Abstract
The aim of this study was to investigate how carbohydrates (glucose or sucrose) affect the characteristics of Enterococcus faecalis (E. faecalis) planktonic and biofilm in vitro. For this study, E. faecalis was cultured in tryptone-yeast extract broth with 0% glucose + 0% sucrose, 0.5% glucose, 1% glucose, 0.5% sucrose, or 1% sucrose. Viability of E. faecalis was examined by colony forming unit counting assays. Biofilm formation was assessed by measuring extracellular DNA (eDNA), a component of the biofilm matrix. Quantitative real-time PCR (qRT-PCR) was performed to investigate the expression of virulence-associated genes. Field emission scanning electron microscopy analysis, confocal laser scanning microscopy analysis, and crystal violet colorimetric assay were conducted to study E. faecalis biofilms. E. faecalis showed the highest viability and eDNA levels in 1% sucrose medium in biofilms. The result of qRT-PCR showed that the virulence-associated genes expressed highest in 1% sucrose-grown biofilms and in 1% glucose-grown planktonic cultures. E. faecalis showed highly aggregated biofilms and higher bacteria and exopolysaccharide (EPS) bio-volume in sucrose than in 0% glucose + 0% sucrose or glucose. The results indicate that the production of eDNA and EPS and expression of virulence-associated genes in E. faecalis are affected by the concentration of carbohydrates in biofilm or planktonic culture.
Similar content being viewed by others
Introduction
Enterococcus faecalis (E. faecalis) is commonly isolated from infected root canals and is regarded as one of the etiological agents associated with failed endodontic treatments1,2,3,4. E. faecalis can be frequently found in failed endodontic treatments with its ability to survive in an environment that lacks nutrients and in the tooth environment that is highly alkaline after treatment with intracanal medicaments4,5,6.
The pathogenicity and difficulty in removal of E. faecalis can be due to its ability to form biofilms since biofilms are related to 65% of bacterial infections and can be 1000-fold more resistant to antibiotics than planktonic cells7,8,9,10. In addition, E. faecalis biofilms formed on medical devices are known to cause some hospital-acquired infections11,12. Accordingly, the role of biofilm components including exopolysaccharides (EPS), proteins, and extracellular DNA (eDNA) in the biofilm matrix has been investigated10,13,14,15,16,17,18. Especially, the significance of eDNA in biofilms has been studied and demonstrated that it promoted and developed biofilm formation by aiding bacterial aggregation and biofilm stability10,19,20. Many researchers have also investigated virulence factors in clinically-isolated E. faecalis and reported that enterococcal surface protein (esp), gelatinase (gelE), aggregation substance (asa1), cytolysin B (cylB), and endocarditis-specific antigen A (efaA) gene, ArgR family transcription factor (ahrC), endocarditis and biofilm-associated pili (ebpA), enterococcal polysaccharide antigen (epal), epal and OG1RF_11715 (epaOX), and (p)ppGpp-synthetase/hydrolase (relA) genes were prevalent21,22,23,24,25,26,27,28,29,30.
One of the factors that affects biofilm formation is nutrient type and availability in the culture environment31. It has been reported that glucose significantly promotes biofilm formation of E. faecalis by inducing Esp synthesis30. However, the effect of glucose and sucrose supplementation on E. faecalis planktonic and biofilm phenotypes is still under-characterized. Therefore, we investigated the effects of different culture conditions on the characteristics of E. faecalis by examining the expression of genes associated with virulence, biofilm formation, and levels of eDNA, a biofilm matrix component.
Results
Enterococcus faecalis viability and eDNA production
The influence of media with or without carbohydrate (0% glucose + 0% sucrose, 0.5% glucose, 1% glucose, 0.5% sucrose, or 1% sucrose) on viability of E. faecalis in planktonic or biofilm conditions was investigated (Fig. 1a,b). E. faecalis showed higher colony-forming units (CFUs) in sucrose than in other media in both culture conditions, whereas it had lower CFUs in glucose in planktonic culture than in other media. The levels of eDNA were also examined since eDNA is one of the major components of the biofilm matrix (Fig. 1c,d). The highest level of eDNA was shown in 1% sucrose followed by 0.5% sucrose, 1% glucose 0.5% glucose, and 0% glucose + 0% sucrose in the biofilm condition (p < 0.05). In the planktonic condition, E. faecalis also had the highest level of eDNA in 0% glucose + 0% sucrose; however, it released more eDNA in glucose than in sucrose.
Analysis of E. faecalis grown in biofilms and planktonic culture. Viability and eDNA levels of ATCC29212 (black) and OG1RF (white) in (a,c) planktonic culture and (b,d) biofilms grown in 0% glucose + 0% sucrose (0%G + 0%S), 0.5% glucose (0.5%G), 1% glucose (1%G), 0.5% sucrose (0.5%S), or 1% sucrose (1%S) were examined. Data represent the mean and standard deviation of 3 independent experiments. Asterisks denote a statistically significant difference compared to 0%G + 0%S (p < 0.05).
Changes in gene expression of E. faecalis virulence-associated genes
In biofilms, genes associated with E. faecalis virulence were expressed at significantly higher levels in sucrose, whereas their expression was upregulated in glucose in the planktonic growth condition (Fig. 2).
Expression of virulence-associated genes by qRT-PCR. RNA was prepared from E. faecalis (a and c) ATCC29212 and (b and d) OG1RF grown in 0% glucose + 0% sucrose (0%G + 0%S), 0.5% glucose (0.5%G), 1% glucose (1%G), 0.5% sucrose (0.5%S), or 1% sucrose (1%S). Data represent the mean and standard deviation of at least 3 independent experiments.
EPS measurement and morphological observation of E. faecalis biofilms
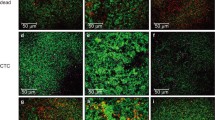
The effect of carbohydrate on E. faecalis biofilms was evaluated via bio-volume of bacteria and EPS using confocal laser scanning microscopy (CLSM) analysis (Fig. 3). E. faecalis showed the lowest level of bacteria and EPS bio-volumes in 0% glucose + 0% sucrose. The bacteria and EPS bio-volumes were higher in biofilms cultured in the presence of sucrose compared to biofilms cultured in glucose. As shown in the scanning electron microscopy (SEM) images in Fig. 4, E. faecalis biofilms aggregated to higher levels in sucrose compared to those in 0% glucose + 0% sucrose or glucose.
CLSM images of E. faecalis (a–e) ATCC29212 and (g–k) OG1RF biofilms. The bacteria were grown in 0% glucose + 0% sucrose (0%G + 0%S), 0.5% glucose (0.5%G), 1% glucose (1%G), 0.5% sucrose (0.5%S), or 1% sucrose (1%S). Bio-volume of bacteria (black) and EPS (white) of ATCC29212 (f) and OG1RF (l) biofilms were quantified by COMSTAT software (COMSTAT2, www.comstat.dk; Technical University of Denmark, Kongens Lyngby, Denmark). Data represent the mean and standard deviation of 3 independent experiments. Asterisks denote a statistically significant difference compared to 0%G + 0%S (p < 0.05).
FE-SEM images of biofilms of E. faecalis (a–e) ATCC29212 and (f–j) OG1RF. E. faecalis biofilms were formed in 0% glucose + 0% sucrose (0%G + 0%S), 0.5% glucose (0.5%G), 1% glucose (1%G), 0.5% sucrose (0.5%S), or 1% sucrose (1%S) on the surface of hydroxyapatite (HA) disks. The images were examined with 30,000× magnification.
Biofilm formation assay
Biofilm formation of E. faecalis ATCC29212, OG1RF and clinical strains was assessed by crystal violet colorimetric assay. As shown in Fig. 5, the strains showed higher biofilm formation in sucrose than in other media. OG1RF had greater biofilm mass than ATCC29212 in 0% glucose + 0% sucrose, 0.5% glucose, 1% glucose, 0.5% sucrose, or 1% sucrose, whereas clinical isolates formed larger biofilms than ATCC29212 and OG1RF in 1% sucrose.
Biofilm formation of E. faecalis strains. Crystal violet staining was used to assay E. faecalis strain biofilm formation. The strains were cultured in 0% glucose + 0% sucrose (0%G + 0%S), 0.5% glucose (0.5%G), 1% glucose (1%G), 0.5% sucrose (0.5%S), or 1% sucrose (1%S). Data represent the mean and standard deviation of 3 independent experiments. Asterisks denote a statistically significant difference compared to 0%G + 0%S (p < 0.05).
Discussion
E. faecalis is a pathogenic bacterium that can form biofilm, which leads to infectious disease, resistance to numerous antibiotics, and endodontic treatment failure4,32,33. Studies have shown that nutrients in culture media may affect bacterial biofilm formation34,35. Glucose was shown to enhance biofilm growth of E. faecalis by inducing Esp synthesis30. However, the results of the current study showed that the expressions of the esp gene along with other biofilm-associated genes such as asa1, cylB, efaA, ahrC, ebpA, epal, epaOX, and relA were higher in biofilms cultured in the presence of sucrose compared to glucose, whereas it showed higher or similar induction in glucose than in sucrose in planktonic culture. The results of this study suggested that E. faecalis could be more virulent in sucrose than in glucose with higher expression of the virulence-associated genes and greater biofilm formation in sucrose since many cases of bacterial infections are related to biofilms with 1000-fold more resistance against antibiotics than planktonic cells7,8. This needs further study.
Interestingly, although CFUs of E. faecalis were lower in sucrose than in 0% glucose + 0% sucrose in planktonic culture, it attached to the surface of HA disks more in 0% glucose + 0% sucrose than in glucose or sucrose in biofilm conditions when the disks were not washed (data not shown). However, when the disks were gently washed, unattached E. faecalis was easily removed from the disks and the result showed that the number of CFUs in 0% glucose + 0% sucrose was lower than that in the other media, whereas there was no significant difference of CFUs in biofilms formed in glucose or sucrose with or without washing. This could be the result of a lack of biofilm components such as eDNA in 0% glucose + 0% sucrose, as shown in Fig. 1d, that could give binding sites for E. faecalis and slower growth rates of bacteria in biofilms than in planktonic culture as demonstrated by Evans et al.17,36. In other words, E. faecalis could grow slowly in biofilms in sucrose since the bacteria aggregated more and formed compact biofilms in sucrose than in 0% glucose + 0% sucrose or glucose (Fig. 4).
Additional studies have investigated the role of eDNA, a component of the biofilm matrix, and demonstrated that it is essential for biofilm formation20,37,38. Thus, eDNA levels were examined and they were higher in biofilms in sucrose than in the other media. This result is in agreement with the result of a study that suggested that the eDNA levels in biofilms were relevant to the CFU numbers in the biofilms39. This could be explained by the higher expression of virulence-associated genes that were upregulated in those media and also could be the result of higher EPS bio-volume that could enmesh the bacteria with biofilm components such as eDNA. Interestingly, although the planktonic growth of E. faecalis in glucose was lower than that in 0% glucose + 0% sucrose or sucrose, their eDNA levels were similar to those collected from 0% glucose + 0% sucrose or sucrose in planktonic culture. This could be the result of higher expression of genes including some virulence-associated genes in glucose than in other media, as shown in Fig. 2. This needs further study.
The effect of sucrose on biofilm formation was extensively studied with Streptococcus mutans (S. mutans), one of the major agents that are associated with dental caries, and demonstrated that S. mutans utilized sucrose to produce glucans, an important component of biofilm matrix, leading to a high incidence of dental caries40,41,42. However, the effect of sucrose had not been studied with E. faecalis. According to the present study, E. faecalis produced higher levels of eDNA in sucrose and expressed more virulence-associated genes in biofilms than in other media. CLSM analysis showed that E. faecalis produced more EPS in sucrose, suggesting that E. faecalis could utilize sucrose and produce EPS. SEM analysis showed that E. faecalis aggregated more in the biofilm matrix in sucrose than in 0% glucose + 0% sucrose or glucose. E. faecalis biofilm formation of the strains was analyzed by crystal violet colorimetric assay and the result showed that E. faecalis formed larger biofilms in sucrose than in 0% glucose + 0% sucrose or glucose. The results of this study suggested that the biofilm formation ability of E. faecalis in sucrose could be significant clinically with greater biofilm formation and higher virulence-associated gene expression than in 0% glucose + 0% sucrose or glucose since infections of many bacteria are related to the ability of biofilm formation that induces more resistance against antibiotics than planktonic bacteria7,8,9,34.
In summary, E. faecalis produced more components of the biofilm matrix such as eDNA and EPS and had more highly expressed virulence-associated genes when grown in biofilms in sucrose relative to 0% glucose + 0% sucrose or glucose in biofilms. In planktonic culture, glucose-induced more eDNA/CFU and expression of virulence-associated genes than in 0% glucose + 0% sucrose or sucrose.
Materials and methods
Bacterial culture conditions
E. faecalis ATCC 29212 and ATCC 47077 (OG1RF) were grown overnight at 37 °C in brain heart infusion (BHI; Difco Laboratories, Detroit, MI, USA) media in 1% glucose as previously described with some modifications39. Briefly, 10 μl of the overnight cultured E. faecalis was cultured in 1 ml of BHI media for 2 h. For planktonic culture, E. faecalis (3 × 106 CFU/mL) was cultured in a 24-well plate (SPL, Daejeon, Korea) in tryptone-yeast extract broth (TY) with 0% glucose + 0% sucrose, 0.5% glucose, 1% glucose, 0.5% sucrose, or 1% sucrose for 24 h without agitation at 37 °C. For biofilm formation, hydroxyapatite (HA) disks (2.93 cm2; Clarkson Chromatography Products, Inc. South Williamsport, PA, USA) were vertically placed in a 24-well plate that contained E. faecalis (3 × 106 CFU/mL) in TY with 0% glucose + 0% sucrose, 0.5% glucose, 1% glucose, 0.5% sucrose, or 1% sucrose for 24 h without agitation at 37 °C. To prepare BHI plates, 1.5% (wt/vol) agar (Difco Laboratories) was added.
Colony forming unit (CFU) assay and eDNA measurement
E. faecalis eDNA and CFUs were measured as previously described with some modifications39. Briefly, E. faecalis grown in planktonic culture was collected carefully and centrifuged at 10,000×g at 4 °C for 10 min. The pellet was resuspended in phosphate buffered saline (PBS) and sonicated using a sonifier (VCX 130 PB; Sonics and Materials Inc., Newtown, CT, USA) for 10 s at 7 W to separate eDNA from the bacteria. To collect E. faecalis in biofilms, HA disks were gently washed twice in PBS and sonicated in an ultrasonic bath (Power Sonic 410; Hwashin Technology Co., Seoul, Korea) for 10 min to break the biofilms in 2 ml PBS. The biofilms were then scraped using a spatula and sonicated with a sonifier for 10 s at 7 W. For CFU counting, an aliquot (0.1 ml) of the homogenized E. faecalis was serially diluted and plated on BHI agar plates and incubated at 37 °C. The remaining cells were centrifuged at 10,000×g at 4 °C for 10 min. The eDNA level was measured using a fluorescence microplate reader (HIDEX, Turku, Finland) at an excitation of 485 nm and emittance of 535 nm.
Quantitative real-time PCR (qRT-PCR)
After 24 h incubation in planktonic culture and in biofilms, the bacteria were collected and sonicated. Then, the homogenized bacteria were treated using Trizol Max Bacterial RNA Isolation Kit (Life Technologies, Carlsbad, CA, USA) to extract RNA according to manufacturer’s instructions. Complementary DNA (cDNA) was amplified using a QuantiTech Reverse Transcription Kit (QIAGEN, Hilden, Germany). qRT-PCR was performed using a Rotor-Gene SYBR Green PCR Kit (QIAGEN, Hilden, Germany) and a Rotor-Gene Q real-time PCR machine (QIAGEN) with a 72-well rotor. The primers used in this study are shown in Table 143. Relative expression was calculated by normalizing each test gene to the E. faecalis 16s RNA reference gene.
Field emission scanning electron microscopy (FE-SEM)
Biofilms formed on the HA disks in 0% glucose + 0% sucrose, 1% glucose, or 1% sucrose were fixed as previously described with some modifications44. In brief, HA disks were in 2.5% glutaraldehyde / 4% paraformaldehyde (Sigma-Aldrich, Saint Louis, MO, USA) in PBS at 4 °C overnight. They were then washed in PBS and a graded series of ethanol (25–100%) was used to dehydrate the samples. The samples were then air-dried, gold-coated, and observed using FE-SEM (Hitachi, Tokyo, Japan). The images were obtained on a Hitachi SU-70 using BSE detector with 10.0 kV voltage acceleration.
Confocal laser scanning microscopy (CLSM)
CLSM analysis was performed to investigate the change of E. faecalis biofilms formed in media with different carbohydrates (0% glucose + 0% sucrose, 1% glucose, or 1% sucrose) as previously described39. E. faecalis (3 × 106 CFU/ml) was cultured in a medium with 1 μM Alexa Fluor 647-labeled dextran conjugate (650/668 nm; Molecular Probes, Eugene, OR, USA), which was incorporated into the EPS matrix for 24 h at 37 °C. Then, E. faecalis on the HA disks were exposed to 2.5 μM SYTO 9 green fluorescent nucleic acid stain (480/500 nm; Molecular Probes) for 20 min at room temperature. Fifteen image stacks (512 × 512 pixel tagged image file format) were obtained using an LSM 510 META microscope (Carl Zeiss, Jena, Germany). The bio-volume of the bacteria and EPS was quantified using. The laser power was 5 mW for Alexa Fluor 647-labeled dextran conjugate (red channel—HeNe laser) and 20 mW for SYTO 9 (green channel—Ar laser). Three individual experiments were performed.
Crystal violet assay
Crystal violet was used to assess the biofilm mass of E. faecalis ATCC29212 and OG1RF including clinical isolates as previously described with some modifications10. Briefly, E. faecalis (3 × 106 CFU/ml) was cultured in a 96 well plate in TY with 0% glucose + 0% sucrose, 0.5% glucose, 1% glucose, 0.5% sucrose, or 1% sucrose for 24 h without agitation at 37 °C. Then the plate was washed with sterile water and added 0.5% crystal violet (Sigma-Aldrich, St. Louis, MO, USA) and kept at room temperature for 10 min. Then the plate was rinsed twice with sterile water, treated with 30% acetic acid (Fisher Scientific, Fair Lawn, NJ, USA), and measured at 590 nm (μQuant, Biotek Instrument, Winooski, VT, USA).
Statistical analysis
All experiments were performed at least three times. The intergroup differences were estimated by one-way analysis of variance (ANOVA). The data are presented as mean and standard deviation (SD). A p value was considered as statistically significant when it was less than 0.05.
Data availability
The data generated and analyzed in this study are available from the corresponding author upon request.
References
Peciuliene, V., Balciuniene, I., Eriksen, H. M. & Haapasalo, M. Isolation of Enterococcus faecalis in previously root-filled canals in a Lithuanian population. J. Endod. 26, 593–595. https://doi.org/10.1097/00004770-200010000-00004 (2000).
Gomes, B. P. et al. Microbiological examination of infected dental root canals. Oral Microbiol. Immunol. 19, 71–76 (2004).
Endo, M. S., Ferraz, C. C. R., Zaia, A. A., Almeida, J. F. A. & Gomes, B. Quantitative and qualitative analysis of microorganisms in root-filled teeth with persistent infection: monitoring of the endodontic retreatment. Eur. J. Dent. 7, 302–309. https://doi.org/10.4103/1305-7456.115414 (2013).
Stuart, C. H., Schwartz, S. A., Beeson, T. J. & Owatz, C. B. Enterococcus faecalis: its role in root canal treatment failure and current concepts in retreatment. J. Endod. 32, 93–98. https://doi.org/10.1016/j.joen.2005.10.049 (2006).
Figdor, D., Davies, J. K. & Sundqvist, G. Starvation survival, growth and recovery of Enterococcus faecalis in human serum. Oral Microbiol. Immunol. 18, 234–239 (2003).
Narayanan, L. L. & Vaishnavi, C. Endodontic microbiology.. J. Conserv. Dent. 13, 233–239. https://doi.org/10.4103/0972-0707.73386 (2010).
Potera, C. Forging a link between biofilms and disease. Science (New York N.Y.) 283(1837), 1999. https://doi.org/10.1126/science.283.5409.1837 (1839).
Hoyle, B. D. & Costerton, J. W. Bacterial resistance to antibiotics: the role of biofilms. Prog. Drug Res. 37, 91–105 (1991).
Ghabraei, S., Marvi, M., Bolhari, B. & Bagheri, P. Minimum intracanal dressing time of triple antibiotic paste to eliminate Enterococcus faecalis (ATCC 29212) and determination of minimum inhibitory concentration and minimum bactericidal concentration: an ex vivo study. J. Dent. (Tehran) 15, 1–9 (2018).
Yu, M. K. et al. Role of extracellular DNA in Enterococcus faecalis biofilm formation and its susceptibility to sodium hypochlorite. J. Appl. Oral Sci. Rev. FOB 27, e20180699. https://doi.org/10.1590/1678-7757-2018-0699 (2019).
Donelli, G. & Guaglianone, E. Emerging role of Enterococcus spp in catheter-related infections: biofilm formation and novel mechanisms of antibiotic resistance. J. Vasc. Access 5, 3–9. https://doi.org/10.1177/112972980400500101 (2004).
Ch’ng, J. H., Chong, K. K. L., Lam, L. N., Wong, J. J. & Kline, K. A. Biofilm-associated infection by enterococci. Nat. Rev. Microbiol. 17, 82–94. https://doi.org/10.1038/s41579-018-0107-z (2019).
Castillo Pedraza, M. C. et al. Extracellular DNA and lipoteichoic acids interact with exopolysaccharides in the extracellular matrix of Streptococcus mutans biofilms. Biofouling 33, 722–740. https://doi.org/10.1080/08927014.2017.1361412 (2017).
Limoli, D. H., Jones, C. J. & Wozniak, D. J. Bacterial extracellular polysaccharides in biofilm formation and function. Microbiol. Spec. 3, 1. https://doi.org/10.1128/microbiolspec.MB-0011-2014 (2015).
Mitrofanova, O., Mardanova, A., Evtugyn, V., Bogomolnaya, L. & Sharipova, M. Effects of Bacillus serine proteases on the bacterial biofilms. BioMed. Res. Int. 2017, 8525912. https://doi.org/10.1155/2017/8525912 (2017).
Ocal, D. N., Dolapci, I., Karahan, Z. C. & Tekeli, A. Investigation of biofilm formation properties of staphylococcus isolates. Mikrobiyol. Bul. 51, 10–19 (2017).
Whitchurch, C. B., Tolker-Nielsen, T., Ragas, P. C. & Mattick, J. S. Extracellular DNA required for bacterial biofilm formation. Science (New York N.Y.) 295, 1487. https://doi.org/10.1126/science.295.5559.1487 (2002).
Li, W., Liu, H. & Xu, Q. Extracellular dextran and DNA affect the formation of Enterococcus faecalis biofilms and their susceptibility to 2% chlorhexidine. J. Endod. 38, 894–898. https://doi.org/10.1016/j.joen.2012.04.007 (2012).
Das, T., Sehar, S. & Manefield, M. The roles of extracellular DNA in the structural integrity of extracellular polymeric substance and bacterial biofilm development. Environ. Microbiol. Rep. 5, 778–786. https://doi.org/10.1111/1758-2229.12085 (2013).
Okshevsky, M. & Meyer, R. L. The role of extracellular DNA in the establishment, maintenance and perpetuation of bacterial biofilms. Crit. Rev. Microbiol. 41, 341–352. https://doi.org/10.3109/1040841x.2013.841639 (2015).
Strateva, T., Atanasova, D., Savov, E., Petrova, G. & Mitov, I. Incidence of virulence determinants in clinical Enterococcus faecalis and Enterococcus faecium isolates collected in Bulgaria. Braz. J. Infect. Dis. 20, 127–133. https://doi.org/10.1016/j.bjid.2015.11.011 (2016).
Lins, R. X. et al. Antimicrobial resistance and virulence traits of Enterococcus faecalis from primary endodontic infections. J. Dent. 41, 779–786. https://doi.org/10.1016/j.jdent.2013.07.004 (2013).
Saffari, F., Sobhanipoor, M. H., Shahravan, A. & Ahmadrajabi, R. Virulence genes, antibiotic resistance and capsule locus polymorphisms in Enterococcus faecalis isolated from canals of root-filled teeth with periapical lesions. Infect. Chemother. 50, 340–345. https://doi.org/10.3947/ic.2018.50.4.340 (2018).
Frank, K. L. et al. Use of recombinase-based in vivo expression technology to characterize Enterococcus faecalis gene expression during infection identifies in vivo-expressed antisense RNAs and implicates the protease Eep in pathogenesis. Infect. Immun. 80, 539–549. https://doi.org/10.1128/iai.05964-11 (2012).
Montealegre, M. C., La Rosa, S. L., Roh, J. H., Harvey, B. R. & Murray, B. E. The Enterococcus faecalis EbpA pilus protein: attenuation of expression, biofilm formation, and adherence to fibrinogen start with the rare initiation codon ATT. mBio 6, e00467-e415. https://doi.org/10.1128/mBio.00467-15 (2015).
Chávez de Paz, L. E., Lemos, J. A., Wickström, C. & Sedgley, C. M. Role of (p)ppGpp in biofilm formation by Enterococcus faecalis. Appl. Environ. Microbiol. 78, 1627–1630. https://doi.org/10.1128/aem.07036-11 (2012).
Dale, J. L., Cagnazzo, J., Phan, C. Q., Barnes, A. M. & Dunny, G. M. Multiple roles for Enterococcus faecalis glycosyltransferases in biofilm-associated antibiotic resistance, cell envelope integrity, and conjugative transfer. Antimicrob. Agents Chemother. 59, 4094–4105. https://doi.org/10.1128/aac.00344-15 (2015).
Zoletti, G. O. et al. Characterization of virulence factors and clonal diversity of Enterococcus faecalis isolates from treated dental root canals. Res. Microbiol. 162, 151–158. https://doi.org/10.1016/j.resmic.2010.09.018 (2011).
Kayaoglu, G. & Ørstavik, D. Virulence factors of Enterococcus faecalis: relationship to endodontic disease. Crit. Rev. Oral Biol. Med. 15, 308–320. https://doi.org/10.1177/154411130401500506 (2004).
Tendolkar, P. M., Baghdayan, A. S., Gilmore, M. S. & Shankar, N. Enterococcal surface protein, Esp, enhances biofilm formation by Enterococcus faecalis. Infect. Immun. 72, 6032–6039. https://doi.org/10.1128/iai.72.10.6032-6039.2004 (2004).
Toyofuku, M. et al. Environmental factors that shape biofilm formation. Biosci. Biotechnol. Biochem. 80, 7–12. https://doi.org/10.1080/09168451.2015.1058701 (2016).
Molander, A., Reit, C., Dahlen, G. & Kvist, T. Microbiological status of root-filled teeth with apical periodontitis. Int. Endod. J. 31, 1–7 (1998).
Fisher, K. & Phillips, C. The ecology, epidemiology and virulence of Enterococcus. Microbiology 155, 1749–1757. https://doi.org/10.1099/mic.0.026385-0 (2009).
Lima, K. C., Fava, L. R. & Siqueira, J. F. Jr. Susceptibilities of Enterococcus faecalis biofilms to some antimicrobial medications. J. Endod. 27, 616–619. https://doi.org/10.1097/00004770-200110000-00004 (2001).
Hancock, V., Witso, I. L. & Klemm, P. Biofilm formation as a function of adhesin, growth medium, substratum and strain type. Int. J. Med. Microbiol. 301, 570–576. https://doi.org/10.1016/j.ijmm.2011.04.018 (2011).
Evans, D. J., Allison, D. G., Brown, M. R. W. & Gilbert, P. Effect of growth-rate on resistance of gram-negative biofilms to cetrimide. J. Antimicrob. Chemother. 26, 473–478. https://doi.org/10.1093/jac/26.4.473 (1990).
Montanaro, L. et al. Extracellular DNA in biofilms. Int. J. Artif. Organs 34, 824–831. https://doi.org/10.5301/ijao.5000051 (2011).
Okshevsky, M., Regina, V. R. & Meyer, R. L. Extracellular DNA as a target for biofilm control. Curr. Opin. Biotechnol. 33, 73–80. https://doi.org/10.1016/j.copbio.2014.12.002 (2015).
Kim, M., Jeon, J. & Kim, J. Streptococcus mutans extracellular DNA levels depend on the number of bacteria in a biofilm. Sci. Rep. 8, 13313. https://doi.org/10.1038/s41598-018-31275-y (2018).
Hong, J., Whelton, H., Douglas, G. & Kang, J. Consumption frequency of added sugars and UK children’s dental caries. Community Dent. Oral Epidemiol. 46, 457–464. https://doi.org/10.1111/cdoe.12413 (2018).
Peres, M. A. et al. Sugar consumption and changes in dental caries from childhood to adolescence. J. Den. Res. 95, 388–394. https://doi.org/10.1177/0022034515625907 (2016).
Duarte, S. et al. Influences of starch and sucrose on Streptococcus mutans biofilms. Oral Microbiol. Immunol. 23, 206–212. https://doi.org/10.1111/j.1399-302X.2007.00412.x (2008).
Salah, R., Dar-Odeh, N., Abu Hammad, O. & Shehabi, A. A. Prevalence of putative virulence factors and antimicrobial susceptibility of Enterococcus faecalis isolates from patients with dental Diseases. BMC Oral Health 8, 17. https://doi.org/10.1186/1472-6831-8-17 (2008).
Plutzer, B., Zilm, P., Ratnayake, J. & Cathro, P. Comparative efficacy of endodontic medicaments and sodium hypochlorite against Enterococcus faecalis biofilms. Aust. Dent. J. 63, 208–216. https://doi.org/10.1111/adj.12580 (2018).
Acknowledgements
This research was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (2018R1A6A3A11041044). E. faecalis OG1RF and clinical strains were kindly provided by Prof. Prasanna Neelakantan in the University of Hong Kong.
Author information
Authors and Affiliations
Contributions
M.A.K. contributed to conception, design, data acquisition, analysis, and drafted manuscript; V.R. and K.S.M. contributed to conception, design, and critically revised the manuscript. All authors gave final approval and agree to be accountable for all aspects of the work.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Kim, MA., Rosa, V. & Min, KS. Characterization of Enterococcus faecalis in different culture conditions. Sci Rep 10, 21867 (2020). https://doi.org/10.1038/s41598-020-78998-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-78998-5
- Springer Nature Limited