Abstract
This study investigated the impact of different ROI placement and analysis methods on the diagnostic performance of simplified IVIM-DWI for differentiating liver lesions. 1.5/3.0-T DWI data from a respiratory-gated MRI sequence (b = 0, 50, 250, 800 s/mm2) were analyzed in patients with malignant (n = 74/54) and benign (n = 35/19) lesions. Apparent diffusion coefficient ADC = ADC(0,800) and IVIM parameters D1′ = ADC(50,800), D2′ = ADC(250,800), f1′ = f(0,50,800), f2′ = f(0,250,800), and D*' = D*(0,50,250,800) were calculated voxel-wise. For each lesion, a representative 2D-ROI, a 3D-ROI whole lesion, and a 3D-ROI from “good” slices were placed, including and excluding centrally deviating areas (CDA) if present, and analyzed with various histogram metrics. The diagnostic performance of 2D- and 3D-ROIs was not significantly different; e.g. AUC (ADC/D1′/f1′) were 0.958/0.902/0.622 for 2D- and 0.942/0.892/0.712 for whole lesion 3D-ROIs excluding CDA at 1.5 T (p > 0.05). For 2D- and 3D-ROIs, AUC (ADC/D1′/D2′) were significantly higher, when CDA were excluded. With CDA included, AUC (ADC/D1′/D2′/f1′/D*') improved when low percentiles were used instead of averages, and was then comparable to the results of average ROI analysis excluding CDA. For lesion differentiation the use of a representative 2D-ROI is sufficient. CDA should be excluded from ROIs by hand or automatically using low percentiles of diffusion coefficients.
Similar content being viewed by others
Introduction
Diffusion-weighted imaging (DWI) is one of the most promising non-contrast techniques that can be readily implemented in standard liver magnetic resonance imaging (MRI) examinations allowing for lesion detection and differentiation1. In routine clinical practice the apparent diffusion coefficient (ADC) is usually calculated with b-values between 0 and 500–1000 s/mm2 assuming a mono-exponential relationship between signal intensity and the b-value2. However the ADC is not only influenced by molecular diffusion, but also by other (pseudo) random motion such as blood flow in small vessels within the tissue (perfusion). According to the intravoxel incoherent motion (IVIM) theory, diffusion and perfusion effects can be separated assuming a bi-exponential behavior of signal intensity, ultimately yielding the diffusion coefficient D, the pseudo-diffusion coefficient D* and the perfusion fraction f3,4,5,6,7. f is associated with microvessel density8,9. D* was negatively correlated with the interstitial fluid pressure (IFP), which influences blood flow10. The problems with IVIM in clinical liver MRI are long acquisition times and limited data quality caused by respiratory and cardiac motion and by low signal-to-noise ratio, which may lead to unstable fitting results, measurement errors and poor reproducibility11,12,13,14. Improved stability can be achieved by segmented fitting approaches, which decrease the degree of freedom by determining the parameters step by step15,16,17,18,19 or by simplified IVIM, which uses numerically stable computation of IVIM parameter estimations from 4 b-values20,21,22,23,24,25,26,27.
For quantitative analysis of ADC and IVIM parameter maps in lesions a region of interest (ROI) based approach is the most commonly used28,29,30. However, there are different ROI-placement and analysis strategies, mostly only investigated for ADC: to place the ROIs into areas with most restricted diffusion (“hot spots”, focused ROIs), to average over multiple small ROIs placed into different regions, to place a large ROI on a central slice of a lesion, or to cover the whole lesion7,21,23. Usually ROI-analysis is done by averaging the voxel values within the ROI (mean). However, in order to address tumor heterogeneity, also histogram-based approaches are employed to subclassify different tumor diffusion and perfusion environments7,31.
The purpose of this study was to investigate whether there are differences in the diagnostic accuracy of ADC and IVIM parameters in the discrimination of liver lesions using different ROI placement and analysis strategies. We compared 2D- and 3D-volume ROIs, inclusion and exclusion of central necrosis, cystic components and scars, and ROI analysis by averaging and histogram metrics.
Materials and methods
Study cohort
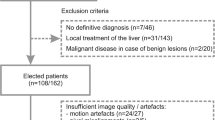
This single-center retrospective study was approved by the ethics committee of the University Hospital of the Rheinische Friedrich-Wilhelms University Bonn, Germany, with a waiver for written informed consent. Data of consecutive patients with focal hepatic lesions ≥ 1 cm undergoing clinical MRI examination of the liver including 4 b-value DWI from 2013 to 2016 were used. A flowchart of patient inclusion and exclusion is given in Fig. 1. Finally, data of 109/73 patients at 1.5/3.0 T were analyzed (Table 1). These two patient groups had already been examined in previous studies21,23. In those studies basic investigations concerning simplified IVIM for liver lesion characterization had been performed. In the present study, the data were used to investigate the influence of different ROI placement and analysis methods concerning diagnostic accuracy.
Diagnosis of liver lesions was undertaken within clinical routine. Cholangiocellular carcinomas (CCCs) were histologically proven. Hepatocellular carcinomas (HCCs) were either histologically proven or diagnosed according to the American Association for the Study for Liver Disease MRI criteria32. Diagnosis of metastasis was based on typical imaging features in combination with histologically proven primary cancer. Diagnosis of focal nodular hyperplasia (FNH) or haemangioma was established on the basis of typical radiological findings on contrast-enhanced MRI and was confirmed by at least one follow-up examination.
Magnetic resonance imaging
Imaging was performed on clinical whole-body 1.5/3.0-T MRI systems (Ingenia, Philips Healthcare; 1.5/3.0-T gradient system: 45/45 mT/m maximum amplitude, 200/200 T/m/s maximum slew rate; 3.0-T system with dual source RF transmission) using 32-channel abdominal coils with a digital interface for signal reception. The standardized imaging protocol included a DWI sequence with a respiratory-triggered single-shot spin-echo echo-planar imaging variant with four b-values (0, 50, 250, 800 s/mm2) before contrast agent administration (Table 2). For each slice, an isotropic diffusion-weighted image was reconstructed from the three images obtained for the different diffusion directions.
Postprocessing
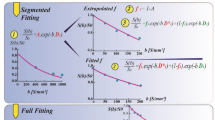
As described previously21,23, two different approximations of D and f were calculated from signal intensities S(b) and S(0) of the acquired b-values, one from b0 = 0, b1 = 50, b3 = 800 and one from b0 = 0, b2 = 250, b3 = 800 s/mm2:
From the four b-values, D* was approximated by using D2′ and f2′ and the reading for b1:
D*′ cannot be determined for all voxels, because some voxels are not affected by perfusion. Voxels with not defined values were excluded from ROI analysis.
Moreover, the conventional ADC was calculated:
Parameter maps and ROI analyses were calculated offline using custom written software in MATLAB (MathWorks, Natick, MA).
Image analysis
Image analysis was performed by a radiologist (N.M.) with 3 years of experience and checked by a radiologist (C.C.P.) with 10 years of experience in abdominal imaging and a physicist (P.M.) with more than 20 years of experience in DWI. All were blinded to clinical information.
One reference lesion per lesion type was analyzed. For each included lesion, 2D- and 3D-volume ROI-based analyses were performed. ROIs were placed as large as possible using DWI with highest contrast between lesion and normal tissue and excluding areas close to the lesion rim to avoid partial-volume effects. After the anatomical position of each ROI had been visually cross-checked for pixel misalignments between images with different b-values, the ROI was analyzed in the related parameter maps.
For 2D-analysis, one hand-drawn ROI was placed centrally in each lesion on a single representative slice (reference slice), which was largely unaffected by motion and susceptibility artifacts and pixel misalignments. For the 3D-volume analysis, a hand-drawn ROI was placed on each slice of the lesion. Slices with artifacts and pixel misalignments as well as the first and the last slice (due to potential partial volume effect) were marked as “bad”. An example of ROI placement is given in Fig. 2. Data from all slices (“good” and “bad”) were combined into a whole-lesion 3D-volume ROI (3DA). Furthermore, a second 3D-volume ROI was calculated including only the “good” slices (3DG). Thus, in each lesion three different ROI-sizes were investigated (2D, 3DA, 3DG).
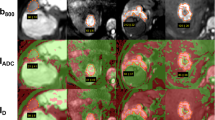
A typical example of 2D and 3D DWI IVIM analysis in a hepatocellular carcinoma at 1.5 T. Original diffusion-weighted images with b = 0, 50, 250, 800 s/mm2 are presented together with conventional ADC maps displayed as color-coded overlays over b800 images. For analysis, on each tumor-containing slice a region of interest (ROI) was selected, where ADC and IVIM parameters (not shown) were analyzed. ADC values are given in units of 10−6 mm2/s. Slices largely unaffected by artifacts were defined as good (“G”), slices close to the lesion’s rim (partial volume) or with images affected by artifacts (see red x) due to motion, susceptibility or pixel misalignments were defined as bad (“B”). One central “good” slice served as reference (“REF”) for the 2D analysis (see green frame), hereby slices in the lower part of the liver should be preferred due to lower motion influences from the heart. For 3D analysis, the voxels of the 2D ROI were combined with voxels of the ROIs on other “good” slices (3DG), voxels of all ROIs was used for whole lesion analysis (3DA).
For lesions with central necrosis, cystic components or scars (centrally deviating areas in DWI), the 2D- and 3D-ROI placements were repeated with exclusion of such areas. Two example analyses are given in Fig. 3. These measurements allowed the evaluation of different ROI sizes as well as of different lesion tissues included to the ROIs.
Typical examples of DWI IVIM analysis comparing in- and exclusion of necrosis in a metastasis of colorectal carcinoma (a) and of liquid in a hemangioma (b) at 1.5 T. For one central slice per lesion, original diffusion-weighted images with b = 0, 50, 250, 800 s/mm2 are presented together with conventional ADC, diffusion sensitive D1′ and D2′ parameter maps, and perfusion sensitive f1′, f2′, D*′ parameter maps. The parameter maps are displayed as color-coded overlays over b = 800. Values of ADC, D1′, D2′ and D*′ are given in units of 10–6 mm2/s, those of f1′ and f2′ in 10−3. If bad data quality led to negative parameter values or to not defined values, these voxels were not colorized. When necrosis/cystic components were excluded (“Without”) from regions of interests (ROIs), the diffusion sensitive parameters were significantly lower compared to inclusion (“With”). Perfusion sensitive parameters remained unchanged because there is only low perfusion in the metastasis and hemangioma anyway.
Finally, a histogram analysis was performed for each 2D-ROI. The following histogram metrics were calculated: median, standard deviation, the 5th, 10th, 25th, 75th, 90th, 95th percentiles, skewness and kurtosis.
Statistical analysis
Statistical analysis was performed using SPSS (Version 24.0, IBM) and pROC package (Version 1.16.2) in R (Version 3.6.1)33. Receiver operating characteristic (ROC) analysis was performed for liver lesions discrimination. Youden’s index was used to determine the optimal cut-off of the ROC curve providing the best trade-off between sensitivity and specificity. DeLong method was used to compare dependent ROC curves34. The area under the curve (AUC) based on mean ROI values was compared for the different ROI variants. Furthermore, it was investigated, whether AUC values can be improved by using one of the histogram metrics instead of the mean value. These investigations were carried out for both types of ROIs, including and excluding centrally deviating areas. In order to investigate whether histogram analyses may replace manual exclusion of such areas, additionally a comparison was performed using ROIs excluding such areas in case of mean values and including them in case of histogram metrics.
Ethical approval and informed consent
The presented study was approved by the institutional review board of the University of Bonn and hence all methods were performed in compliance with the ethical standards set in the 1964 Declaration of Helsinki as well as its later amendments. Written informed consent was waived.
Results
At 1.5/3.0 T, 74/54 malignant and 35/19 benign liver lesions were analyzed (Table 1). Mean volume of malignant lesions was 96.6/76.6 cm3 (range: 1.3–1715.7/1.2–521.2 cm3) and of benign lesions 72.1/20.4 cm3 (range: 0.9–856.3/1.1–118.3 cm3). Of these 109/73 lesions, 36/11 had centrally deviating areas. In total, 1333 ROIs were placed. The mean values of ADC and IVIM parameters for the benign and malignant lesion group together with the ROC analyses results for lesion differentiation are presented in Table 3. In Fig. 4 an overview to the obtained AUC values are given. In general, the values of diffusion and perfusion sensitive parameters were lower in malignant lesions than in benign lesions.
The highest AUC values for lesion differentiation were found for ADC (0.967–0.911) and D1′ (0.941–0.857) followed by D2′ (0.919–0.816), f2′ (0.731–0.656), f1′ (0.673–0.616), and D*′ (0.563–0.515). For all parameters, diagnostic performance was compared for the different 2D- and 3D-ROI variants, for ROIs in- and excluding centrally deviating areas, and for mean values and histogram metrics.
Comparison of 2D- and 3D-ROIs
In Table 4 the results of the AUC value comparisons with respect to the different ROI types (2D, 3DG, 3DA) are presented. No significant differences were found in any of the comparisons, neither for ROIs that include centrally deviating areas, nor for those excluding such areas. The only exceptions were that AUC values for 3DA ROIs compared to those for 3DG ROIs were slightly larger in case of f1′ and f2′ at 1.5 T (for ROIs including centrally deviating areas: 0.712 vs 0.620 with p = 0.049 and 0.761 vs 0.675 with p = 0.031, respectively; for ROIs excluding those areas: 0.712 vs 0.622 with p = 0.055 and 0.773 vs 0.688 with p = 0.029, respectively), and in case of D2′ at 3.0 T, but only for ROIs including centrally deviating areas (0.895 vs 0.825 with p = 0.029).
Comparison of ROIs with included and excluded central necrosis, cystic components or scars
Table 5 summarizes the results of AUC value comparison with respect to included tissue. Exclusion of centrally deviating areas from ROIs yields larger AUC values of ADC, D1′, and D2′, for all 2D- and 3D-ROI variants. Improvements were significant at 1.5 T, at 3 T, however, sometimes only by tendency, potentially due to fewer cases with centrally deviating areas. For 2D-ROIs at 1.5 T for example, AUC values of ADC improved from 0.925 to 0.958 (p = 0.01), of D1′ from 0.866 to 0.902 (p = 0.0081), and of D2′ from 0.822 to 0.864 (0.00089). Perfusion parameters did not show any differences. Typical examples of DWI IVIM analysis comparing in- and exclusion of centrally deviating areas are presented in Fig. 3.
Comparison of mean values versus histogram analysis
Table S1 gives the mean values and values of histogram metrics for the benign and malignant lesion group together with the ROC analyses results for lesion differentiation using 2D-ROIs. In Table S2 the results of the different AUC value comparisons are given.
At 1.5 T, the 5th and 10th percentiles of ADC and D1′ and the 25th percentiles of ADC, D1′ and D2′ lead to significantly higher AUC values than the mean values for ROIs including centrally deviating areas. For example, by using the 10th percentile instead of mean value, AUC values could be improved for ADC from 0.925 to 0.969 (p = 0.018), for D1′ from 0.866 to 0.926 (p = 0.0042), and for D2′ from 0.822 to 0.856 (p = 0.074). For ROIs excluding centrally deviating areas, these improvements were observed to a lesser degree. For example, by using the 10th percentile instead of mean value, AUC values could only be improved for ADC from 0.958 to 0.975 (p = 0.13) and for D1′ from 0.902 to 0.935 (p = 0.038) and not for D2′. The additional comparison using ROIs excluding centrally deviating areas in case of mean value analysis and including such areas in case of histogram analysis, no significant differences were found for ADC, D1′ and D2′. This means, that the use of low percentiles can replace the elaborate exclusion of centrally deviating areas by hand without reducing the diagnostic accuracy. At 3.0 T, where there were fewer cases with centrally deviating areas, similar results were obtained but with higher p-values.
At both field strengths, the 5th and 10th percentiles of D*′ lead to significantly higher AUC values than the mean values, regardless of whether centrally deviating areas were included or excluded or excluded only in case of mean value analysis. For example, by using the 5th percentile instead of the mean value, AUC values could be improved from 0.515 to 0.646 (p = 0.00085) at 1.5 T and from 0.559 to 0.717 (p = 0.0079) at 3.0 T for ROIs excluding centrally deviating areas. This behavior also tended to be observed for f1′. For example, by using the 5th percentile instead of the mean value, AUC values could be improved from 0.622 to 0.708 (p = 0.034) at 1.5 T and from 0.661 to 0.681 (p = 0.74) at 3.0 T for ROIs excluding centrally deviating areas. All other histogram metrics including skewness and kurtosis performed with lower or not significantly different AUC values compared to the ROI mean values.
Discussion
The main findings of the present study were: (1) No significant differences in diagnostic performance were found between 2D- and 3D-ROIs even if only slices with good image quality were included. (2) Differentiation was more accurate when centrally deviating areas were excluded from ROIs. (3) When such areas were included, diagnostic accuracy of diffusion sensitive parameters was improved by histogram analysis of the ROIs using low percentiles instead of mean values. (4) Diagnostic accuracy of perfusion parameters, especially of D*′ was improved by histogram analysis using low percentiles instead of mean values, regardless of whether centrally deviating areas were in- or excluded.
To our knowledge, to date no systematic evaluation of different ROI placement and analysis methods for liver lesion analysis by IVIM-derived DWI parameters has been performed. However, it is important for potential clinical use of IVIM DWI techniques for lesion characterization to establish an appropriate ROI placement and analysis strategy as simple as possible that leads to highest possible diagnostic accuracy.
The technically simplest way for ROI placement in clinical practice is to draw a single 2D-ROI on a representative slice encompassing the whole lesion including centrally deviating areas. In scientific studies, however, 3D-volume ROIs are often used e.g. together with automated segmentation software. In the present work we performed comparisons with respect to ROI-type (2D on a reference slice, 3DA for whole-tumor volume, 3DG considering only “good” slices) and tumor tissue by inclusion and exclusion of centrally deviating areas. For different ROI-types, we did not find significant differences in diagnostic accuracy of ADC and IVIM parameters. Compared to 3D-whole-lesion ROIs (3DA), the inclusion of only “good” slices (3DG) or the selection of a ROI on a reference slice (2D) was expected to improve diagnostic accuracy due to less influence of artifacts, pixel misalignments and partial volume effects. However, this effect was hard to find. One reason might be that in case of whole-tumor 3DA volumes negative influences by “bad” slices were compensated by improved statistics due to higher number of included voxels compared to 3DG and 2D. More voxel averaging and thus a better noise robustness was noticeable especially in small lesions (see Table S3). A previous study on prostate cancer also yielded no improved diagnostic performance using 3D-ROIs instead of 2D-ROIs35. Although further studies on a larger population with liver lesions are needed to confirm the finding of this study, the analysis of a central representative slice of “good” image quality seems to be sufficient for reliable lesion discrimination and is applicable in clinical practice and less time consuming.
The exclusion of centrally deviating areas significantly improves the diagnostic accuracy of diffusion parameters, as was to be expected. For perfusion parameters no differences were found. A previous study on breast lesions, also found improved accuracy of differential diagnosis for ADC in ROIs including only viable tissue instead of whole tumor29. Necrosis, cystic areas and scars increase the diffusion coefficient of a lesion at random due to the admixture of varied proportions of high values. Especially in case of necrosis, the malignancy of tumors may be masked by measurement of a higher ADC due to varying amounts of necrotic tissue. Perfusion parameters, in contrast, are low in necrosis which further reduces the already small values in malignant tumors. In liver metastases, a correlation was found between diffusion parameters and liver tumor necrosis, but not for perfusion parameters36.
For lesion assessment, the exclusion of centrally deviating areas is more time consuming and, therefore, not a routine clinical practice and can be challenging for unexperienced radiologists. Thus, automated segmentation would be helpful. In this respect, histogram analysis can provide additional quantitative metrics beyond the mean value of a ROI, which reflect the heterogeneity of pathologic changes without additional imaging7. In our study, histogram analysis of ROIs including centrally deviating areas showed that low percentiles led to similar diagnostic accuracy for ADC and diffusion coefficients than mean value analysis of ROIs without such areas. Thus, this method may be of use to automatically determine voxels of viable tumor for ADC and IVIM analysis. In some other studies, it was also shown that diagnostic accuracy of ADC and D in whole-lesion ROI analysis was improved when low percentiles were used instead of mean values, e.g. in predicting microvascular invasion of hepatocellular carcinoma37, differentiation of malignancy in breast and testicular lesions31,38, differentiating of different grades of prostate cancer39, and gliomas40,41,42.
Furthermore, of special interest is the finding that for the perfusion parameters, especially D*, diagnostic accuracy in lesion discrimination was significantly improved by the use of low percentiles instead of mean values regardless of whether centrally deviating areas were included or excluded or excluded only in case of mean value analysis. Because D* depends on blood flow velocity and length of microvessel segments3,4, this may indicate that differences between benign and malignant lesions exist especially for small vessels. Other studies investigating histogram analysis for IVIM perfusion parameters in liver lesions are rare. There is one other study investigating hepatocellular carcinoma with and without microvascular invasion, but no significant differences were found for parameters D* and f, neither for mean values nor for low percentiles37.
This study has several limitations. First, it was a retrospective study with inherent methodological limitations. For example, due to the lack of raw data, no motion correction of the individual images43 could be performed before averaging. Second, although the total number of lesions included was relatively large, only common lesion types were analyzed, which may affect the generalizability of the results. Also, there was a relatively small number of patients who underwent MRI examination at 3.0 T MRI system and, therefore, statistical power was lower compared to 1.5 T. We included a typical clinical patient cohort of a large tertiary reference center so that not only large lesions were included. Therefore, a study including more large lesions may show differences between 2D- and 3D-volume measurements. On the other hand, not even tendencies concerning differences of 2D- and 3D-ROIs were found in the present study.
In conclusion, using representative 2D-ROIs seems to be sufficient for reliable liver lesion discrimination in routine clinical practice. Central necrosis, cystic components or scars should be excluded from ROIs either by hand or by computing low percentiles of diffusion coefficients instead of mean values.
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Shenoy-Bhangle, A., Baliyan, V., Kordbacheh, H., Guimaraes, A. R. & Kambadakone, A. Diffusion weighted magnetic resonance imaging of liver: Principles, clinical applications and recent updates. World J. Hepatol. 9, 1081–1091 (2017).
Thoeny, H. C. & Ross, B. D. Predicting and monitoring cancer treatment response with diffusion-weighted MRI. J. Magn. Reson. Imaging 32, 2–16 (2010).
Le Bihan, D. et al. Separation of diffusion and perfusion in intravoxel incoherent motion MR imaging. Radiology 168, 497–505 (1988).
Koh, D. M., Collins, D. J. & Orton, M. R. Intravoxel incoherent motion in body diffusion-weighted MRI: Reality and challenges. Am. J. Roentgenol. 196, 1351–1361 (2011).
Koh, D. M. & Collins, D. J. Diffusion-weighted MRI in the body: Applications and challenges in oncology. Am. J. Roentgenol. 188, 1622–1635 (2007).
Koh, D. M. Science to practice: Can intravoxel incoherent motion diffusion-weighted MR imaging be used to assess tumor response to antivascular drugs?. Radiology 272, 307–308 (2014).
Padhani, A. R. et al. Diffusion-weighted magnetic resonance imaging as a cancer biomarker: Consensus and recommendations. Neoplasia 11, 102–125 (2009).
Meyer, H.-J. et al. Associations between IVIM histogram parameters and histopathology in rectal cancer. Magn. Reson. Imaging 77, 21–27 (2021).
Lee, H. J. et al. Tumor perfusion-related parameter of diffusion-weighted magnetic resonance imaging: Correlation with histological microvessel density. Magn. Reson. Med. 71, 1554–1558 (2014).
Kim, S. et al. Interstitial fluid pressure correlates with intravoxel incoherent motion imaging metrics in a mouse mammary carcinoma model. NMR Biomed. 25, 787–794 (2012).
Lee, Y. et al. Intravoxel incoherent motion diffusion-weighted MR imaging of the liver: Effect of triggering methods on regional variability and measurement repeatability of quantitative parameters. Radiology 274, 405–415 (2015).
Cho, G. Y. et al. A versatile flow phantom for intravoxel incoherent motion MRI. Magn. Reson. 67, 1710–1720 (2012).
Kakite, S. et al. Hepatocellular carcinoma: Short-term reproducibility of apparent diffusion coefficient and intravoxel incoherent motion parameters at 3.0T. J Magn Reson Imaging 41, 149–156 (2015).
Andreou, A. et al. Measurement reproducibility of perfusion fraction and pseudodiffusion coefficient derived by intravoxel incoherent motion diffusion-weighted MR imaging in normal liver and metastases. Eur Radiol 23, 428–434 (2013).
Wang, M. et al. Evaluation of hepatic tumors using intravoxel incoherent motion diffusion-weighted MRI. Med. Sci. Monit. 22, 702–709 (2016).
Colagrande, S. et al. Focal liver lesion classification and characterization in noncirrhotic liver: A prospective comparison of diffusion-weighted magnetic resonance-related parameters. J. Comput. Assist. Tomogr. 37, 560–567 (2013).
Doblas, S. et al. Determination of malignancy and characterization of hepatic tumor type with diffusion-weighted magnetic resonance imaging: Comparison of apparent diffusion coefficient and intravoxel incoherent motion-derived measurements. Invest. Radiol. 48, 722–728 (2013).
Ichikawa, S. et al. Intravoxel incoherent motion imaging of focal hepatic lesions. J. Magn. Reson. Imaging 37, 1371–1376 (2013).
Luo, M., Zhang, L., Jiang, X. & Zhang, W. Intravoxel incoherent motion diffusion-weighted imaging: Evaluation of the differentiation of solid hepatic lesions. Transl. Oncol. 10, 831–838 (2017).
Coenegrachts, K. et al. Evaluation of true diffusion, perfusion factor, and apparent diffusion coefficient in non-necrotic liver metastases and uncomplicated liver hemangiomas using black-blood echo planar imaging. Eur. J. Radiol. 69, 131–138 (2009).
Mürtz, P. et al. Accurate IVIM model-based liver lesion characterisation can be achieved with only three b-value DWI. Eur. Radiol. 28, 4418–4428 (2018).
Mürtz, P. et al. Intravoxel incoherent motion model-based analysis of diffusion-weighted magnetic resonance imaging with 3 b-values for response assessment in locoregional therapy of hepatocellular carcinoma. OncoTargets Ther. 9, 6425–6433 (2016).
Mürtz, P. et al. Is liver lesion characterisation by simplified IVIM DWI also feasible at 3.0 T?. Eur. Radiol. 29, 5889–5900 (2019).
Pieper, C. C. et al. Incidence and risk factors of early arterial blood flow stasis during first radioembolization of primary and secondary liver malignancy using resin microspheres: An initial single-center analysis. Eur. Radiol. 26, 2779–2789 (2016).
Pieper, C. C. et al. Evaluation of a simplified intravoxel incoherent motion (IVIM) analysis of diffusion-weighted imaging for prediction of tumor size changes and imaging response in breast cancer liver metastases undergoing radioembolization: A retrospective single center analysis. Medicine (Baltimore) 95, 1–9 (2016).
Pieper, C. et al. The value of intravoxel incoherent motion model-based diffusion-weighted imaging for outcome prediction in resin-based radioembolization of breast cancer liver metastases. OncoTargets Ther. 9, 4089–4098 (2016).
Pieper, C. C., Sprinkart, A. M., Kukuk, G. M. & Mürtz, P. Short-term measurement repeatability of a simplified intravoxel incoherent motion (IVIM) analysis for routine clinical diffusion-weighted imaging in malignant liver lesions and liver parenchyma at 1.5 T. RöFo Fortschritte Auf Dem Geb. Röntgenstrahlen Bildgeb. Verfahr. 191, 199–208 (2019).
Ma, C. et al. Effect of region of interest size on ADC measurements in pancreatic adenocarcinoma. Cancer Imaging 17, 13 (2017).
Gity, M., Moradi, B., Arami, R., Arabkheradmand, A. & Kazemi, M. A. Two different methods of region-of-interest placement for differentiation of benign and malignant breast lesions by apparent diffusion coefficient value. Asian Pac. J. Cancer Prev. 19, 2765–2770 (2018).
Bickel, H. et al. Diffusion-weighted imaging of breast lesions: Region-of-interest placement and different ADC parameters influence apparent diffusion coefficient values. Eur. Radiol. 27, 1883–1892 (2017).
Suo, S. et al. Characterization of breast masses as benign or malignant at 3.0T MRI with whole-lesion histogram analysis of the apparent diffusion coefficient. J. Magn. Reson. Imaging 43, 894–902 (2016).
Bruix, J. & Sherman, M. Management of hepatocellular carcinoma: An update. Hepatology 53, 1020–1022 (2011).
Robin, X. et al. pROC: An open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics 12, 1–8 (2011).
DeLong, E. R., DeLong, D. M. & Clarke-Pearson, D. L. Comparing the areas under two or more correlated receiver operating characteristic curves: A nonparametric approach. Biometrics 44, 837–845 (1988).
Tamada, T. et al. Apparent diffusion coefficient values of prostate cancer: Comparison of 2D and 3D ROIs. Am. J. Roentgenol. 210, 113–117 (2018).
Chiaradia, M. et al. Intravoxel incoherent motion (IVIM) MR imaging of colorectal liver metastases: Are we only looking at tumor necrosis?. J Magn Reson Imaging 39, 317–325 (2014).
Li, H. et al. Preoperative histogram analysis of intravoxel incoherent motion (IVIM) for predicting microvascular invasion in patients with single hepatocellular carcinoma. Eur. J. Radiol. 105, 65–71 (2018).
Fan, C. et al. Discrimination between benign and malignant testicular lesions using volumetric apparent diffusion coefficient histogram analysis. Eur. J. Radiol. 126, 108939 (2020).
Donati, O. F. et al. Prostate cancer aggressiveness: Assessment with whole-lesion histogram analysis of the apparent diffusion coefficient. Radiology 271, 143–152 (2014).
Horvath-Rizea, D. et al. The value of whole lesion ADC histogram profiling to differentiate between morphologically indistinguishable ring enhancing lesions-comparison of glioblastomas and brain abscesses. Oncotarget 9, 18148–18159 (2018).
Lu, S. S. et al. Histogram analysis of apparent diffusion coefficient maps for differentiating primary CNS lymphomas from tumefactive demyelinating lesions. Am. J. Roentgenol. 204, 827–834 (2015).
Kang, Y. et al. Gliomas: Histogram analysis of apparent diffusion coefficient maps with standard- or high-b-value diffusion-weighted MR imaging—Correlation with tumor grade. Radiology 261, 882–890 (2011).
Pathak, R. et al. Considering tumour volume for motion corrected DWI of colorectal liver metastases increases sensitivity of ADC to detect treatment-induced changes. Sci. Rep. 9, 3828 (2019).
Funding
Open Access funding enabled and organized by Projekt DEAL.
Author information
Authors and Affiliations
Contributions
N.M.: Conception and design of the study, data acquisition, analysis and interpretation, drafted the manuscript. P.M.: Conception and design of the study, data acquisition, analysis and interpretation, drafted the manuscript. A.M.S.: Data analysis and interpretation, critical revision of the manuscript. W.B.: Data analysis and interpretation, critical revision of the manuscript. J.A.L.: Data interpretation, critical revision of the manuscript. U.A.: Data interpretation, critical revision of the manuscript. C.C.P.: Conception and design of the study, data acquisition, analysis and interpretation, critical revision of the manuscript. All authors: Approval of final version of submitted manuscript. P.M. and C.C.P. are guarantors of integrity of the entire study.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Mesropyan, N., Mürtz, P., Sprinkart, A.M. et al. Comparison of different ROI analysis methods for liver lesion characterization with simplified intravoxel incoherent motion (IVIM). Sci Rep 11, 22752 (2021). https://doi.org/10.1038/s41598-021-01108-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-01108-6
- Springer Nature Limited