Abstract
In human gliomas, anti-tumor T cell responses are inhibited through induction of local and systemic immunosuppression. Immune checkpoint blockade is proving to be a success in several types of cancers. However, many studies reported that the treatment of glioblastoma patients with anti-CTLA-4 or anti-PD-1 has no survival benefit compared to standard chemotherapy. This study aimed to investigate the expression and role of VISTA, a newly described immune checkpoint regulator, in human gliomas. mRNA expression was assessed in a total of 87 samples from glioma patients. 57 glioma tissues were taken at different grades. 20 peripheral blood mononuclear cells (PBMC) samples were taken before surgery and ten after surgery, all from the same set of patients. As for the control, ten specimens of PBMC were taken from healthy donors. Protein expression using immunohistochemistry was performed for 30 patients. The Cancer Genome Atlas (TCGA) data set, was also used to investigate VISTA expression through analysis of RNA-seq data of 667 glioma patients. In the Moroccan cohort, VISTA gene expression was significantly upregulated in glioma tissues related to PBMC of healthy donors. This high expression was specific to patient tissues since VISTA expression in PBMC was low when assessed either before or after surgery. Besides, VISTA exhibited higher expression levels in grade III/IV relative to grade I/II glioma patients. Interestingly, VISTA correlated positively with PD-1 expression. PD-1 also showed elevated expressions in higher glioma grades. The TCGA cohort corroborated these observations. Indeed, VISTA was also found to be strongly expressed in high grades. It was positively correlated with other critical immune checkpoints. Finally, increased VISTA transcript levels were associated with weak overall survival of glioma patients. Our study highlighted a correlation between high levels of VISTA expression and poor prognosis in glioma patients. VISTA might be involved in glioma progression and could be considered as a possible new therapeutic target, especially in advanced gliomas.
Similar content being viewed by others
Introduction
Gliomas are the most frequent and violent primary brain tumors in adults1. Among glioma types, glioblastoma (GBM) is the most common and invasive type. Despite the multimodal-conventional therapy such as neurosurgical resection and radical or chemotherapy, this pathology remains a significant cause of death in human cancer, with a median survival of only 14.6 months2. For a decade, several studies on molecular markers and targeted drugs, generated restricted effect in extending life of glioma patients. The discovery of intracranial lymphatic system has brought a new theoretical basis and a new hope for brain tumor immunotherapy3.
In the past few years, a collection of data has clarified the crucial role of the immune checkpoints in regulating the immune response in different cancer types. However, research on immunotherapy of glioma has extended in an exponential manner4,5,6,7. The majority of gliomas is obstinate to usual immunotherapies. Most of glioma patients did not respond to the blockade of habitual immune checkpoints pathways (CTLA-4 and PD1/PD-L1)8,9,10. This has heightened our interest in finding novel immune checkpoints whose targeting could be beneficial for glioma patients.
Programmed Death 1 (PD-1) protein is a co-inhibitory receptor which is expressed on activated T cells, B cells, macrophages, dendritic cells (DC) and monocytes11,12. It has been demonstrated that PD-1 inhibits adaptive and innate immune responses when coupling to its ligands PDL-1 and PD-L2, which are expressed mainly by tumor cells11,13. PD-L1 can block the cytolytic activity of PD-1+ tumor-infiltrating CD4+ and CD8+ T cells and cytokine production12. Besides, the latest clinical trials showed that neutralizing monoclonal antibodies (mAbs) against PD-1 or PD-L1 resulted in an impressive anti-tumor effect in various types of solid tumors with complete regression in some patients14.
V-domain Immunoglobulin suppressor of T cell activation (VISTA) is a new Immunoglobulin (Ig) superfamily ligand, which was recently discovered15. VISTA (also known as c10orf54 or PD-1H) shares sequence homology with PD-1 and PD-L1 and can act as a receptor on T lymphocytes or a ligand on antigen-presenting cells16.
VISTA expression is observed mostly in most immune cells, including CD4+ and CD8+ T cells, NK cells, macrophages, DCs and neutrophils, but not B cells. However, VISTA-knockout (KO) mice developed a spontaneous accumulation of activated T cells in multiple organs17. Also, VISTA–KO mice are resistant to the growth of GL261 glioma18.
Here, we reported a significant expression profile of VISTA in advanced versus primary glioma grades. VISTA, whose expression correlated with CD8 T cell presence in glioma patients, appeared to be one of the most highly expressed immune checkpoints in this tumor microenvironment. Interestingly, it was found that high VISTA expression is associated with poor patient’s outcome, which strongly suggests that VISTA could be considered as a new potential therapeutic target in advanced gliomas.
Materials and methods
Patients and samples
mRNA expression was assessed in a total of 87 samples from glioma patients. There were 57 glioma tissues at different grades:
-
24 grade IV (Glioblastoma), 6 grade III (3 Astrocytomas and 3 Ependymomas), 9 grade II (1 Astrocytoma, 4 Ependymomas, 3 Oligodendrogliomas and 1 Xantoastrocytoma), 18 grade I (16 Astrocytomas and 2 Gangliogliomas).
-
20 peripheral blood mononuclear cell (PBMC) samples taken before surgery and 10 (PBMC) after surgery, all from the same set of patients, at the Ibn Rochd University Hospital, neurosurgery department (Casablanca, Morocco). As for the control, ten specimens of PBMC were taken from healthy donors, at Regional Blood Transfusion Center (Casablanca, Morocco). The inclusion criteria were adopted as follows: informed consent to participate in the study protocol, full documentation of the study. Patients had been previously diagnosed with glioma.
Selected patients have been already diagnosed with glioma and not been undergone any therapy before tumor resection. However, the exclusion criteria were: incomplete documentation of the study, no informed consent available to participate in the study protocol.
The samples were recruited from May 2016 until June 2019. All glioma tissues were graded according to the World Health Organization (WHO) 2007 and 2016, clinical information, including gender, age and smoking status was obtained from the medical records of the patients.
TCGA data analysis
The RNA-seq and clinicopathological characteristics data from 667 glioma samples were collected from The Cancer Genome Atlas (TCGA) dataset, graded according to the World Health Organization (WHO) from grade II to grade IV, were analyzed in our study (http://cancergenome.nih.gov). The Inclusion criteria adopted were full RNA-seq and clinicopathological characteristics information of each sample. The exclusion criteria were the lack of RNA-seq and clinicopathological characteristics information.
To confirm the generated results, data analysis and statistical tests were carried out by two different scientists in the lab. Gene expression profiling data were log- converted before data analysis.
Peripheral blood mononuclear cell isolation
Mononuclear cells were isolated by density gradient centrifugation according to Ficoll’s protocol as previously described19. 5 ml of human peripheral blood was first mixed with 5 ml of saline solution (0.9% NaCl). This mixture was then added to 5 ml of Ficoll medium (Biowest, France). The total was then centrifugated at 350g for 10 min. The layer corresponding to mononuclear cells was collected and washed twice in 0.9% NaCl.
Total RNA isolation and reverse transcription (RT)
Total RNA was extracted from PBMCs, and frozen glioma samples using TRIzol reagent (Invitrogen, France) as previously described20. RNA concentration and quality were measured using the NanoVueTM Plus Spectrophotometer (GE Healthcare, UK). According to the manufacturer’s instructions, cDNA first was synthesized using Tetro Reverse Transcriptase Enzyme (Bioline, France) from 0.5 μg of total RNA in a 20 μl reaction mixture with 1 μl Random Hexamer Primer 25 µg (Bioline, France) and 4 μl of RNase-Free Water added and incubated at 70 °C for 5 min to break the secondary structure of RNA. Then, the mixture was maintained on ice. 4 μl Tetro Reverse Transcriptase buffer, 4 μl of dNTP (10 mM), 0.5 μl of RNase Inhibitor (Invitrogen, France), 0.5 μl Tetro Reverse Transcriptase Enzyme (Bioline, France) and 1 μl of RNase-Free Water were added and incubated at 25 °C for 10 min, then at 45 °C for 30 min and then at 85 °C for 5 min.
Real-time RT-PCR assays
Relative quantification of gene expression was analyzed by real-time PCR in the presence of the fluorescent dye SYBR Green PCR Master Mix (Thermo Fischer). β-Actin was used as an internal control to evaluate the relative expression of VISTA and PD-1. Experiments were performed in a 20 μl reaction volume with specific primer pairs used at 10 µM for all genes.
PCR was programmed as follows: 10 min at 95 °C for polymerase activation and sample denaturation, then 40 cycles of 15 s at 95 °C and 60 s at 60 °C for annealing and extension. Fluorescence readings, at the end of the extension phase of each cycle, were used to estimate the values for the threshold cycles (Ct). The Ct values for each gene were converted into relative quantification (2−ΔCt).
Primer pairs were as follows:
-
β-Actin Forward: 5′-TGGAATCCTGTGGCATCCATGAAAC-3′.
Reverse: 5′-TAAAACGCAGCTCAGTAACAGTCCG-3′.
-
VISTA Forward: 5-TGTAGACCAGGAGCAGGATG-3′.
Reverse: 5-ATGCACCATCCAACTGTGTG-3′.
-
PD-1 Forward: 5′-GCTGGATTTCCAGTGGCGA-3′.
Reverse: 5′-ATGAGGTGCCCATTCCGCTA-3′.
Immunohistochemistry assays (IHC)
30 paraffin-embedded human glioma tissues (13 low grade and 17 high-grade cases) were sectioned (thickness of 3–4 µm). First, samples were incubated at 65 °C for 1 h and then at 37 °C overnight before being deparaffinized and rehydrated. For antigen retrieval step the water bath method was performed, using PT Link (Dako, Denmark) and a low pH (pH = 6) retrieval solution (EnVision Flex target retrieval solution low PH (× 50) 30 ml, Dako, Denmark) at 98 °C for 20 min.
For blocking the endogenous peroxidase activity, samples were immersed in 3% hydrogen peroxide (EnVision flex peroxidase-blocking reagent, Dako, Denmark) for 10 min at room temperature followed by incubation in wash buffer (EnVision flex wash buffer, Dako, Denmark). Two times for 2 min each to reduce non-specific binding. Slides then were incubated with a primary polyclonal rabbit anti-human VISTA antibody at 2 μg/ml (MyBioSource, San Diego, CA, USA). For each case, a second slide was used as a negative control with Rabbit IgG Isotype Control at 1:200 dilution (Clinisciences, France) at room temperature for 45 min.
After rinsing in wash buffer two times for 2 min for each, slides were incubated with a secondary horseradish peroxidase-conjugated goat anti-rabbit anti-mouse IgG (EnVision Flex/HRP, Dako, USA) for 20 min at room temperature. Slides were then rinsed thoroughly in wash buffer, two times for 2 min each, before incubation with diaminobenzidine solution (EnVision DAB + CHROMOGEN, Dako, USA) to develop color for 10 min at room temperature. Finally, slides were counterstained with hematoxylin solution at room temperature for 1 min dehydrated and then mounted to being examined under an Olympus light microscope (Olympus, Tokyo, Japan).
Evaluation of immunostaining
Intensity, percentage, the intracellular distribution of stained tumor cells (TC), the amount of positive immune cells and positivity of endothelial cells were evaluated separately by the pathologist. The intensity of immunostaining of tumor cells was graded as negative (0), weak (1+), moderate (2+) or strong (3+). VISTA-positive immune cells were counted in intratumoralhotspot regions. Immunostaining of endothelial and immune cells was graded as present or absent21.
Ethics approval and consent to participate
The current study was approved by the Ethical Board of the Ibn Rochd University Hospital of Casablanca. Written informed consent was obtained from glioma patients (from parents and/or legal guardian for the subject under 18 years old), and healthy donors involved in this study. Methods were carried out by relevant guidelines and regulations.
Statistical analysis
In this study, statistical analysis was performedusing GraphPad Prism 6.0 (GraphPad Software, Inc., La Jolla, CA, USA) or R (version 3.3.1, Auckland, NZ).
The paired t-test was used for paired samples to make a statistical comparison of gene expression between groups. The Mann–Whitney–Wilcoxon test were used for ranked data as appropriate. The two-sided p value less than 0.05 was considered as statistically significant for all statistical analyses.
The prognostic value of VISTA was investigated by Kaplan–Meier analysis using GraphPad Prism 6. Additionally, multivariate survival analysis was performed using the Cox proportional hazards regression model.
Results
VISTA expression according to characteristics of glioma patients
In total, 57 glioma tissues in the Moroccan cohort (33 men and 24 women) and 667 glioma cases in the TCGA cohort were recruited in the current study. The characteristics of the enrolled Moroccan patients were described in Table 1. The expression profile of VISTA was found to be associated with glioma grades (p = 0.0422) but not with other characteristics such as the gender (p = 0.1295), histological type (p = 0.058), smoking status (p = 0.5573) and age (p = 0.364). However, in the TCGA data set (Table 2), VISTA mRNA expression was significantly associated with glioma grades (p < 0.0001), histological type (p < 0.0001), and molecular subtype (p = 0.0002), but not with otherparameters such as gender (p = 0.647), age (p = 0.4028), Karnofsky score (p = 0.055) and IDH mutation status (p = 0.0702).
VISTA gene expression was upregulated in glioma tissues relative to PBMC of healthy donors
To evaluate the association between VISTA gene expressionand the pathogenesis of glioma, a Moroccan cohort, of 87 samples and 10 PBMCof healthy donors, were analyzed. The levels of expression of VISTA were evaluatedby Real-Time RT-PCR. VISTA expression was significantly increased in high grade versus lowgrade glioma tissues (p = 0.042) (Fig. 1a). VISTA mRNA expression was significantly elevated in glioma grade IV compared to grade II (p = 0.0494) (Fig. 1b). Tovalidate VISTAgene expression results observed in our glioma cases, PD-1 (a known checkpointmolecule) mRNA expression was analyzed in 47 glioma samples. Indeed, as expected, the expression level of PD-1 was also found to be significantly higher in grades III and IV compared to low grades (I and II) (p = 0.015) (Fig. 1c).
VISTA gene expression was upregulated in glioma tissues relative to PBMC of healthy donors (Moroccan cohort). VISTA and PD-1 transcripts expression were performed using RT-PCR analysis. (a) VISTA gene strongly expressed in grade III–IV compared with grade I–II of glioma patients. (b) VISTA mRNA expressionwas significantly increased in advanced glioma (grade IV) compared to grade II. (c) PD-1was highly expressed in advanced glioma grading (grade III–IV). (d) Elevated expression of VISTA gene in glioma tissues (G-Tissues) compared to PBMC of healthy donors (H-PBMC) and PBMC of glioma patients before (pre-G PBMC) and after surgery (post-G PBMC). (e) VISTA was upregulated in high grade glioma tissues (grade III–IV) relative to PBMC of the same patients. Paired t-test was used to make a statistical comparison of VISTA and PD-1 expression between groups. p value less than 0.05 was considered as statistically significant for all statistical analyses.
To assess whether there is a relationship between VISTA expression in the tumor microenvironment versus patient PBMCs, compared with PBMCs of healthy donors, 57 glioma tissuesand 30 PBMC samples of the same glioma patients(20 PBMCs before and 10 after surgery) were analyzed. Expression levels ofVISTA transcripts were elevated in glioma tissues compared to both PBMCs of healthy donors (p = 0.03) and PBMCs from glioma patients before surgery (p = 0.012). However, the difference was not significant when compared to PBMCs collected after surgery (p = 0.08) (Fig. 1d). When patients were stratified according to glioma grades, we detected a significantly elevated expression of VISTA in high-grade glioma tissues than PBMCs collected from the same patients before surgery (p = 0.04). In contrast, in low-grade gliomas, no significant difference was observed between VISTA expression in tumor tissues versus PBMCs collected again from the same patients before surgery (p = 0.12) (Fig. 1e). These observations indicated that the elevated expression levels of VISTA were noticed in high versus low glioma grade. This expression was specific to tumor tissues.
VISTA protein exhibited elevated expression levels inhigh-grade gliomas
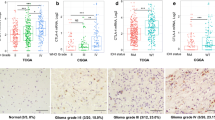
In order to corroborate the results of VISTA gene expression obtained in the transcript level (Fig. 1), VISTA protein analysis was performed on 30 glioma cases (13 low grade (I/II) and 17 high grade (III/IV) cases) of the same patient samples by immunohistochemistry assay. The IgG isotype control was assessed in glioma tissue (used as a negativecontrol for VISTA protein expression) (Fig. 2a). Interestingly, VISTAstaining was negative in the majority (9 out of 13 cases, 69.23%) of low-grade gliomas (Fig. 2b). Furthermore, a significant VISTA staining was detected in 82.35% (14 out of 17 cases) ofhigh-grade gliomas (Fig. 2c). VISTA also exhibitedsignificantly higher expression in advanced versus low-grade gliomas (Fig. 2g, p = 0.0004). Notably, VISTAprotein was detected on glioma cells, especially in glioblastoma cases, wheresignificant staining was observed (Fig. 2e). Besides, about 20%of glioma samples showed positive staining of VISTA on immune and endothelialcells (Fig. 2d,f). Altogether, our dataindicate that VISTA was highly expressed at both mRNA and protein levels in patients with high grade compared to lowgrade gliomas.
Immunohistochemical staining of VISTA revealed elevated expression levels in high grade gliomas. Representative staining intensity of VISTA protein was detected in human glioma tissues (Moroccan cohort) using immunohistochemistry assay. (a) Negative control staining in glioma case with Rabbit IgG Isotype Control (magnification × 20). (b) Negative staining of VISTA in low grade glioma (grade I). (c) Positive staining of VISTA in high grade glioma (Glioblastoma IV) (magnification × 20). (d) Positive staining of VISTA on endothelial cells (magnification × 20). (e) Positive staining of VISTA on glioma cells (magnification × 40). (f) Positive staining of VISTA on immune cells in a glioblastoma case (magnification × 20). (g) Expression of VISTA according to glioma grades. Statistical analysis was performed by using a t-test to compare the expression of VISTA between different grades of glioma patients. p value less than 0.05 was considered as statistically significant for all statistical analyses.
VISTA transcripts strongly expressed in high glioma grades in the TCGA cohort
To assess the expression of VISTA in a distinct cohort, we evaluated the RNA-sequencing data of gliomas from the TCGA database.667 samples were analyzed and graded according to the WHO grading system. Compared to low grade gliomas, glioblastomas present a significantly higher VISTA expression (p < 0.0001) (Fig. 3a). Further analysisshowed an elevated expression of VISTA in Astrocytomacompared to Oligoastrocytoma (p = 0.0089) and Oligodendroglioma (p < 0.0001) (Fig. 3b). However, Classical andProneural molecularsubtypes revealed lower VISTA gene expressioncompared to Neural (p = 0.0010 and p = 0.0023, respectively) (Fig. 3c).
VISTA transcripts strongly expressed in high glioma grades in the TCGA cohort and positively correlated with critical immune checkpoint regulators. RNAseq of 667 glioma patients of different grades were analyzed using TCGA dataset. (a) VISTA mRNA evaluation revealed high expression in advanced gliomas (grade IV). (b) Astrocytomas showed elevated expression of VISTA compared to oligoastrocytomas and oligodendrogliomas. (c) Mesenchymal and Neural glioma subtypes presented high VISTA expression in comparison with classical and proneural. (d) PD-1, Tim-3 and LAG-3 were upregulated in high grade glioma (glioblastoma).T-test was applied to compare gene expression between different grades and groups of glioma patients. p value less than 0.05 was considered as statistically significant for all statistical analyses.
VISTA positively correlated with critical immune checkpoint regulators in glioma patients
The expression pattern of VISTA was compared, using the TCGA dataset, to the expression of three critical immune checkpoints (Tim-3, LAG-3, PD-1) known for being highly expressed in advanced versus low glioma grades22,23,24. Indeed, high glioma grades showed significantly higher PD-1, Tim-3 and LAG-3 expressions compared to low grades, exhibiting a similar expression profile to VISTA (p < 0.0001) (Fig. 3d).
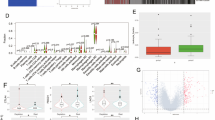
A correlation study was conducted between VISTA expression and the same immune checkpoints (PD-1, Tim-3 and LAG-3). VISTA was positively correlated with PD-1 (p = 0.0048, r = 0.112), Tim-3 (p < 0.0001, r = 0.682) and LAG-3 (p < 0.0001, r = 0.2) gene expression (Fig. 4a), suggesting that tumor cells may likely use VISTA gene in the same way as these three immune checkpoints to escape the immune system. At last, VISTA mRNA expression levels appeared to be the highest in all glioma cases (low and high grade) when compared to those of other critical immune checkpoints, Tim-3, PD-1, CTLA-4, LAG-3 and TIGIT (p < 0.0001) (Fig. 4b).
VISTA expression positively correlated with critical immune checkpoint regulators. (a) VISTA expression was positively correlated with PD-1, Tim-3 and LAG-3. (b) VISTA gene exhibited the highest expression in comparison with other immune checkpoints (Tim-3, PD-1, CTLA-4, LAG-3, TIGIT). Spearman correlation test was used to examine the association of relative gene expression levels between VISTA and other immune checkpoints. Also, the one-way ANOVA test was used to determine the statistical significance of gene expression among different groups of glioma patients. p value less than 0.05 was considered as statistically significant for all statistical analyses.
Anti-tumoral genes were inhibited in glioma microenvironment
To clarify the relationship between VISTA expression profile and the occurrence of different immune cell populations in the tumor microenvironment, we have created a binary clustering of patients (TCGA cohort), using the median as a cut off for patient stratification. One group with a high expression of VISTA and the second one lower. Then, we assessed the expression of CD4 and CD8 genes. CD4 and CD8 mRNA expression were both elevated in high versus low VISTA expression (p < 0.0001). CD4 mRNA levels were also higher in the elevated VISTA gene expression group compared to CD8 (p < 0.0001) (Fig. 5a), suggesting that CD8 and CD4 cells are present within the tumor microenvironment but with limited effector function because of the higher expression level of VISTA. As for T lymphocyte-related cytokines, we assessed the expression of two distinct sets. TGF-β/IL-10 genes, known to play critical roles in inhibiting CD8 and CD4 T cell functions and IL-2/IFNγ genes, which are known to boost the anti-tumor immune response25. TGF-β and IL-10 gene expression was upregulated in glioma patients who present an elevated expression of VISTA. However, IFNγalso exhibited a significantly higher expression (p = 0.0002). For IL-2 gene expression, no significant difference has been observed (Fig. 5b).
Increased VISTA transcripts level associated to a poor prognosis of glioma patients in the TCGA dataset. A binary clustering of patients has been performed, using the median as a cut off for patient stratification: one group with a high expression of VISTA and a second one with lower expression. (a) CD4 and CD8 mRNA expression were both elevated in high versus low VISTA expression. (b) The immunomodulatory genes (TGF-β, IL-10 and IFNγ) showed high expression in glioma patients with higher levels of VISTA gene expression. (c) High VISTA expression levels associated with a bad overall survival. (d) Elevated expression of both VISTA and PD-1 correlated with a weak survival. Paired t-test was used to make statistical comparison of gene expression between groups. The prognostic value of VISTA gene was investigated by Log-rank test using the Kaplan–Meier plot. p value less than 0.05 was considered as statistically significant for all statistical analyses.
Increased VISTA transcript levels associated with weak overall patient survival
To examine the impact on patient survival, we evaluated the prognostic value of VISTA in the TCGA dataset. Survival data were available for 666 human glioma patients. As showed using Kaplan–Meier curves, patients with lower VISTA expression had prolonged survival compared to patients with higher expression of VISTA (p = 0.0085) (Fig. 5c). In addition, glioma patients with elevated expression of PD-1 showed a poor overall survival (Fig. 5d). Remarkably, patients have increased expression levels of both VISTA and PD-1 showed worse survival compared with those presenting low expression of both genes (p < 0.0001) (Fig. 5e). These results indicated that VISTA could be considered as a negative prognostic factor in glioma. In order to identify the effects of confounding variables on glioma patients’ survival, the clinical characteristics were selected from the TCGA dataset a variables for multivariate cox regression analysis. Results revealed that in glioma patients (high and low grades), histological type, age and grade were associated with the worse glioma patients’ survival ([HR] = 1.27697, p = 0.0132; [HR] = 2.63324, p = 2.23 e − 09; and [HR] = 2.76359, p = 2.67e − 08), respectively. However, no significant correlation was found between Sex and VISTA gene expression with clinical outcome (Table S1). Additionally, multivariate cox regression analysis has also been performed on two separate bases, one containing low-grade gliomas and the other high-grade gliomas, in order to evaluate the effect of clinical parameters and VISTA gene expression on patient’s survival. In this case, histological type, grade (II, III), age and Karnofsky score were associated with bad patients’ survival ([HR] = 1.3740, p = 0.0201; [HR] = 3.0360, p = 6.32e − 05; [HR] = 3.0914, p = 2.02e − 05; ([HR] = 2.2554, p = 3.57e − 05) (Table S2). Surprisingly, in high grade glioma patients, our results indicated a correlation between patients’ sex and VISTA gene expression with patients’ poor survival ([HR] = 1.981e + 00, p = 0.00764 and [HR] = 1.727e + 00, p = 0.03192, respectively) (Table S3).
Discussion
Gliomas are the most frequent and fatal brain tumors in adults1. Despite treating glioblastoma patients with conventional therapies such as surgical resection with subsequent radiation and chemotherapy, the prognosis for glioma patients is still poor2.
In the last few years, immunotherapy has brought new hope as a potential novel therapeutic approach for glioma patients26. However, the majority of glioma patients did not respond to the blockade of usual immune checkpoints pathways (CTLA-4 and PD1/PD-L1)11,12,13.
This has increased our interest in exploring the role of other immune checkpoint molecules, including the recently discovered one, VISTA15,16.
Thus, the main objective of this work was to investigate the role of VISTA in human gliomas. The study revealed that: (1) VISTA gene expression was upregulated in high versus low glioma grades, ((2) VISTA protein exhibited elevated expression levels in high-grade glioma, (3) VISTA expression positively correlated with other critical immune checkpoint regulators, (4) genes, which are known to be linked to an anti-tumor signature, were inhibited in the glioma microenvironment, (5) elevated VISTA transcript levels negatively correlated with the IDH mutation in patients, and (6) high VISTA transcript levels associated to weak overall patient survival. To the best of our knowledge. This is the first exploration of the role of VISTA in clinically resected glioma tumors. It is also the largest and most comprehensive study describing the expression pattern of VISTA in human glioma samples using two cohorts (TCGA and Moroccan cohort). VISTA expression has, however, been investigated in other cancer tissues including colorectal carcinoma27, human hepatocellular carcinoma28, gastric cancer21, human oral squamous cell carcinoma29, pancreatic cancer30, oesophagal adenocarcinoma31 and prostate cancer21. In these reports, and consistent with the present work. It has also been shown that VISTA expression is upregulated in higher versus lower grades of the disease.
Besides, protein level assessment using immunohistochemistry assay confirmed the VISTA expression pattern initially observed at the mRNA level.
In gastric cancer, using immunohistochemistry in a large cohort of 464 samples and 14 corresponding liver metastases, it was revealed that VISTA expression was observed in tumor and immune cells, but not in non-neoplastic gastric epithelium. Also, this expression varies with tumor progression 21. In the same context using the immunofluorescence method, 28 clinical colorectal cancer specimens were used to evaluate VISTA protein expression showing that VISTA is expressed in normal colorectal samples, in para-tumors and tumors cases, with elevated expression levels in the tumors. However, VISTA was revealed to be expressed at high levels in different subsets of myeloid cells in the tumors compared to PBMCs27. Otherwise, VISTA expression was suggested to be linked to PD-L1 expression in gastric cancer, suggesting that VISTA cooperates with PD-L1 in the mechanism underlying immune evasion21. A recent study revealed that pancreatic tumors with high cytolytic activity have increased the expression of immune checkpoint genes such as CTLA-4, TIGIT, TIM-3, and VISTA32. In contrast, others have reported that there was no correlation between VISTA and other checkpoint-markers such as PD-L1 and LAG-3 in oesophagal adenocarcinoma cohort, indicating that VISTA might function separately28. Chraa et al. reviewed the tumor microenvironment and its infiltration by distinct T lymphocyte subpopulations33. This report explained the association of these different subtypes with cancer progression33,34. However, treatment with several cytokines, including IFN-γ, IL-2, IL-4, and IL-6, did not raise VISTA expression in tumor cells35. Kondo et al. demonstrated that VISTA blockade efficiently converted resting and exhausted T cells into functionally differentiated effector CD8+ T cells, indicating that monotherapy using VISTA might have the ability to enhance multifunctionality of CD8+ T cells in squamous cell carcinoma16. Wang et al. showed that in the presence of TGF-ß, VISTA Ig promoted partially the differentiation of iTreg and that this effect could be observed in both murine and human CD4+ T cells35,36. Furthermore, VISTA is necessary for the generation of iTreg from naïve T cells, which enhances a microenvironment, which is favorable for the expansion of tumor cells37.
Interestingly, blocking VISTA in tumor cells extended mice survival that was inoculated with ovarian cancer cells overexpressing VISTA, although combined therapy using anti-PD-1 and anti-VISTA did not further improve mice survival compared to anti-VISTA treatment alone38. Latest studies on CTLA-4 and PD-1 blockade showed an upregulation of VISTA expression in prostate cancer and melanoma treated patients. This observation suggested the importance of considering a potential VISTA blockade for these patients.
In summary, our data revealed a correlation between VISTA expression and glioma progression in patients. This study also indicated that VISTA is a negative prognostic factor in glioma, and pinpoints VISTA as a possible new therapeutic target, particularly in advanced glioma stages.
Abbreviations
- PD-1:
-
Programmed cell death 1
- VISTA:
-
V-domain Immunoglobulin suppressor of T cell activation
- PDL-1:
-
Programmed death-ligand 1
- PDL-2:
-
Programmed death-ligand 2
- LAG-3:
-
Lymphocyte-activation gene 3
- Tim-3:
-
T-cell immunoglobulin and mucin-domain containing-3
- CTLA-4:
-
Cytotoxic T-lymphocyte-associated protein 4
- TIGIT:
-
T cell immunoreceptor with Ig and ITIM domains
- GBM:
-
Glioblastoma multiforme
- TCGA:
-
The Cancer Genome Atlas
- PBMC:
-
Peripheral blood mononuclear cell
- mRNA:
-
Messenger RNA
- WHO:
-
World Health Organization
- PCR:
-
Polymerase chain reaction
- IL-2:
-
Interleukin-2
- IL-10:
-
Interleukin-10
- IL-6:
-
Interleukin-6
- IL-4:
-
Interleukin-4
- TGFβ:
-
Transforming growth factor β
- IFNγ :
-
Interferon-gamma
References
Van Meir, E. G. et al. Exciting new advances in neuro-oncology: The avenue to a cure for malignant glioma. CA Cancer J. Clin. 60(3), 166–193. https://doi.org/10.3322/caac.20069 (2010).
Stupp, R. et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N. Engl. J. Med. 352(10), 987–996. https://doi.org/10.1056/NEJMoa043330 (2005).
Louveau, A. et al. Structural and functional features of central nervous system lymphatic vessels. Nature 523(7560), 337–341. https://doi.org/10.1038/nature14432 (2015).
Wang, Z. et al. Molecular and clinical characterization of PD-L1 expression at transcriptional level via 976 samples of brain glioma. Oncoimmunology. https://doi.org/10.1080/2162402X.2016.1196310 (2016).
Ghouzlani, A. et al. The promising IgSF11 immune checkpoint is highly expressed in advanced human gliomas and associates to poor prognosis. Front. Oncol. 10, 608609. https://doi.org/10.3389/fonc.2020.608609 (2021).
Ghouzlani, A. et al. High expression levels of Foxp3 and VISTA in advanced human gliomas and impact on patient’s prognosis. Arch Clin. Biomed. Res. 4(6), 691–703 (2020).
Ghouzlani, et al. Immune checkpoint inhibitors in human glioma microenvironment. Front. Immunol. https://doi.org/10.3389/fimmu.2021.679425 (2021).
Carter, T., Shaw, H., Cohn-Brown, D., Chester, K. & Mulholland, P. Ipilimumab and bevacizumab in glioblastoma. Clin. Oncol. R. Coll. Radiol. G. B. 28(10), 622–626. https://doi.org/10.1016/j.clon.2016.04.042 (2016).
Hu, J. L., Sharma, P., Yu, J., Black, K. L. & Rudnick, J. D. Ipilimumab for recurrent glioblastoma: A single-institution case series. J. Clin. Oncol. 32(15_suppl), e13010. https://doi.org/10.1200/jco.2014.32.15_suppl.e13010 (2014).
Reardon, D. A. et al. OS10.3 randomized phase 3 study evaluating the efficacy and safety of nivolumab vs bevacizumab in patients with recurrent glioblastoma: CheckMate 143. Neuro Oncol. 19(suppl_3), iii21. https://doi.org/10.1093/neuonc/nox036.071 (2017).
Dong, H., Zhu, G., Tamada, K. & Chen, L. B7–H1, a third member of the B7 family, co-stimulates T-cell proliferation and interleukin-10 secretion. Nat. Med. 5(12), 1365–1369. https://doi.org/10.1038/70932 (1999).
Taube, J. M. et al. Colocalization of inflammatory response with B7–h1 expression in human melanocytic lesions supports an adaptive resistance mechanism of immune escape. Sci. Transl. Med. 4(127), 127ra37. https://doi.org/10.1126/scitranslmed.3003689 (2012).
Dong, H. et al. Tumor-associated B7–H1 promotes T-cell apoptosis: A potential mechanism of immune evasion. Nat. Med. 8(8), 793–800. https://doi.org/10.1038/nm730 (2002).
Brahmer, J. R. et al. Safety and activity of anti-PD-L1 antibody in patients with advanced cancer. N. Engl. J. Med. 366(26), 2455–2465. https://doi.org/10.1056/NEJMoa1200694 (2012).
Wang, L. et al. VISTA, a novel mouse Ig superfamily ligand that negatively regulates T cell responses. J. Exp. Med. 208(3), 577–592. https://doi.org/10.1084/jem.20100619 (2011).
Flies, D. B., Wang, S., Xu, H. & Chen, L. Cutting edge: A monoclonal antibody specific for the programmed death-1 homolog prevents graft-versus-host disease in mouse models. J. Immunol. Baltim. Md 1950 187(4), 1537–1541. https://doi.org/10.4049/jimmunol.1100660 (2011).
Wang, L. et al. Disruption of the immune-checkpoint VISTA gene imparts a proinflammatory phenotype with predisposition to the development of autoimmunity. Proc. Natl. Acad. Sci. USA 111(41), 14846–14851. https://doi.org/10.1073/pnas.1407447111 (2014).
Flies, D. B. et al. Coinhibitory receptor PD-1H preferentially suppresses CD4+ T cell-mediated immunity. J. Clin. Invest. 124(5), 1966–1975. https://doi.org/10.1172/JCI74589 (2014).
Moutia, M. et al. Allium sativum L. regulates in vitro IL-17 gene expression in human peripheral blood mononuclear cells. BMC Complement. Altern. Med. 16(1), 377. https://doi.org/10.1186/s12906-016-1365-9 (2016).
Moutia, M. et al. Capparis spinosa L. promotes anti-inflammatory response in vitro through the control of cytokine gene expression in human peripheral blood mononuclear cells. BMC Immunol. 17(1), 26. https://doi.org/10.1186/s12865-016-0164-x (2016).
Böger, C., Behrens, H.-M., Krüger, S. & Röcken, C. The novel negative checkpoint regulator VISTA is expressed in gastric carcinoma and associated with PD-L1/PD-1: A future perspective for a combined gastric cancer therapy?. Oncoimmunology 6(4), e1293215. https://doi.org/10.1080/2162402X.2017.1293215 (2017).
Liu, Z. et al. Expression of the galectin-9-Tim-3 pathway in glioma tissues is associated with the clinical manifestations of glioma. Oncol. Lett. 11(3), 1829–1834. https://doi.org/10.3892/ol.2016.4142 (2016).
Harris-Bookman, S. et al. Expression of LAG-3 and efficacy of combination treatment with anti-LAG-3 and anti-PD-1 monoclonal antibodies in glioblastoma. Int. J. Cancer 143(12), 3201–3208. https://doi.org/10.1002/ijc.31661 (2018).
Dejaegher, J. et al. Characterization of PD-1 upregulation on tumor-infiltrating lymphocytes in human and murine gliomas and preclinical therapeutic blockade. Int. J. Cancer 141(9), 1891–1900. https://doi.org/10.1002/ijc.30877 (2017).
Lamrani, M. et al. TLR4/IFNγ pathways induce tumor regression via NOS II-dependent NO and ROS production in murine breast cancer models. OncoImmunology. 5(5), e1123369. https://doi.org/10.1080/2162402X.2015.1123369 (2016).
Han, S. J., Zygourakis, C., Lim, M. & Parsa, A. T. Immunotherapy for glioma: Promises and challenges. Neurosurg. Clin. N. Am. 23(3), 357–370. https://doi.org/10.1016/j.nec.2012.05.001 (2012).
Xie, S. et al. Expression of the inhibitory B7 family molecule VISTA in human colorectal carcinoma tumors. Cancer Immunol. Immunother. CII. https://doi.org/10.1007/s00262-018-2227-8 (2018).
Zhang, M. et al. VISTA expression associated with CD8 confers a favorable immune microenvironment and better overall survival in hepatocellular carcinoma. BMC Cancer 18, 511. https://doi.org/10.1186/s12885-018-4435-1 (2018).
Wu, L. et al. Expression of VISTA correlated with immunosuppression and synergized with CD8 to predict survival in human oral squamous cell carcinoma. Cancer Immunol. Immunother. CII. 66(5), 627–636. https://doi.org/10.1007/s00262-017-1968-0 (2017).
Blando, J. et al. Comparison of immune infiltrates in melanoma and pancreatic cancer highlights VISTA as a potential target in pancreatic cancer. Proc. Natl. Acad. Sci. 116(5), 1692–1697. https://doi.org/10.1073/pnas.1811067116 (2019).
Loeser, H. et al. The expression of the immune checkpoint regulator VISTA correlates with improved overall survival in pT1/2 tumor stages in esophageal adenocarcinoma. OncoImmunology 8(5), e1581546. https://doi.org/10.1080/2162402X.2019.1581546 (2019).
Balli, D., Rech, A. J., Stanger, B. Z. & Vonderheide, R. H. Immune cytolytic activity stratifies molecular subsets of human pancreatic cancer. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 23(12), 3129–3138. https://doi.org/10.1158/1078-0432.CCR-16-2128 (2017).
Chraa, D., Naim, A., Olive, D. & Badou, A. T lymphocyte subsets in cancer immunity: Friends or foes. J. Leukocyte Biol. https://doi.org/10.1002/JLB.MR0318-097R (2019) (consulté le juill. 10, 2019).
Ait Ssi, S. et al. Prognostic gene expression signature in patients with distinct glioma grades. Front. Immunol. https://doi.org/10.3389/fimmu.2021.685213 (2021).
Lines, J. L. et al. VISTA is an immune checkpoint molecule for human T cells. Cancer Res. 74(7), 1924–1932. https://doi.org/10.1158/0008-5472.CAN-13-1504 (2014).
Mercier, I. L. et al. VISTA regulates the development of protective antitumor immunity. Cancer Res. 74(7), 1933–1944. https://doi.org/10.1158/0008-5472.CAN-13-1506 (2014).
Wang, Q., He, J., Flies, D. B., Luo, L. & Chen, L. Programmed death one homolog maintains the pool size of regulatory T cells by promoting their differentiation and stability. Sci. Rep. 7(1), 6086. https://doi.org/10.1038/s41598-017-06410-w (2017).
Mulati, K. et al. VISTA expressed in tumour cells regulates T cell function. Br. J. Cancer. 120(1), 115. https://doi.org/10.1038/s41416-018-0313-5 (2019).
Acknowledgements
We would like to thank all members of the Neurosurgery department, including, Pr. A. El azhari, Pr. S. Hilmani, Pr. K. Ibahioin, Pr. A. Bertal, Pr. A. Naja, Pr. A. Challaoui, Dr. A. Bocco, Dr. T. Mesbahi, Dr. Z. Ennhaili, Dr. M. Fatihi, Dr. O. Benhayoun, Dr. M. Haous, Dr. D. Kaba, Dr. I. Mahazou, Dr. A. Jehri, Dr S. Stitou, and nurses (S. Khayam, S. Watti, H. Bchira, Fz. Remiany) for their help. Special thanks to Dr. M. Benlahfid for his help throughout this study by training for the use of statistical analysis software and the management of bibliographic references.
Funding
This work was supported by the Moroccan Ministry of Higher Education and Research and The National Center for Scientific and Technical Research (CNRST) through “PPR1” and Al-khawarizmi projects coordinated by A.B. A.G. was supported by a “CNRST” fellowship.
Author information
Authors and Affiliations
Contributions
A.G. collected, analyzed, and interpreted data; wrote the manuscript; S.R. collected, analyzed data; A.L. collected and analyzed data; M.K. analyzed and interpreted data; A.B. designed research, analyzed and interpreted data, wrote the manuscript and supervised the study.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Ghouzlani, A., Lakhdar, A., Rafii, S. et al. The immune checkpoint VISTA exhibits high expression levels in human gliomas and associates with a poor prognosis. Sci Rep 11, 21504 (2021). https://doi.org/10.1038/s41598-021-00835-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-00835-0
- Springer Nature Limited
This article is cited by
-
From glioma gloom to immune bloom: unveiling novel immunotherapeutic paradigms-a review
Journal of Experimental & Clinical Cancer Research (2024)
-
Bridging the divide: unveiling mutual immunological pathways of cancer and pregnancy
Inflammation Research (2024)
-
Immune related biomarkers for cancer metastasis to the brain
Experimental Hematology & Oncology (2022)
-
IGSF11 and VISTA: a pair of promising immune checkpoints in tumor immunotherapy
Biomarker Research (2022)