Abstract
The desert pioneer plant Stipagrostis pennata plays an important role in sand fixation, wind prevention, and desert ecosystem recovery. An absence of reference genes greatly limits investigations into the regulatory mechanism by which S. pennata adapts to adverse desert environments at the molecular and genetic levels. In this study, eight candidate reference genes were identified from rhizosheath development transcriptome data from S. pennata, and their expression stability in the rhizosheaths at different development stages, in a variety of plant tissues, and under drought stress was evaluated using four procedures, including geNorm, NormFinder, BestKeeper, and RefFinder. The results showed that GAPDH and elF were the most stable reference genes under drought stress and in rhizosheath development, and ARP6 and ALDH were relatively stable in all plant tissues. In addition, elF was the most suitable reference gene for all treatments. Analysis of the consistency between the reverse transcription-quantitative PCR (RT-qPCR) and RNA sequencing data showed that the identified elF and GAPDH reference genes were stable during rhizosheath development. These results provide reliable reference genes for assuring the accuracy of RT-qPCR and offer a foundation for further investigations into the genetic responses of S. pennata to abiotic stress.
Similar content being viewed by others
Introduction
Drought is a critical environmental factor that significantly affects plant growth and crop yield. Xinjiang is an extensive arid and semi-arid region comprising the most widely distributed area of desertified land in China. The region is subject to land and wind erosion and is characterized by shifting sand dunes, which seriously hampers normal agricultural production and human development. Plants growing in this area have adapted to the extreme environment. Stipagrostis pennata is a desert plant distributed in the Gurbantünggüt Desert in Xinjiang that mainly grows on mobile and semi-mobile sand dunes, having important roles in wind prevention, sand fixation, and the protection of the desert ecosystem1. This plant has a typical rhizosheath structure around the roots, which allows the roots to resist the unfavorable external conditions of the arid desert environment and endows the plant with sand fixation ability2. The rhizosheaths are also the key factor by which the plants tolerate drought conditions3.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) is a powerful method for detecting gene expression profiles and has been widely used in plants due to its good repeatability and high sensitivity4,5,6,7. The accuracy of RT-qPCR assays depends greatly on the suitability of the reference genes used8. Typically, the ideal reference genes should be stably expressed under different treatment conditions and in various types of cells or tissues. Many studies have shown that reference genes are not universal and that gene expression stability is relatively constant under a certain type of cell or experimental factor7,9,10,11. In plants, the candidate internal reference genes for RT-qPCR are usually housekeeping genes, including elongation factor-1α (EF-1α), glyceraldehyde-3-phosphate dehydrogenase (GAPDH), actin (ACT), cyclophilin (CYP), phosphoglycerate kinase gene (PGK), 18SrRNA, S-adenosylmethionine synthase (SAMS), tubulin (TUB), and ubiquitin (UBQ)12,13. However, the stability of these genes is inconsistent under different conditions, plant species, tissues, growth and development stages, and experimental treatments12,14,15,16,17.
Reference gene stability has not yet been explored in S. pennata. This greatly limits further analysis of the functional and mechanistic elucidation of genes, thereby hindering research into the molecular basis of adaptation to abiotic stress. In the present study, transcriptome data for rhizosheath development based on RNA sequencing (RNA-Seq) were screened from the rhizosheaths of 30, 60, 90 days post-germination (DPG) plants (R30, R60, R90), and non-rhizosheath roots of 90 DPG plants (R90F). Additionally, the roots of 60 DPG plants subjected to osmotic stress were also sampled. Based on the results of three independent algorithms, eight candidate reference genes were discovered, and their expression stability was evaluated. Based on a comprehensive evaluation of their ranking, the genes of GAPDH and eukaryotic translation initiation factor (elF) under drought stress, actin related protein (ARP6) and aldehyde dehydrogenase (ALDH) in the plant tissues, elF and GAPDH in rhizosheath development, as well as ARP6 and elF in all samples, were identified as the most stable reference genes. Our results provide reliable reference genes for RT-qPCR and further genetic function studies in S. pennata.
Results
Identification of candidate reference genes in different stages of rhizosheath development
The conditions of |log2FoldChange| < 1, q-value ≥ 0.05, and FPKM (Fragments per Kilobase of exon model per Million mapped reads) ≥ 6 were used to screen the RNA-Seq data, and the relatively low coefficient of variation (CV) of FPKM was set as a high standard at all sampling points, generating a total of eight candidate reference genes (Table 1). Detailed information of these genes, including unigene name, gene symbol, homologue locus, and E-value, via comparison with the homologous genes in rice is listed in Supplementary Table S1. The FPKM-based heatmap of the eight genes during different stages of rhizosheath development is provided in Fig. 1 and indicates that GAPDH, α-TUB, TIP41, Histone H3 (HIS-3), elF, and ARP6 exhibited stable expression in different stages of rhizosheath development, whereas ALDH and protein phosphatase 2A (PP2A) showed relative unsteady expression.
Heatmap of eight candidate reference genes based on Fragments per Kilobase of exon model per Million mapped reads (FPKM) value of transcriptome of rhizosheath development. R30, R60, R90, and R90F indicate the tissues of 30-, 60-, and 90-DPG rhizosheaths and 90-DPG rhizosheath-free roots, respectively. The screening conditions were q-value ≥ 0.05, FPKM ≥ 6, and |log2FoldChange| < 1. The eight candidate genes were glycolide-3-phosphate dehydrogenase (GAPDH), aldehyde dehydrogenase (ALDH), eukaryotic translation initiation factor (elF), actin related protein (ARP6), tonoplast intrinsic protein, (TIP41), α-Tubulin (α-TUB), protein phosphotase 2A (PP2A), and Histone H3 (HIS-3).
Verification of primer specificity and PCR amplification efficiency
Specific primers for the eight candidate reference genes were designed and amplified by RT-qPCR to verify the specificity using cDNA from the roots as a template. Referring to MIQE (Minimum Information for Publication of Quantitative Real-Time PCR Experiments) guidelines, we have provided information about the specific primers of the eight reference genes and RT-qPCR experiments (Supplementary Figs. S1, S2)18. According to the slope of the standard curve, the amplification efficiency and R2 of the RT-qPCR assays were calculated, which indicated that the amplification efficiency ranged from 90.8% (TIP41) to 95.4% (PP2A), and the R2 value ranged from 0.988 (PP2A) to 0.999 (HIS-3) (Table 1). These results indicated that all the primers of the eight genes had high specificity and amplification efficiency and were thus suitable for further analysis.
Expression profiles of the eight candidate reference genes in S. pennata
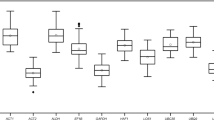
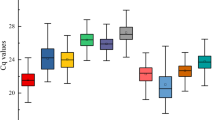
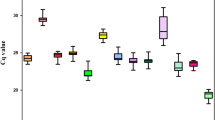
RT-qPCR assays were performed with the designed specific primers for the eight candidate reference genes using the cDNA as template extracted from different tissues and treated materials of S. pennata. Based on the Ct values obtained from RT-qPCR, a box diagram was generated to reflect the differences in the expression levels of the eight candidate genes (Fig. 2). The average Ct values of the reference genes ranged between 16.89 and 29.12, which indicated that the expression levels of these reference genes differed in S. pennata. The results showed that ALDH had the smallest variation, followed by GAPDH, elF, and PP2A, while α-TUB had the largest variation. Therefore, ALDH, GAPDH, elF, and PP2A were identified as relatively stable genes.
Distribution of RT-qPCR Ct values for the eight candidate reference genes across all S. pennata samples. The S. pennata materials of rhizosheaths of different development stages, roots, leaves, flowers, nodes, seeds, and PEG-treated roots were used for RNA extraction and cDNA synthesis that were then utilized for RT-qPCR reactions. Each RT-qPCR Ct value is the average of three independent experiments. The median (horizontal line), upper and lower quartiles (box), and maximum and minimum values (whisker) of each gene are displayed.
Expression stability analysis of the eight candidate reference genes
NormFinder, geNorm, BestKeeper, and RefFinder were used to assess the expression stability of the eight candidate reference genes. NormFinder calculates the stable value (M-value) of each gene based on analysis of the variance to select the most suitable reference gene, with a low value representing high stability. The results identified elF and HIS-3 for rhizosheath development with an M-value of 0.12 and 0.189, GAPDH and elF under drought stress with an M-value of 0.06 and 0.179, ARP6 and ALDH in the tissues with an M-value of 0.392 and 0.58, and ARP6 and ALDH in all samples with an M-value of 0.604 and 0.622 as the two most stable genes for each different treatment (Table 2).
The geNorm program evaluates the expression stability of reference genes by calculating the M-value, with a high value representing low stability. The results showed that the M-values of the eight reference genes were all lower than 1.5 under drought stress, indicating high expression stability of these genes. In addition, TIP41 and elF (M-value of 0.221) under drought conditions, TIP41 and ALDH (M-value of 0.286) in all plant tissues, TIP41 and GAPDH (M-value of 0.212) in rhizosheath development, and elF and GAPDH (M-value of 0.623) in all samples were identified as the two most stable reference genes (Fig. 3A–D). The geNorm software was also used to calculate the paired variation value Vn/Vn+1 (n represents the reference gene number) to determine the optimal number of reference genes for RT-qPCR standardization. The results showed that the Vn/Vn+1 values of the different sample groups were all lower than 0.15, suggesting that two reference genes were sufficient to complete the RT-qPCR normalization in S. pennata (Fig. 3E–H).
Stability ranking of the eight candidate reference genes based on M value and paired variation (Vn/Vn+1) value calculated by geNorm. M values represent the expression stability of each gene, with lower M value to indicate higher stability. The ratio of Vn/Vn+1 was used to determine the optimal reference number of multiple reference genes. M values (A–D) and Vn/Vn+1 values (E–H) for candidate reference genes in each group were calculated.
The BestKeeper procedure was utilized to analyze the expression stability of the eight candidate reference genes, with high R-values and low CV ± SD values denoting better stability19. As shown in Table 3, HIS-3 in rhizosheath development (R = 0.803, CV ± SD = 2.92 ± 0.73) and GAPDH under drought stress (R = 0.984, CV ± SD = 6.43 ± 1.01) were identified as the most stable reference genes. ARP6 was relatively stable in the tissues (R = 0.973, CV ± SD = 9.73 ± 2.78) and all samples (R = 0.944, CV ± SD = 6.65 ± 1.76).
As shown above, evaluation by these three procedures produced different results for stable reference genes from those reported in some studies20,21,22,23. To provide a comprehensive assessment of the eight candidate reference genes in different conditions, we performed further analysis by RefFinder24 according to the geometric mean of the reference genes20,25 to generate a final comprehensive ranking of the expression stability of the reference genes. The results showed that elF and GAPDH in rhizosheath development, GAPDH and elF under drought stress, and ARP6 and ALDH in the tissues were the most stable genes. For all samples, ARP6 and elF were the two most stable reference genes (Table 4).
Validation of the reference genes by RT-qPCR
To verify the expression stability of the identified reference genes, two genes, namely CL9727 and U3887, which are closely related to rhizosheath development, were selected for RT-qPCR to analyze the similarity between the RT-qPCR results and the RNA-Seq data. The two stable genes of elF and GAPDH and two unstable genes of ALDH and α-TUB screened by the comprehensive assessment ranking under the condition of rhizosheath development were used as reference genes for normalization to calculate the relative expression levels of CL9727 and U3887. The results indicated that the expression levels of CL9727 and U3887 detected by RT-qPCR were highly consistent with the RNA-Seq data. When the unstable reference genes of ALDH and α-TUB were selected for standardization, the expression levels of CL9727 and U3887 detected by RT-qPCR were significantly different from the RNA-Seq data (Fig. 4). Additionally, correlation analysis of the relative expression levels of CL9727 and U3887 normalized by elF and GAPDH between RT-qPCR and RNA-Seq was performed, indicating a strong positive correlation between the RT-qPCR results and RNA-Seq data (R2 = 0.6061–0.9257), once again validating the stability and reliability of the two reference genes.
Validation of identified candidate reference genes as internal controls for normalizations of target genes of CL9729 and U3887. Two stable reference genes of GAPDH and elF (A, B) and two unstable reference genes of ALDH and α-TUB (C, D) were used as internal controls to detect the expression levels of CL9729 and U3887 during different rhizosheath development stages. R30, R60, R90, and R90F indicate the tissues of 30-, 60-, and 90-DPG rhizosheaths and 90-DPG rhizosheath-free roots, respectively.
Discussion and conclusion
Reverse transcription qPCR has become a common method for gene expression analysis due to its high sensitivity, good repeatability, high specificity, and high throughput, using reliable internal reference genes for the correction of target gene data to obtain accurate results25,26,27,28. Due to the lack of reference gene information in S. pennata, the reliability and accuracy of related gene expression detection significantly limits further gene function and genetic studies. Housekeeping genes such as GAPDH and TUB have typically been used as internal controls for the normalization of RT-qPCR, obtaining high effectiveness and reliability in other plant species12,13. However, there is great variability in the expression stability of some reference genes as a result of differences among conditions, species, tissues, growth and development stages, and experimental treatments12,14,15,16,17. In this study, a total of eight candidate reference genes, including ARP6, elF, PP2A, ALDH, GAPDH, TIP41, HIS-3, and α-TUB, were screened for suitability in S. pennata (Table 1).
The algorithms of geNorm, NormFinder, BestKeeper, and RefFinder are commonly utilized to assess the expression stability of candidate reference genes to identify the optimal reference genes, such as in soybean4, potato29, Plukenetia volubilis L.30, and banana31. Using these four programs, the eight screened reference genes showed varying expression stability under different conditions, with the different programs obtaining different results. The most stably expressed genes in the present study included GAPDH by NormFinder and BestKeeper under drought stress, elF and TIP41 by geNorm under drought stress, ARP6 by NormFinder and BestKeeper in all tissues, ALDH and TIP41 by geNorm in all tissues, elF by NormFinder and geNorm in rhizosheath development, and HIS-3 by BestKeeper in rhizosheath development (Tables 2, 3; Fig. 3). The 18S rRNA and 25S rRNA genes have been widely used as internal controls for the normalization of RT-qPCR in rice under stress conditions. In NaCl- and mannitol-treated rice seedlings, 18S rRNA and 25S rRNA showed the most stable expression levels of all reference genes14. It was also found that the 26S rRNA gene in Arabidopsis thaliana and other herbaceous plants was stably expressed under 10% PEG treatment32. In this study, GAPDH and elF were identified as the most stable reference genes under PEG treatment (Table 4).
Different reference genes have been used in different plants and tissues14,33,34,35. The polyubiquitin genes UBQ4 and UBQ10 demonstrated the most stable expression in different tissues of Brachypodium distachyon17. GAPDH showed the best expression stability in different tissues and organs of Saccharum sp.36. GAPDH and EF1α exhibited better expression stability and were selected as optimal internal reference genes during fruit development in Lycium barbarum L. EF1α and TUA were used as two internal reference genes for gene expression analysis during fruit development in Amomum villosum Lour37. Our comprehensive analysis identified ARP6 and ALDH in the tissues, elF and GAPDH in rhizosheath development, and ARP6 and elF in all samples as suitable reference genes in S. pennata (Table 4). In Vitis amurensis Rupr. berries, the expression of GAPDH was more stable than in Triticum aestivum L.38,39, while ACT and UBI exhibited better expression stability in wheat but worse expression stability in Solanum lycopersicum L.39,40. Comparative analysis of the correlation between the RNA-Seq data and RT-qPCR results using the screened stable genes of elF and GAPDH in rhizosheath development, which was done to quantify the target gene expression of CL9727 and U3887, indicated their high stability (Figs. 4 and 5), validating that these screened genes constituted reliable and effective internal reference genes for the normalization of RT-qPCR in S. pennata.
Correlation analysis of the relative expression levels of CL9727 and U3887 between RT-PqCR results and RNA-Seq data. X-axis represented the FPKM value in RNA-Seq data. Y-axis denoted Log2 (the ratio of RT-qPCR) using the method 2−ΔΔCt to calculate the relative expression level. The correlation analysis between RNA-Seq data and RT-qPCR results was applied through detection of the relative expression levels of CL9727 and CL3887 normalized by elF (A, B) and GAPDH (C, D).
In conclusion, we screened eight candidate reference genes for RT-qPCR normalization based on transcriptome datasets for S. pennata. Using the three procedures of geNorm, NormFinder, and BestKeeper, the comprehensive assessment analysis by RefFinder, as well as comparison and correlation analysis between the RT-qPCR results and RNA-Seq data, we identified reliable and suitable internal reference genes for RT-qPCR normalization under various conditions, thereby providing a foundation for further investigations of the genetic functions and regulatory mechanisms at the molecular level in S. pennata.
Materials and methods
Plant materials
The S. pennata plants were collected from the area of Mosuowan Reservoir in Shihezi City, Xinjiang, and were identified by Professor Ping Yan, vice president of the Xinjiang Botanical Society and member of the plant taxonomy and phylogeny Committee of the Chinese Botanical Society. The seeds and seedlings of wild S. pennata are presently permitted for scientific research. A specimen was stored in the Herbarium of Shihezi University (SHI: 2018013).
The S. pennata seeds were grown in sand at 37 °C with a photoperiod of 18 h light and 6 h dark in a climate chamber until experimentation. The roots, nodes, and leaves of 60 DPG plants, the rhizosheaths of 30 (R30), 60 (R60), 90 (R90) DPG plants, and 90-DPG non-rhizosheaths root (R90F), as well as the flowers, mature seeds, were collected. Additionally, the roots of 60-DPG S. pennata plants treated with different concentrations (0%, 5%, 10%, 15%, 20%, and 30%) (w/v) of PEG 6000 (Solarbio, Beijing, China) for 36 h were also collected.
Total RNA extraction and cDNA synthesis
The total RNA of the collected S. pennata materials was extracted using the RNAprep Pure Plant Kit (Tiangen, Beijing, China) according to the manufacturer’s instructions. The purity of the extracted RNA was detected by a NanoPhotometer spectrophotometer (IMPLEN, Calabasas, USA). The cDNA was synthesized from 200 ng RNA using the PrimeScript (TM) RT Reagent Kit with a gDNA Eraser (TaKaRa, Dalian, China) according to the manufacturer’s instructions.
Selection of candidate reference genes and design of primers
Based on the S. pennata rhizosheath development transcriptome data obtained from DNBseq (BGI, Shenzhen, China), a total of eight reference genes were identified using the filter conditions of q-value ≥ 0.05, FPKM value ≥ 6, and |log2FoldChange| < 1. The primers used for RT-qPCR were designed according to the nucleic acid sequences by Primer (Version 5.0) software (Premier Biosoft International, Palo Alto, USA). All primers were synthesized by Sangon Biotech (Shanghai, China).
RT-qPCR analysis
The RT-qPCR was performed on a LightCycler® 480 real-time PCR system (Roche Diagnostics, Mannheim, Germany) using the SYBR Green-based PCR assay. The total reaction volume was 20 µL, which included 10 µL 2× SuperReal PreMix Plus (SYBR Green, TANGEN BIOTECH, Beijing, China), 0.5 µL each of 10 µM forward and reverse gene-specific primer, 3 µL template (first-strand cDNA), and 6 µL ddH2O2. Amplifications were performed using the following program: initial denaturation at 94 °C for 2 min, followed by a cycling procedure of 30 s denaturation at 94 °C, 30 s annealing at 55 °C, 30 s extension at 72 °C, and then a final extension at 72 °C for 10 min. The RT-qPCR analysis was tested in three biological replicates. Relative gene expression levels were calculated using the 2−ΔΔCt method41.
Determination and validation of the expression stability of the reference genes
The geNorm25, NormFinder42, BestKeeper43, and RefFinder44 tools are used to screen stable reference genes for data analysis based on raw quantification cycle (Cq) values. The geNorm program selects the stable reference gene by calculating the M-value of each reference gene and determines the number of optimal reference genes according to the Vn/Vn+1 value. The default value of V is 0.15. If Vn/Vn+1 < 0.15, the number of optimal reference genes is n, if Vn/Vn+1 > 0.15, the number of optimal internal reference genes is n + 1. The calculation principle of NormFinder is similar to that of GeNorm, and the most suitable internal parameter gene is selected according to the stability value, with the most suitable gene being the lowest stability value. The correlation coefficient (R), standard deviation (SD), and coefficient of variation (CV) of pairing between each gene can be calculated by BestKeeper, with a larger R value or smaller SD and CV values denoting better stability of the reference gene. We used the online RefFinder software (https://github.com/fulxie/RefFinder) for the comparative analysis of ΔCt. The geometric mean of the Ct values of all candidate reference genes was analyzed to rank the expression stability.
Validation of the expression stability of the reference genes
The CL9729 and U3887 genes in the transcriptome data showed a close connection with rhizosheath development. In order to verify the analysis results of the candidate genes, two stably expressed (GAPDH and elF) and two unstably expressed (ALDH and α-TUB) reference genes were selected for RT-qPCR validation for these two target genes (CL9729 and U3887).
Abbreviations
- ACT:
-
Actin
- ALDH:
-
Aldehyde dehydrogenase
- ARP:
-
Actin related protein
- Cq:
-
Quantification cycle
- CV:
-
Coefficient of variation
- CYP:
-
Cyclophilin
- DPG:
-
Day post germination
- EF-1α:
-
Elongation factor-1α
- elF:
-
Eukaryotic translation initiation factor
- FPKM:
-
Fragments per Kilobase of exon model per Million mapped reads
- GAPDH:
-
Glyceraldehyde-3-phosphate dehydrogenase
- HIS-3:
-
Histone H3
- PEG:
-
Polyethylene glycol
- PGK:
-
Phosphoglycerate kinase gene
- PP2A:
-
Protein phosphotase 2A
- RNA-Seq:
-
RNA sequencing
- RT-qPCR:
-
Reverse transcription quantitative polymerase chain reaction
- SAMSC:
-
S-Adenosylmethionine synthase
- SD:
-
Standard deviation
- TIP:
-
Tonoplast intrinsic protein
- TUB:
-
Tubulin
- UBQ:
-
Ubiquitin
References
Tian, Y. et al. Relationship between microbial diversity and nitrogenase activity of Stipagrostis pennata rhizosheath. J. Cell. Biochem. 120(8), 13501–13508 (2019).
Gong, B. H., Wang, A. Y., Zhao, H. Y., Zhu, J. B. & Zhang, Y. X. Study on the drought-resistant mechanism of San Miscanthus through the analysis of root sheath structure. J. Desert Res. 30(3), 556–559 (2010).
Qiu, D., Wu, N., Zhang, Y. M. & Pan, H. X. Ecological regulation of root sheath microhabitat on the adaptability of feather needles and sands. J. Desert Res. 6, 1647–1654 (2012).
Hu, R. B., Fan, C. M. & Fu, Y. F. Reference gene selection in plant real-time quantitative reverse transcription PCR (qRT-PCR). J. Agric. Sci. Technol. 11(6), 30–36 (2009).
Liu, L. et al. Selection and validation of the optimal panel of reference genes for RT-qPCR analysis in the developing rat cartilage. Front. Genet. 16(11), 590124 (2020).
Yan, J. W., Yuan, F. R., Long, G. Y., Qin, L. & Deng, Z. N. Selection of reference genes for quantitative real-time RT-PCR analysis in citrus. Mol. Biol. Rep. 39(2), 1831–1838 (2012).
Zhang, Y. F., Zhao, L. J. & Zen, Y. L. Selection and application of internal reference genes in gene expression research. Plant Physiol. Commun. 50(8), 1119–1125 (2014).
Guo, R. et al. Evaluation of reference genes for RT-qPCR analysis in wild and cultivated Cannabis. Biosci. Biotechnol. Biochem. 82(11), 1902–1910 (2018).
Radoni, A. et al. Guideline to reference gene selection for quantitative real-time PCR. Biochem. Biophys. Res. Commun. 313(4), 856–862 (2004).
Andersen, C. L., Jensen, J. L. & θrntoft, T. F. Normalization of real-time quantitative reverse transcription-PCR data a model-based variance estimation approach to identify gene suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 64(15), 5245–5250 (2004).
Die, J. V., Román, B., Nadal, S. & González-Verdejo, C. I. Evaluation of candidate reference genes for expression studie in Pisumsativum under different experimental conditions. Planta 232(1), 145–153 (2010).
Gutierrez, L. et al. The lack of a systematic validation of reference genes: A serious pitfall undervalued in reverse transcription-polymerase chain reaction RT-PCR analysis in plants. Plant Biotechnol. J. 6, 609–618 (2008).
Zheng, T. et al. Reference gene selection for qRT-PCR analysis of flower development in Lagerstroemia indica and L. speciosa. PLoS ONE 13(3), e0195004 (2018).
Jain, M., Nijhawan, A., Tyagi, A. K. & Khurana, P. J. Validation of housekeeping genes as internal control for studying gene expression in rice by quantitative real-time PCR. Biochem. Biophys. Res. Commun. 345(2), 646–651 (2006).
Gao, Z. H., Wei, J. H., Yang, Y., Zhang, Z. & Zhao, W. T. Selection and validation of reference genes for studying stress-related agarwood formation of Aquilaria sinensis. Plant Cell Rep. 31, 1759–1768 (2012).
Rubie, C. et al. Housekeeping gene variability in normal and cancerous colorectal, pancreatic, esophageal, gastric and hepatic tissues. Mol. Cell Probes. 19, 101–109 (2005).
Hong, S. Y., Seo, P. J., Yang, M. S., Xiang, F. & Park, C. M. Exploring valid reference genes for gene expression studies in Brachypodium distachyon by real-time PCR. BMC Plant Biol. 8, 112 (2008).
Taylor, S., Wakem, M., Dijkman, G., Alsarraj, M. & Nguyen, M. A practical approach to RT-qPCR—Publishing data that conform to the MIQE guidelines. Methods 50, S1–S5 (2010).
Pfaffl, M. W., Tichopad, A., Prgomet, C. & Neuvians, T. P. Determination of stable housekeeping genes, diferentially regulated target genes and sample integrity: BestKeeper-Excel-based tool using pair-wise correlations. Biotechnol. Lett. 26, 509–515 (2004).
Saddhe, A. A., Malvankar, M. R. & Kumar, K. Selection of reference genes for quantitative real-time PCR analysis in halophytic plant Rhizophora apiculata. Peer J. 6, 5226 (2018).
Xia, W. et al. Analysis of multiple transcriptomes of the African oil palm (Elaeis guineensis) to identify reference genes for qRT-PCR. Biotechnol. 184, 63–73 (2014).
Fausto, A. K. S., Silva, T. D. F., Romanel, E. & Vaslin, M. F. S. microRNAs as reference genes for quantitative PCR in cotton. PLoS ONE 12(4), e0174722 (2017).
Liu, X. et al. Reference gene selection for qRT-PCR assays in Stellera chamaejasme subjected to abiotic stresses and hormone treatments based on transcriptome datasets. Peer J. 6, e4535 (2018).
Shakeel, M., Rodriguez, A., Rodriguez, A., Tahir, U. B. & Jin, F. L. Gene expression studies of reference genes for quantitative real-time PCR: An overview in insects. Biotechnol. Lett. 40, 227–236 (2018).
Vandesompele, J. et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3, 7 (2002).
Yoo, W. G. et al. Reference genes for quantitative analysis on Clonorchis sinensis gene expression by real-time PCR. Parasitol. Res. 104(2), 321–328 (2009).
Quacknbush, J. Microarray data normalization and transformation. Nat. Genet. 32, 496–501 (2002).
Nolan, T., Hands, R. E. & Bustin, S. A. Quantification of mRNA using realtime RT-PCR. Nat. Protoc. 1(3), 1559–1582 (2006).
Nico, T. N. et al. Reference gene selection for RT-qPCR normalization in potato during biotic and abiotic stress. J. Exp. Bot. 56(421), 7–14 (2005).
Niu, L. et al. Selection of reliable reference genes for gene expression studies of a promising oilseed crop, plukenetia volubilis, by real-time quantitative PCR. Int. J. Mol. Sci. 16(6), 12513–12530 (2014).
Chen, L. et al. Validation of reference genes for RT-qPCR studies of gene expression in banana fruit under different experimental conditions. Planta 234(2), 377–390 (2011).
Singh, K. et al. 26S rRNA based internal control gene primer pair for reverse transcription polymerase chain reaction based quantitative expression studies in diverse plant species. Anal. Biochem. 335(2), 330–333 (2004).
Tong, Z. G., Gao, Z., Wang, F., Zhou, J. & Zhang, Z. Selection of reliable reference genes for gene expression studies in peach using real-time PCR. BMC Mol. Biol. 10, 71 (2009).
Maroufi, A., Bockstaele, E. V. & Loose, M. D. Validation of reference genes for gene expression analysis in chicory (Cichorium intybus) using quantitative real-time PCR. BMC Mol. Biol. 11, 15 (2010).
Ding, J. Y. et al. Validation of a rice specific gene, sucrose phosphate synthase, used as the endogenous reference gene for qualitative and real-time quantitative PCR detection of transgenes. Agric. Food Chem. 52(11), 3372–3377 (2004).
Iskandar, H. M. et al. Comparison of reference genes for quantitative real-time polymerase chain reaction analysis of gene expression in sugarcane. Plant Mol. Biol. Rep. 22, 325–337 (2004).
Yu, A. M. et al. Screening of reference gene for real-time fluorescence quantitative PCR in Amomum villosum Lour., Guangzhou Zhongyiyao Daxue Xuebao. J. Guangzhou Univ. Traditional Chin. Med. 31(5), 814–820 (2014).
Reid, K. E., Olsson, N., Schlosser, J., Peng, F. & Lund, S. T. An optimized grapevine RNA isolation procedure and statistical determination of reference genes for real-time RT-PCR during berry development. BMC Plant Biol. 6, 27 (2006).
Long, X. Y. et al. Genome-wide identification and evaluation of novel internal control genes for Q-PCR based transcript normalization in wheat. Plant Mol. Biol. 74, 307–311 (2010).
Mascia, T., Santovito, E., Gallitelli, D. & Cillo, F. Evaluation of reference genes for quantitative reverse transcription polymerase chain reaction normalization in infected tomato plants. Mol. Plant Pathol. 11, 805–816 (2010).
Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(− Delta Delta C(T)) method. Methods 25, 402–408 (2001).
Claus, L. A., Jens, L. J. & Torben, F. R. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Can. Res. 64(15), 5245–5250 (2004).
Michael, W. P., Ales, T., Christian, P. & Tanja, P. N. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: Best Keeper-Excel-based tool using pair-wise correlations. Biotech. Lett. 26(6), 509–515 (2004).
Zhou, L., Chen, F., Ye, J. R. & Pan, H. Selection of reliable reference genes for qRT-PCR analysis of Bursaphelenchus mucronatus gene expression from different habitats and developmental stages. Front. Genet. 9, 269 (2018).
Acknowledgements
We thank Prof. Paul Pieter Dijkwel from School of Fundamental Sciences of Massey University for the beneficial suggestion to this work.
Funding
This research was funded by the International Science and Technology Cooperation Project of Bingtuan [Grant number 2020BC002], the National Natural Science Foundation of China [Grant number 32060082], and the Scientific and Technological project of Shihezi University [Grant number XJ2020003201].
Author information
Authors and Affiliations
Contributions
Conceptualization, R.L. and H.L.; data curation, F.W., R.L. and K.C.; formal analysis, H.S. and L.Z.; funding acquisition, R.L. and H.L.; investigation, R.L. and K.C.; methodology, R.L. and K.C.; project administration, X.J. and H.L.; resources, F.W. and H.L.; software, Q.X.; supervision, H.L.; validation, S.X. and X.C.; visualization, R.L. and A.C.; writing—original draft preparation, R.L. and H.L.; Writing—review and editing, H.L.; All authors have read and agreed to the published version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Li, R., Cui, K., Xie, Q. et al. Selection of the reference genes for quantitative gene expression by RT-qPCR in the desert plant Stipagrostis pennata. Sci Rep 11, 21711 (2021). https://doi.org/10.1038/s41598-021-00833-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-00833-2
- Springer Nature Limited
This article is cited by
-
Evaluation and validation of reference genes for RT-qPCR gene expression in Naegleria gruberi
Scientific Reports (2023)
-
Analysis of controlling genes for tiller growth of Psathyrostachys juncea based on transcriptome sequencing technology
BMC Plant Biology (2022)
-
Validation of suitable reference genes by various algorithms for gene expression analysis in Isodon rubescens under different abiotic stresses
Scientific Reports (2022)