-specific helicase (LSH), and methyl-CpG-binding protein 2 (MeCP2), and mammalian DNA methylation patterns are mainly generated by DNMT3A and DNMT3B28. We determined that the mRNA expression level of DNMT3A, but not that of DNMT1, DNMT3B, LSH or MeCP2, was selectively downregulated in AIBD and DM compared to that in HC and SLE (HC vs. AIBD, P < 0.05; HC vs. DM, P < 0.001) (Fig. 1C and S1A). Collectively, these results suggested that hypomethylation of the LINE-1 promoter by reduced expression of DNMT3A is likely to upregulate the expression of retrotransposons in DM and AIBD.
Different regulation of retrotransposons, IFNs, and ISGs among autoimmune diseases. (A) Retrotransposons are upregulated in AIBD and DM but not in SLE. The mRNA levels of retrotransposons, such as LINE-1, HERVK14C, and SVA, in the whole blood cells from HCs and patients with AIBD, SLE, or DM were analyzed by qPCR. (B) Reduced LINE-1 promoter methylation in AIBD and DM. Methylation in the LINE-1 promoter region was analyzed by pyrosequencing. (C) Reduced expression of DNMT3A in AIBD and DM. The mRNA levels of DNMT3A in blood cells were analyzed by qPCR. (D) Different induction of IFNs in autoimmune diseases. The mRNA levels of IFN-α2, IFN-β1, IFN-γ, and IFN-λ1 were quantified by qPCR. (A–D) Data are shown as the means ± SD by Mann–Whitney test. HC (n = 10), AIBD (n = 14), SLE (n = 19), and DM (n = 24). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, ns; not significant. (E) Increased expression of type I IFN-inducible proteins in skin lesions from autoimmune patients. Specimens from HC and skin lesions obtained from pretreated AIBD, SLE, and DM patients were stained with anti-MxA and anti-ISG15 antibodies. Bars = 100 μm.
Differential expression of IFNs in autoimmune diseases
To investigate the relationship between retrotransposons and interferonopathy, we next assessed the mRNA levels of IFN-α2, IFN-β1 (type I), IFN-γ (type II), and IFN-λ1 (type III) in the patients’ peripheral blood cells (Fig. 1D and S1B). Notably, the mRNA levels of the type I-III IFNs examined were significantly upregulated in the DM patients (IFN-α2, HC vs. DM, P < 0.01; IFN-α4, HC vs. DM, P < 0.05; IFN-β1, HC vs. DM, P < 0.0001; IFN-γ, HC vs. DM, P < 0.01; and IFN-λ1, HC vs. DM, P < 0.05). In contrast, only type II IFN (IFN-γ) was upregulated in the SLE patients (IFN-γ, HC vs. SLE, P < 0.01), and only type I IFNs (IFN-α2 and IFN-β1) were upregulated in AIBD (IFN-α2, HC vs. AIBD, P < 0.05, IFN-β1, HC vs. AIBD, P < 0.05). Thus, these results clearly indicate that the induction spectra of IFN genes are distinct among DM, SLE, and AIBD patients.
To further assess the expression of ISGs in patients’ pretreated skin lesions (erythema, vesicle or erosion), we performed immunohistochemistry to detect type I IFN-inducible proteins such as MxA and ISG15. The MxA protein was clearly observed in the keratinocytes of the SLE (10/11), AIBD (7/9) and DM (3/3) samples but not in the skin samples of the HCs (0/3) (Fig. 1E and Table S1). The signal from the ISG15 protein was also strong in the cytoplasm of keratinocytes in the SLE (10/10), AIBD (9/9), and DM (3/3) samples. In contrast, skin samples of the HCs showed only nuclear staining of ISG15 (Fig. 1E and Table S1). These results indicated that the expression of IFN-inducible proteins is upregulated in the skin lesions of pretreated patients with DM, SLE, and AIBD. Although little is known about the roles of type I IFN in AIBD, the increased expression of IFN-inducible proteins in the pretreated skin lesions of AIBD patients suggests that IFNs may play suppressive or promoting roles in blister formation.
Differential induction of IFN signaling factors in autoimmune diseases
To clarify the gene expression in the IFN receptor signaling pathway, we performed qPCR analyses of JAK1 and JAK2, TYK2, STAT1 and STAT2, and IRF1. Compared with that of HCs, the mRNA level of JAK2 was decreased in AIBD patients (HC vs. AIBD, P < 0.0001), and the levels of JAK1 and JAK2 were also decreased in SLE patients (JAK1, HC vs. SLE, P < 0.0001; JAK2, HC vs. SLE, P < 0.05). In contrast, JAK2 upregulation and TYK2 downregulation were detected in DM samples (JAK2, HC vs. DM, P < 0.01; TYK2, HC vs. DM, P < 0.05) (Fig. 2A). Moreover, the expression levels of IRF1, STAT1, and STAT2 were significantly increased in the SLE samples (IRF1, HC vs. SLE, P < 0.05; STAT1, HC vs. SLE, P < 0.001; STAT2, HC vs. SLE, P < 0.05), although an increase in STAT2 expression alone was detected in the DM samples (HC vs. DM, P < 0.01) (Fig. 2B). We further examined the expression of downstream IFN-stimulated genes (ISGs), such as ddx58, IFIH1, IFIT1, IFI27, IFI44L, IRF7, ISG15, Mx1, and OAS1, as well as IFN-γ-specific downstream chemokines CXCL9/MIG and CXCL10/IP-10, by qPCR (Fig. 2C and S1C). Notably, the expression levels of all ISGs were significantly increased in the SLE patients compared to those of the HCs, while increased expression levels of IFIH1, IFIT, IFI44L, and CXCL-10 were detected in the DM patients compared to the HCs. In contrast, no significant induction of ISGs was detected in the AIBD patients.
Diversity in the expression of IFN signaling factors in autoimmune diseases. (A) The expression level of nRTKs is upregulated in DM. The mRNA levels of nRTKs, such as JAK1, JAK2, and TYK2, in the whole blood cells from HCs and patients with autoimmune diseases were analyzed by qPCR. (B) Transcription factors are highly expressed in SLE. The mRNA levels of STAT1, STAT2, and IRF1 were analyzed by qPCR. (C) ISGs are upregulated in SLE and DM but not in AIBD and HC. The mRNA levels of ISGs were analyzed by qPCR. (A–C) Data are shown as the means ± SD by Mann–Whitney test. HC (n = 10), AIBD (n = 14), SLE (n = 19), and DM (n = 24). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, ns; not significant.
Collectively, the data showed that type I-III IFNs are activated in DM. In contrast, in SLE, only type II IFN is activated, and transcription factors (TFs) and ISGs in type II IFN signaling pathway are also upregulated.
Expression profiles of retrotransposons and IFNs in patients
Since distinct IFN expression was detected among the DM, SLE, and AIBD patients, we analyzed the expression patterns of retrotransposons and IFN pathways. We categorized retrotransposons and IFN-related genes into four categories: retrotransposons and IFNs, nonreceptor tyrosine kinases (nRTKs), TFs, and ISGs (Fig. S2). Patients SLE03, SLE04, DM03, DM05, DM05, DM23, DM24, and AIBD04 showed overexpression of retrotransposons and type I and/or II IFNs but no consistent upregulated expression of nRTKs, TFs or ISGs. In addition, high induction of TFs, particularly STAT1 and STAT2, was detected in the SLE patients, whereas STAT2 was selectively induced in the DM patients (Fig. S2). Although high ISG induction was commonly found in —SLE patients, several patients, including AIBD06, AIBD13, DM02, DM17, DM18, DM21, and DM22, also showed high expression of some ISGs (Fig. S2). Taken together, these data showed that there was no significant correlation between IFNs and ISGs.
Correlation of IFNs and retrotransposons with downstream signaling
To evaluate the relationship between retrotransposons and IFNs, we performed Spearman’s rank correlation analysis. The correlations of LINE-1 with IFNs, nRTKs, TFs, and ISGs were analyzed in patients with DM, SLE or AIBD and HCs. First, we tested the correlation of retrotransposons with type I-III IFNs (Fig. 3A). Significant positive correlations between LINE-1 and type I IFNs were detected in DM samples (IFN-α2, ρ = 0.895, P < 0.0001; IFN-β1, ρ = 0.965, P < 0.0001), SLE samples (IFN-α2, ρ = 0.993 P < 0.0001; IFN-β1, ρ = 0.963, P < 0.0001), and AIBD samples (IFN-α2, ρ = 0.939, P < 0.0001; IFN-β1, ρ = 0.943, P < 0.0001). LINE-1 also showed positive correlations with IFN-λ1 in the HCs, AIBD patients and DM patients but not in the SLE patients. In contrast, LINE-1 showed no correlation with type II IFN-γ in patients with autoimmune diseases or HCs. Furthermore, we detected coordinated relationships among retrotransposons, such as LINE-1 and HERVK14C or SVA, in each type of autoimmune disease and HC sample (Fig. 3B). Similar to the correlation of LINE-1 and IFN-λ1, the correlation between STAT1 and ddx58 showed significant positive correlations in the DM, AIBD, and HC samples but not in the SLE samples (Fig. 3C). Among ISGs, both ddx58 with IFIT1 and ISG15 with OAS1 showed significant positive correlations in all types of autoimmune disease and HC samples (Fig. 3D).
Correlation of mRNA levels of retrotransposons, IFNs, STAT1, and ISGs in autoimmune diseases. (A) Type I and III IFNs, but not type II IFN, are correlated with LINE-1. The correlation of mRNA levels between LINE-1 and IFNs, such as IFN-α2, IFN-β1, IFN-γ, and IFN-λ1, was analyzed. (B) Synchronized expression of LINE-1 and retrotransposons. The correlations of the mRNA levels between LINE-1 vs. HERVK14C and SVA retrotransposons are shown. (C) There was no positive correlation between STAT1 and ddx58 in SLE. The correlation of mRNA levels between STAT1 and ddx58, an ISG, was examined. (D) Positive correlation in ISGs. The correlations of mRNA levels between ISGs, such as ddx58 with IFIT1 and ISG15 with OAS1, were analyzed. (A–D) Representative correlation coefficients are shown between two genes in each group, and ρ-values were determined by nonparametric Spearman’s rank correlation test. The numbers of samples: HC (n = 10), SLE (n = 19), AIBD (n = 14), and DM (n = 24).
The correlation of each gene in the autoimmune disease and HC samples is summarized in a correlation heat map (Fig. 4 and Tables S2–S5). Retrotransposons and type I IFN showed a clear, positive correlation regardless of disease. On the other hand, the correlation of retrotransposons and nRTKs was different depending on the disease. Retrotransposons showed significant positive correlations with JAK1 and TYK2 in DM samples and only JAK1 in SLE samples, not in AIBD or HC samples (Tables S2–S5). Although retrotransposons did not correlate with TFs or ISGs, they showed coordinated expression with each gene, especially those expressed in HC and DM (Fig. 4). These results strongly suggested that retrotransposons and type I IFN production are positively synchronized; however, retrotransposon expression is not positively correlated with IFN receptor downstream signaling, except for that of some nRTKs.
Correlation heat map of transcripts in HCs and patients with autoimmune diseases. A hierarchically clustered heat map matrix showing the correlation between the expression of each gene. The ρ-values, which were calculated by the nonparametric Spearman’s rank correlation test, are represented as colored bars.
Mutual regulation of retrotransposons and IFNs in cultured cells
Finally, we investigated the correlation between IFNs and retrotransposons in vitro using four established human adherent cell lines, NP-2, A431, G361, and HaCaT cells. Interestingly, enhanced expression levels of LINE-1, HERVK14C, IFN-α2, and IFN-β1 were detected in A431 cells (Fig. 5A). Furthermore, relatively high expression levels of TFs and ISGs were detected in HaCaT and A431 cells but not in NP-2 or G361 cells. The expression of LINE-1 and IFN-α2 or IFN-β1 was positively correlated in HaCaT and A431 cells (Fig. 5B). The methylation level of the LINE-1 promoter, which was analyzed by pyrosequencing, was significantly reduced in A431 cells compared to that in NP-2 and HaCaT cells (Fig. 5C).
Correlated expression of LINE-1 and type I IFN in cell lines. (A) Retrotransposons and type I IFNs are upregulated in A431 cells. The mRNA levels of retrotransposons, TFs, and ISGs in the indicated cell lines were examined by qPCR. (B) Positive correlation of LINE-1 and IFNs in cell lines. The correlation of the relative mRNA levels between LINE-1 vs. IFN-α2 or IFN-β1 was analyzed, and Pearson correlation coefficients (r-values) are shown. (C) Methylation in the LINE-1 promoter was reduced in A431 cells. The methylation levels in three sites of the LINE-1 promoter were analyzed by pyrosequencing. (D) Poly(I:C) induces the expression of LINE-1 and IFN-β1. A431 and NP-2 cells were treated with 30 ng/ml poly(I:C) for 12 and 24 h, and the relative mRNA expression levels of LINE-1 and IFN-β1 were examined by qPCR. (E) Hypomethylation enhances LINE-1 and IFN-β1. A431 and NP-2 cells were treated with 100 μM 5AzaC for 24 h, and the expression levels of LINE-1 and IFN-β1 were examined by qPCR. Data are shown as the mean ± SEM by one-way ANOVA with Tukey’s post hoc test (A,C) or Student’s t-test (D,E), n = 3. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, ns; not significant.
Poly(I:C), a synthetic analog of double-stranded RNA, is a potent inducer of type I IFNs29,30. Upon stimulation with poly(I:C), the expression of LINE-1, as well as IFN-β1, was remarkably increased in A431 and NP-2 cells (Fig. 5D). 5AzaC, a demethylation agent, causes hypomethylation and induction of LINE-131. Treatment with 5AzaC elevated the mRNA levels of both LINE-1 and IFN-β1 in A431 and NP-2 cells (Fig. 5E). These results revealed the mutual regulation of LINE-1 and type I IFN in cultured cells. Finally, the coordinated expression of retrotransposons and type I IFN was observed not only in the blood cells of patients receiving treatment but also in adherent culture cells in vitro.
Discussion
In this study, we detected elevated levels of type I IFN mRNA in blood samples of patients with DM and AIBD. Interestingly, the expression of retrotransposons and type I IFNs showed synchronicity regardless of disease, suggesting that various pathways of type I IFN activation may be mutually correlated with retrotransposon activation. In DM and AIBD, the type I IFN pathway may be continuously activated at a low level in patients undergoing treatment (Figs. 1D and 2). Our results suggested that the altered expression of retrotransposons may induce constitutive type I IFN signaling prematurely, and additional factors, such as viral infections, may further upregulate the type I IFN pathway32, leading to the development of autoimmune diseases. In contrast, type II IFN activation was not correlated with retrotransposon expression (Figs. 3 and 4), indicating that IFN-γ expression is regulated by a different mechanism than that involved in upregulated retrotransposon expression. In addition to IFNs, the expression of various ISGs was coordinated; however, their expression was not correlated with that of IFNs (Fig. 4). A time lag between IFN production and receptor binding or prolonged expression of ISGs due to unphosphorylated STAT133 or positive feedback stimulated by STAT1 expression34 may account for this discrepancy. In addition, IFN-independent constitutive upregulation of STAT1/2 and IRF1 expression has been reported35. Phosphorylation was not required to realize this increase, but only protein expression was observed. A protective role against viral infection has also been reported for STAT1/2 and IRF136; these findings have not yet been reported in SLE or DM.
We showed the mutual regulation of LINE-1 and IFN using cultured cells, and this finding was compatible with the data obtained with the clinical samples. The number of LINE-1 mRNA transcripts was increased in the kidneys of patients with lupus nephritis and in the minor salivary glands of patients with primary Sjögren’s syndrome, in concert with type I IFN expression23. Moreover, LINE-1 RNA contributes to the production of type I IFN through both the MDA5 and RIG-I pathways26, and LINE-1 induces type I IFN expression, which in turn regulates the activity and propagation of LINE-137. Thus, LINE-1 and type I IFN seem to be mutually regulated.
We showed the distinct expression patterns of retrotransposons and IFN-related genes between SLE and DM patients. The significant positive correlation between retrotransposons and type I IFN and the absence of a positive correlation between retrotransposons and IFN receptor downstream signaling, except for some nRTKs, are common to both SLE and DM. The difference between SLE and DM relates to INF expression; only type II IFN and IFN-γ expression was upregulated in SLE, and in contrast, the expression of all types of IFNs was upregulated in DM (Figs. 1D and 6A). In downstream signaling induced by the IFN receptor, retrotransposons showed significant positive correlations with JAK1 and TYK2 expression in DM and only with JAK1 expression in SLE. The expression of TFs and ISGs was upregulated in SLE, but only STAT2 and some ISGs (IFIH1, IFIT1, and IFI44 L) expression was upregulated in DM. In contrast, comparing the correlation of each gene, larger numbers of positive correlations were seen among genes in DM (Fig. 6B, Tables S4 and S5). ISGs were generally correlated with TFs in DM. Although STAT1 and IRF1 expression was were upregulated in SLE, no significant correlations were seen between STAT1 and ISGs or between IRF1 and ISGs (Fig. 6B). High STAT1 expression is a characteristic of SLE38. In accordance with our findings, Kartonitsch et al. suggested that the increased level of the STAT1 protein was involved in IFN-γ responsiveness and resulted in chronically activated IFN-γ signaling in SLE39. Subsequently, IFN-γ and downstream chemokines, including CXCL9 and CXCL10, are dysregulated prior to disease flares in SLE patients40,41,42,43. However, our results show no dysregulation in the expression of CXCL9 or CXCL10 from mRNA level in whole blood cells.
Scheme of retrotransposon-mediated IFN responses in SLE and DM. (A) The activation level of the IFN production pathway in patients with SLE and DM differed. Genes with significantly elevated and suppressed expression compared with those in the control group are highlighted in red and blue, respectively. (B) The correlation among genes. Genes with significantly elevated and suppressed expression compared with those in the control are highlighted in red and blue, respectively. The connected lines show the significant positive correlations between two genes calculated by a nonparametric Spearman’s rank correlation test. TE; transposable element, nRTK;: nonreceptor tyrosine kinase, TF; transcription factor.
The expression of JAK1 and JAK2 in SLE and TYK2 in DM was significantly downregulated, which might be due to the negative feedback of upregulated TFs. On the other hand, upregulation of LINE-1 and type I IFNs but no significant upregulation of the downstream components of the IFN pathway were observed in AIBD, which may reflect the fact that AIBD is an organ-specific autoimmune disease.
There are several clinical implications of this study. We noticed that the SLE flare was more frequent in the high LINE-1 and IFN groups (SLE Pt 3, 4, 7, 18 in Fig. S2 and Table S7), while it was not clear in the DM group. This result is consistent with the high prevalence of p40, encoded by LINE-1, in serum correlating with flares and high type I IFN scores in SLE44. On the other hand, SLE and DM samples showed similar but distinct cutaneous manifestations. The triggers for skin lesions depend on the disease: in DM, external stimuli (Köbner’s phenomenon) and, in SLE, sun exposure cause inflammation of the skin and infiltration of peripheral blood leukocytes that may secrete IFNs. The epidermis is thought to respond to these IFNs and produce ISGs. As shown in Fig. 1E, MxA expression was especially prominent in the basal layer of the epidermis, suggesting that peripheral blood cells infiltrating the epidermal-dermal interface influence MxA expression in the epidermis. Although we found no clear association between the expression of LINE-1/IFNs and cutaneous manifestations, we speculate that circulating peripheral blood cells may be involved in the skin rash: peripheral blood cells in SLE and DM secrete different cytokines and may cause different skin lesions.
Recently, antibody drugs such as anifrolumab against IFNAR and BIIB059 against the BDCA-2 antigen in pDCs have been developed to treat SLE45. Anifrolumab reduced the glucocorticoid dose and decreased the severity of skin disease in SLE patients46. Since STAT1/2 are significantly upregulated in SLE samples, an inhibitor of the JAK-STAT pathway is another promising therapeutic candidate. In addition, the expression of multiple IFNs, STAT2 and retrotransposons was upregulated in DM (Figs. 1A,D, 2B, and 6A), and JAK2 expression was high in DM and strongly associated with downstream TFs and ISGs (Fig. 6), suggesting that JAK2 inhibitors may be effective in DM. In this context, tofacitinib, a pan-JAK inhibitor, seems to be a possible therapeutic option for use against DM47,48. Although the mechanism by which retrotransposons induce the production of type I IFNs remains to be elucidated, inhibitors targeting retrotransposons and IFN interactions may be preferable drugs to treat DM.
In conclusion, our analyses of peripheral blood cells from SLE and DM patients receiving treatment revealed distinct expression patterns of retrotransposons and IFNs. The expression levels of retrotransposons and type I and III IFNs were generally correlated, and the upregulation of these genes may be partly responsible for the development of DM. Further investigations will clarify these relationships.
Materials and methods
Patients
Nineteen patients with SLE and 24 patients with DM were recruited from Gunma University. For comparison, we also investigated 14 patients with AIBD (bullous pemphigoid, 10; pemphigus vulgaris, 3; pemphigus foliaceus, 1) and 10 unaffected healthy controls. There was no racial difference among participants. Detailed information on the patients is shown in Supplementary Tables 6–8. We extracted RNA from peripheral blood cells from patients receiving treatment. The DM and healthy control samples (HC) were the same as those described previously32. We also collected biopsied skin samples from the lesions (erythema, vesicle and erosion) of 11 SLE patients, 9 AIBD patients, 3 DM patients, and 3 healthy individuals. Most of the patients under treatment had well controlled and stable disease. Informed consent was obtained from all subjects and/or their legal guardian(s). Ethics approval for this study was granted by the ethics commission of Gunma University (reference number: 1349) in accordance with the Declaration of Helsinki protocols.
DNA and RNA extraction from peripheral blood cells
DNA was extracted from the patients’ whole blood cells using a Wizard Genomic DNA Purification kit (Promega) according to the manufacturer’s instructions. RNA was extracted from the patients’ peripheral blood cells as follows: whole blood collection was performed with EDTA-treated tubes. After washing the blood cells three times with phosphate-buffered saline (PBS), the red blood cells were lysed with ammonium–chloride–potassium (ACK) lysing buffer. RNA was extracted from white blood cells using an RNeasy Mini Kit (Qiagen). Complementary DNA (cDNA) was synthesized using GoScript Reverse Transcriptase (Promega).
Detection of retrotransposons
LINE-1, human endogenous retrovirus (HERVK14C), and SVA were assessed using cDNA prepared from the patients’ blood samples by quantitative polymerase chain reaction (qPCR), as previously described26,49. To examine the expression of methylation enzymes, including IFNs, JAK family members, TFs, and ISGs, qPCR was performed using specific primer pairs (Table S9). In brief, qPCR was performed using a THUNDERBIRD Probe and SYBR qPCR Mix (Toyobo). The expression level relative to that of a housekeeping gene was used in the analyses. Real-time PCR was performed using an ABI 7300 PCR thermal cycler (Applied Biosystems) under the following conditions: 10 min at 95 °C, 40 cycles of 15 s at 95 °C, and 1 min at 60 °C. The data were analyzed using the ΔΔCt method.
Immunohistochemistry
Immunohistochemistry (IHC) was performed as described50,51. Paraffin embedded biopsy skin specimens from 11 SLE patients, 9 AIBD patients, and 3 DM patients before treatment and 3 healthy individuals were available. These samples were obtained from the same individuals whose peripheral whole blood samples were studied. Deparaffinized and rehydrated sections were autoclaved in 5 mM citrate buffer for 10 min at 121 °C for antigen retrieval. Endogenous peroxidase activity was blocked with peroxidase blocking solution (DAKO) for 5 min. The sections were incubated overnight at 4 °C with an anti-MxA antibody (1/100, sc-166412, Santa Cruz Biotechnology) and an anti-ISG15 antibody (1/50, ab131119, Abcam), followed by incubation for 1 h with a secondary antibody solution (anti-mouse K4000 and anti-rabbit K4002 antibodies, respectively, DAKO). Nuclear staining was performed using hematoxylin. The specificity of staining was confirmed using a matched isotype normal IgG as the first antibody. Immunoreactivity was evaluated in comparison with the staining patterns of skin specimens from normal individuals. The proportion (%) of positive epidermal cells was evaluated in each sample, and the mean proportion was assessed as follows: > 50%, 3; 50–20%, 2; < 20%,1; -,0. The intensity was expressed semiquantitatively using a four-level ordinal scale: 0, negative; 1, low; 2, intermediate; and 3, high. The scoring was performed independently by two observers (Y.K. and A.S.) (Table S1).
Quantification of L1 promoter methylation by pyrosequencing
A pyrosequencing-based methylation assay was used to assess the three CpG sites in the promoter region of LINE-1. To analyze LINE-1 promoter methylation, genomic DNA was extracted from peripheral whole blood cells and modified with sodium bisulfite using an EpiTect bisulfite kit (Qiagen). PCR and subsequent pyrosequencing for LINE-1 were performed with a PyroMark kit (Qiagen). The PCR products were immobilized and converted to the single-stranded form to act as templates in the pyrosequencing reaction using the Pyrosequencing Vacuum Workstation (Qiagen). The amount of C relative to the sum of the amounts of C and T at each CpG site was calculated. The average percentages of the relative amounts of C in the three CpG sites of LINE-1 were used as the overall LINE-1 methylation levels. The position was from 331 to 318 in GenBank accession number X58075. Samples were analyzed with a PyroMark Q24 system (Qiagen).
Cell culture, real-time PCR, and reagents
A431 (epidermoid carcinoma), G361 (malignant melanoma), NP-2 (glioma), and HaCaT (epidermal keratinocytes) cells were cultured in DMEM containing 10% fetal bovine serum, 100 IU/ml penicillin G and 100 μg/ml streptomycin at 37 °C in a 5% CO2 atmosphere. Cellular RNA was extracted from each cell line using an RNeasy mini kit (Qiagen). Complementary DNA was prepared with GoScript Reverse Transcriptase (Promega) using random primers. qPCR was performed by using THUNDERBIRD Probe and SYBR qPCR Mix (Toyobo), as described above. The cells were treated with 100 μM 5-azacytidine (5AzaC) (Cayman Chemical) or 30 ng/ml poly(I:C) (InvivoGen) for 12 and 24 h, and RNA was then extracted.
Statistical analyses
Two-group comparisons of continuous data were assessed using the Mann–Whitney U test because the data were not normally distributed. The correlations between the gene expression levels were determined using a nonparametric Spearman’s rank correlation test. The data are shown as the means ± SEM from experiments performed in triplicate, and p values were calculated using Student’s t-test or one-way ANOVA followed by post hoc Tukey HSD test for in vitro experiments. The correlations between the gene expression levels were determined using Pearson's correlation coefficient of in vitro experiments. P values < 0.05 were considered statistically significant.
References
Theofilopoulos, A. N., Kono, D. H. & Baccala, R. The multiple pathways to autoimmunity. Nat. Immunol. 18, 716–724. https://doi.org/10.1038/ni.3731 (2017).
Didier, K. et al. Autoantibodies associated with connective tissue diseases: What meaning for clinicians?. Front Immunol. 9, 541. https://doi.org/10.3389/fimmu.2018.00541 (2018).
Kiriakidou, M. & Ching, C. L. Systemic lupus erythematosus. Ann. Intern. Med. 172, Itc81–Itc96. https://doi.org/10.7326/aitc202006020 (2020).
Vera-Recabarren, M. A., García-Carrasco, M., Ramos-Casals, M. & Herrero, C. Comparative analysis of subacute cutaneous lupus erythematosus and chronic cutaneous lupus erythematosus: Clinical and immunological study of 270 patients. Br. J. Dermatol. 162, 91–101. https://doi.org/10.1111/j.13652133.2009.09472.x (2010).
Fujimoto, M., Watanabe, R., Ishitsuka, Y. & Okiyama, N. Recent advances in dermatomyositis specific autoantibodies. Curr. Opin. Rheumatol. 28, 636–644. https://doi.org/10.1097/BOR.0000000000000329 (2016).
Baum, S., Sakka, N., Artsi, O., Trau, H. & Barzilai, A. Diagnosis and classification of autoimmune blistering diseases. Autoimmun. Rev. 13, 482–489. https://doi.org/10.1016/j.autrev.2014.01.047 (2014).
Kershenovich, R., Hodak, E. & Mimouni, D. Diagnosis and classification of pemphigus and bullous pemphigoid. Autoimmun. Rev. 13, 477–481. https://doi.org/10.1016/j.autrev.2014.01.011 (2014).
Liao, A. P. et al. Interferon β is associated with type 1 interferon-inducible gene expression in dermatomyositis. Ann. Rheum. Dis. 70, 831–836. https://doi.org/10.1136/ard.2010.139949 (2011).
Postal, M. et al. Type I interferon in the pathogenesis of systemic lupus erythematosus. Curr. Opin. Immunol. 67, 87–94. https://doi.org/10.1016/j.coi.2020.10.014 (2020).
Baccala, R., Kono, D. H. & Theofilopoulos, A. N. Interferons as pathogenic effectors in autoimmunity. Immunol. Rev. 204, 9–26. https://doi.org/10.1111/j.0105-2896.2005.00252.x (2005).
Theofilopoulos, A. N., Baccala, R., Beutler, B. & Kono, D. H. Type I interferons (alpha/beta) in immunity and autoimmunity. Annu. Rev. Immunol. 23, 307–336. https://doi.org/10.1146/annurev.immunol.23.021704.115843 (2005).
Hillyer, P. et al. Expression profiles of human interferon-alpha and interferon-lambda subtypes are ligand- and cell-dependent. Immunol. Cell Biol. 90, 774–783. https://doi.org/10.1038/icb.2011.109 (2012).
Pestka, S., Krause, C. D. & Walter, M. R. Interferons, interferon-like cytokines, and their receptors. Immunol. Rev. 202, 8–32. https://doi.org/10.1111/j.0105-2896.2004.00204.x (2004).
Siegal, F. P. et al. The nature of the principal type 1 interferon-producing cells in human blood. Science 284, 1835–1837. https://doi.org/10.1126/science.284.5421.1835 (1999).
Schroder, K., Hertzog, P. J., Ravasi, T. & Hume, D. A. Interferon-gamma: An overview of signals, mechanisms and functions. J. Leukoc. Biol. 75, 163–189. https://doi.org/10.1189/jlb.0603252 (2004).
O’Shea, J. J. et al. The JAK-STAT pathway: Impact on human disease and therapeutic intervention. Annu. Rev. Med. 66, 311–328. https://doi.org/10.1146/annurev-med-051113-024537 (2015).
Platanitis, E. et al. A molecular switch from STAT2-IRF9 to ISGF3 underlies interferon-induced gene transcription. Nat. Commun. 10, 2921. https://doi.org/10.1038/s41467-019-10970-y (2019).
Schindler, C., Levy, D. E. & Decker, T. JAK-STAT signaling: From interferons to cytokines. J. Biol. Chem. 282, 20059–20063. https://doi.org/10.1074/jbc.R700016200 (2007).
Darnell, J. E. Jr., Kerr, I. M. & Stark, G. R. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science 264, 1415–1421. https://doi.org/10.1126/science.8197455 (1994).
Schneider, W. M., Chevillotte, M. D. & Rice, C. M. Interferon-stimulated genes: A complex web of host defenses. Annu. Rev. Immunol. 32, 513–545. https://doi.org/10.1146/annurev-immunol-032713-120231 (2014).
Chiche, L. et al. Modular transcriptional repertoire analyses of adults with systemic lupus erythematosus reveal distinct type I and type II interferon signatures. Arthritis Rheumatol. 66, 1583–1595. https://doi.org/10.1002/art.38628 (2014).
Munroe, M. E. et al. Altered type II interferon precedes autoantibody accrual and elevated type I interferon activity prior to systemic lupus erythematosus classification. Ann. Rheum. Dis. 75, 2014–2021. https://doi.org/10.1136/annrheumdis-2015-208140 (2016).
Mavragani, C. P. et al. Expression of long interspersed nuclear element 1 retroelements and induction of Type I interferon in patients with systemic autoimmune disease. Arthritis Rheumatol. 68, 2686–2696. https://doi.org/10.1002/art.39795 (2016).
Benihoud, K. et al. Unusual expression of LINE-1 transposable element in the MRL autoimmune lymphoproliferative syndrome-prone strain. Oncogene 21, 5593–5600. https://doi.org/10.1038/sj.onc.1205730 (2002).
Ray, D. A. & Batzer, M. A. Reading TE leaves: New approaches to the identification of transposable element insertions. Genome Res. 21, 813–820. https://doi.org/10.1101/gr.110528.110 (2011).
Zhao, K. et al. LINE1 contributes to autoimmunity through both RIG-I- and MDA5-mediated RNA sensing pathways. J. Autoimmun. 90, 105–115. https://doi.org/10.1016/j.jaut.2018.02.007 (2018).
Hata, K. & Sakaki, Y. Identification of critical CpG sites for repression of L1 transcription by DNA methylation. Gene 189, 227–234. https://doi.org/10.1016/s0378-1119(96)00856-6 (1997).
Okano, M., Bell, D. W., Haber, D. A. & Li, E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99, 247–257. https://doi.org/10.1016/s0092-8674(00)81656-6 (1999).
Field, A. K., Tytell, A. A., Lampson, G. P. & Hilleman, M. R. Inducers of interferon and host resistance: II: Multistranded synthetic polynucleotide complexes. Proc. Natl. Acad. Sci. U S A 58, 1004–1010. https://doi.org/10.1073/pnas.58.3.1004 (1967).
Kato, H. et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature 441, 101–105. https://doi.org/10.1038/nature04734 (2006).
Weber, B., Kimhi, S., Howard, G., Eden, A. & Lyko, F. Demethylation of a LINE-1 antisense promoter in the cMet locus impairs Met signalling through induction of illegitimate transcription. Oncogene 29, 5775–5784. https://doi.org/10.1038/onc.2010.227 (2010).
Kuriyama, Y. et al. The synchronized gene expression of retrotransposons and type I interferon in dermatomyositis. J. Am. Acad. Dermatol. https://doi.org/10.1016/j.jaad.2020.05.051 (2020).
Cheon, H. & Stark, G. R. Unphosphorylated STAT1 prolongs the expression of interferon induced immune regulatory genes. Proc. Natl. Acad. Sci. U S A 106, 9373–9378. https://doi.org/10.1073/pnas.0903487106 (2009).
Yuasa, K. & Hijikata, T. Distal regulatory element of the STAT1 gene potentially mediates positive feedback control of STAT1 expression. Genes Cells 21, 25–40. https://doi.org/10.1111/gtc.12316 (2016).
Michalska, A., Blaszczyk, K., Wesoly, J. & Bluyssen, H. A. R. A positive feedback amplifier circuit that regulates interferon (IFN)-stimulated gene expression and controls Type I and Type II IFN responses. Front Immunol. 9, 1135. https://doi.org/10.3389/fimmu.2018.01135 (2018).
Wang, W. et al. Unphosphorylated ISGF3 drives constitutive expression of interferon-stimulated genes to protect against viral infections. Sci. Signal https://doi.org/10.1126/scisignal.aah4248 (2017).
Yu, Q. et al. Type I interferon controls propagation of long interspersed element-1. J. Biol. Chem. 290, 10191–10199. https://doi.org/10.1074/jbc.M114.612374 (2015).
Goropevšek, A., Holcar, M. & Avčin, T. The role of STAT signaling pathways in the pathogenesis of systemic lupus erythematosus. Clin. Rev. Allergy Immunol. 52, 164–181. https://doi.org/10.1007/s12016-016-8550-y (2017).
Karonitsch, T. et al. Activation of the interferon-gamma signaling pathway in systemic lupus erythematosus peripheral blood mononuclear cells. Arthritis Rheum. 60, 1463–1471. https://doi.org/10.1002/art.24449 (2009).
Kong, K. O. et al. Enhanced expression of interferon-inducible protein-10 correlates with disease activity and clinical manifestations in systemic lupus erythematosus. Clin. Exp. Immunol. 156, 134–140. https://doi.org/10.1111/j.1365-2249.2009.03880.x (2009).
Ruffilli, I. MIG in cutaneous systemic lupus erythematosus. Clin. Ter. 170, e71–e76. https://doi.org/10.7417/ct.2019.2111 (2019).
Munroe, M. E. et al. Pathways of impending disease flare in African-American systemic lupus erythematosus patients. J. Autoimmun. 78, 70–78. https://doi.org/10.1016/j.jaut.2016.12.005 (2017).
Munroe, M. E. et al. Proinflammatory adaptive cytokine and shed tumor necrosis factor receptor levels are elevated preceding systemic lupus erythematosus disease flare. Arthritis Rheumatol. 66, 1888–1899. https://doi.org/10.1002/art.38573 (2014).
Carter, V. et al. High prevalence and disease correlation of autoantibodies against p40 encoded by long interspersed nuclear elements in systemic lupus erythematosus. Arthritis Rheumatol. 72, 89–99. https://doi.org/10.1002/art.41054 (2020).
Chaichian, Y., Wallace, D. J. & Weisman, M. H. A promising approach to targeting type 1 IFN in systemic lupus erythematosus. J. Clin. Invest. 129, 958–961. https://doi.org/10.1172/JCI127101 (2019).
Morand, E. F. et al. Trial of anifrolumab in active systemic lupus erythematosus. N. Engl. J. Med. 382, 211–221. https://doi.org/10.1056/NEJMoa1912196 (2020).
Moghadam-Kia, S., Charlton, D., Aggarwal, R. & Oddis, C. V. Management of refractory cutaneous dermatomyositis: Potential role of Janus kinase inhibition with tofacitinib. Rheumatology (Oxford) 58, 1011–1015. https://doi.org/10.1093/rheumatology/key366 (2019).
Paik, J. J. et al. Study of tofacitinib in refractory dermatomyositis (STIR): An open label pilot study of 10 patients. Arthritis Rheumatol. https://doi.org/10.1002/art.41602 (2020).
Tie, C. H. et al. KAP1 regulates endogenous retroviruses in adult human cells and contributes to innate immune control. EMBO Rep. https://doi.org/10.15252/embr.201745000 (2018).
Kato, M. et al. An autosomal recessive mutation of DSG4 causes monilethrix through the ER stress response. J. Invest. Dermatol. 135, 1253–1260. https://doi.org/10.1038/jid.2015.12 (2015).
Kuriyama, Y., Shimizu, A., Kanai, S. & Ishikawa, O. Expression of long interspersed nuclear elements-1 as a surrogate marker for global genomic hypomethylation of malignant skin tumors. J. Dermatol. Sci. 99, 213–215. https://doi.org/10.1016/j.jdermsci.2020.07.008 (2020).
Acknowledgements
The authors thank Dr. Ishibuchi, Dr. Sekiguchi and Dr. Kishi for their help in contacting the participants for enrollment and Ms. Okada for her wonderful support in designing the figures.
Funding
This study was supported by MEXT/JSPS KAKENHI (19K08766 to A.S.).
Author information
Authors and Affiliations
Contributions
All authors were involved in drafting the article or revising it critically for important intellectual content, and all authors approved the final version to be published. Study conception and design: Y.K., A.S., S.M., F.T., O.I. Acquisition of data: Y.K., D.O., S.K. Analysis and interpretation of data: Y.K., A.S., D.O., S.M., F.T., O.I.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Kuriyama, Y., Shimizu, A., Kanai, S. et al. Coordination of retrotransposons and type I interferon with distinct interferon pathways in dermatomyositis, systemic lupus erythematosus and autoimmune blistering disease. Sci Rep 11, 23146 (2021). https://doi.org/10.1038/s41598-021-02522-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-02522-6
- Springer Nature Limited